 Food and Nutrition Sciences, 2013, 4, 1-9 http://dx.doi.org/10.4236/fns.2013.47A001 Published Online July 2013 (http://www.scirp.org/journal/fns) Quantification of Antibiotic Residues and Determination of Antimicrobial Resistance Profiles of Microorganisms Isolated from Bovine Milk in Lebanon Kassaify Zeina1*, Abi Khalil Pamela1, Sleiman Fawwak2 1Department of Nutrition and Food Sciences, American University of Beirut, Beirut, Lebanon; 2Department of Animal and Veteri- nary Sciences, American University of Beirut, Beirut, Lebanon. Email: *zk18@aub.edu.lb, pamabkh@gmail.com, sleimanf@aub.edu.lb Received March 18th, 2013; revised April 18th, 2013; accepted April 28th, 2013 Copyright © 2013 Kassaify Zeina et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT The rapid growth of dairy sectors in the Middle East, particularly in Lebanon, led to extensive use of antibiotics to en- hance the health and productivity of animals. Prolonged usage may lead to antibiotic residues in foods of animal origin; hence, the emergence of antimicrobial resistant microorganisms. Accurate data on the antibiotic usage in livestock treatment, antibiotic residues and antimicrobial resistances in raw milk in Lebanon are lacking. This study aimed to in- vestigate the types and usages of antibiotics in cattle, their residual levels and the potential microbial resistances in raw milk samples. A questionnaire-based survey identified Gentamicin and Streptomycin as the most frequently used anti- biotics. Selected raw milk samples from main dairy farms were then analyzed in duplicate by quantitative ELISA for the antibiotics residual levels. The mean residual levels of Gentamicin and Streptomycin were 90 and 80 μg/L, respec- tively; which are below the allowable maximum residue limit of 200 μg/L as set by the FAO/WHO. Staphylococcus aureus, Listeria monocytogenes, E. coli and total aerobic microorganisms isolated from the milk samples were then tested for resistance against Gentamicin and Streptomycin by the disc agar diffusion method. All the S. aureus, E. coli, and L. monocytogenes isolates showed high resistance to Gentamicin. However, 95% of S. aureus, 60% of E. coli and 58% of L. monocytogenes isolates were resistant to Streptomycin. The obtained results provide evidence that antimicro- bial resistant strains of the above pathogens have become remarkably widespread in raw milk. This requires better management for antibiotic usages among livestock farmers to control sources of food contamination and reduce the health risks associated with the development of resistant microbial strains. Keywords: Milk; Antibiotics; Residues; Resistant Pathogens 1. Introduction Antibiotics, the microbiologically produced compounds, are used in humans to treat or prevent certain diseases caused by infectious agents. However, the major antibi- otics used for humans either belong to the same general classes or have the same mode of action as those used for animals [1]. A questionnaire-based survey conducted preliminarily by the authors in this study showed that the most commonly used antibiotics in livestock among ma- jor Lebanese dairy farmers are Gentamicin and Strepto- mycin which belong to the Aminoglycoside group of an- tibiotics. Furthermore, a limited survey conducted by Choueiri [2] on 17 main dairy farms in Lebanon as part of her thesis work also found that Streptomycin and Gentamicin are used in all the surveyed farms. It is also worth mentioning that the exact amounts of antibiotics used by farmers in livestock production in Lebanon are not known since they are not regulated. Streptomycin is mainly used to treat plague and infrequently Brucellosis. It is also combined with penicillin in treating enterococ- cal and Listeria monocytogenes infections. Gentamicin sulfate is quite effective against several types of bacterial infections mainly those caused by gram-negative bacteria. It is mainly used against Pseudomonas, E. coli, Entero- bacteria, Proteus and Serratia [3]. These antibiotics, despite their effectiveness, can leave residues in the treated animal and contaminate its edible parts; the muscle meat and milk [4]. Environmental and *Corresponding author. Copyright © 2013 SciRes. FNS  Quantification of Antibiotic Residues and Determination of Antimicrobial Resistance Profiles of Microorganisms Isolated from Bovine Milk in Lebanon 2 human health risks are associated with these residues. They range from direct toxicity on consumers exhibiting allergic reactions to indirect problems through the gen- eration of resistant strains of pathogenic bacteria and the residual contamination of manures used in crop produc- tions [5]. To ensure consumer safety, worldwide regula- tory authorities have set MRL’s (Maximum Residual Li- mit) for several veterinary drugs [6,7]. These MRL’s, are expected to regulate the maximum permitted levels of the drug residue for each antibiotic which is considered safely acceptable in food of animal origin [8]. The major worldwide public concern and health hazard associated with antibiotic residues is the development of the antimicrobial resistant bacterial strains of animal origin and its consequent effect on human health [9-13] regarding the efficacy of antimicrobial therapy [14]. As the pathogens have become more resistant to the used doses of antibiotics, incidences of morbidity have increased, therapeutic effi- ciency has failed, healthcare costs are on the increase, and the antimicrobial doses have increased with scientists oc- cupied searching for alternatives to relieve the burden [15]. According to Prescott and Baggot [16], microbial resis- tance to aminoglycosides, particularly Streptomycin, Neo- mycin, and Kanamycin is very common and pathogens present in the milk mainly S. aureus, E. coli O157:H7 and L. monocytogenes may easily develop antimicrobial resis- tance [17-19]. This study aimed to determine the residual levels of the two most commonly used antibiotics, Gentamicin and Streptomycin, in milk samples collected from 24 random dairy farms in Lebanon. The enzyme linked immunosor- bant assay (ELISA) being user friendly, sensitive and can be economical when many samples need to be analyzed [20] was the method of choice. Another main objective of this work was to determine and link the resistance prevalence of pathogenic Staphylococcus aureus, Escherichia coli and L. monocytogenes isolated from the same milk samples to Streptomycin and Gentamicin residues. 2. Materials and Methods 2.1. Questionnaire-Based Survey on Major Farms A Questionnaire-based survey was conducted on twenty six Lebanese dairy cattle farms to identify the most com- monly used antibiotics, their dosage, timing of use and the practiced withholding times prior to dispatch. In this survey, farms located only in the Bekaa Valley and Mount Lebanon were visited because the southern region was not accessible. Between July and August, several farms were contacted, only 26 agreed to answer the sur- vey divided as 54.1% in the Bekaa Valley and 45.8% in Mount Lebanon. Two of the farms were for meat produc- tion; therefore, the collected data were eliminated. The remaining 24 were divided as nine farms for both milk and meat production and 15 for only milk production. Cattle farms varied in capacity; two were large-scale farms with >1000 cows, six were medium-scale with cattle capacity 100 - 400 and 16 were small-scale with cattle capacity <100. Interviews were conducted with the owners and/or the farmers and the questions were close- ended. For instance, farmers were asked about the milk- ing techniques used and their corresponding schedule, the sanitary conditions of the flock, percentage mortality and the feed (composition was important for the medi- cated feed). In addition, farmers were asked about any treatment administered including the brand name or ac- tive compound of the medication, treatment period (date of beginning and end of treatment), withdrawal time and identification number of the veterinary prescription. 2.2. The Controlled Study A 10 days controlled study was performed at the Agri- cultural Research and Education Center (AREC), Faculty of Agricultural and Food Sciences of the American Uni- versity of Beirut in the Bekaa of Lebanon. In the study, four Holstein cows aged between 34 and 39 months, that yielded approximately the same daily amount of milk (16 - 18 kg) were selected. The cows weighed around 450 - 500 kg and had no signs of disease when inspected. They tested negative for mastitis and were not exposed to any antibiotic treatment for a minimum of eight weeks. Each cow was placed in a labeled separate pen and were all given the same amount of water and feed. The two anti- biotics Gentamicin and Streptomycin were selected for this study based on the data acquired from the survey that was performed during this study and described above. Two cows were treated with the antibiotic Gentamicin (Gentaprim, Invesa, Veterinary Industry, Barcelona, Spain) at a concentration of 4 ml/100kg three times a day for 3 days, and the other two were treated with the antibiotic Streptomycin (Pen and Strep, Norbrook Laboratories Li- mited, Newry, BT35 6JP) at a concentration of 1 ml/25kg once a day for five days. Treatments were administered by deep intramuscular injection using sterile disposable syringes (20 ml syringe, Luer-Lok and BD Microlance 18 G needles) to prevent possible contamination. Milking was performed twice per day; at 2:00 am and 2:00 pm. The milk samples used for analysis consisted of a mix- ture of both milking times and were collected in 100 ml sterile plastic containers in duplicate. Aseptic measures were taken during sample collection by cleaning and drying the udders before milking and sanitizing the milk buckets before and between every milking session. All samples were analyzed for antibiotic residues and antim- icrobial susceptibility. Copyright © 2013 SciRes. FNS  Quantification of Antibiotic Residues and Determination of Antimicrobial Resistance Profiles of Microorganisms Isolated from Bovine Milk in Lebanon 3 2.3. The Experimental Study In order to validate the information provided by the farm- ers in the Questionnaire-based survey with respect to the withholding periods and the proper dosage of antibiotics used, analyses were conducted on random milk samples collected from the different farms. The 26 farms that were visited for the survey were contacted again and only 22 farms had agreed to provide samples to be analyzed. These farms varied from large-scale (9%) to medium- scale (27%) and small-scale (64%). Microbiological, an- tibiotic residues and antimicrobial susceptibility analyses were performed on all the samples in duplicate. 2.4. Analysis of Antibiotic Residues Enzyme linked immunosorbant assay (ELISA) test kits were used to quantitatively analyze both the control and the experimental milk samples for the presence of the two most commonly used antibiotics by the farmers, the aminoglycoside antibiotics Streptomycin and Gentamicin (BIO SCIENTIFIC Austin, TX 78744 USA. ELISA kits 1014 and 1027, respectively). Reagents and samples were prepared according to the ELISA kits instructional man- ual. Six standard antibiotic concentrations of the Gen- tamicin (negative control, 0.25 ng/ml, 0.75 ng/ml, 1.5 ng/ml, 3.0 ng/ml and 15 ng/ml) and 42 prepared milk test samples (20 samples from the control study and 22 ex- perimental samples collected from various farms) were pipetted in duplicate into different wells. ELISA proce- dure was followed according to the provided manufac- turers’ instructional material. Absorbance was read on a micro titer plate reader with 450 nm wavelength. The same procedure was followed for Streptomycin analyses but the standards provided were; negative control, 0.5 ng/ml, 1 ng/ml, 2.5 ng/ml, 5 ng/ml and 10 ng/ml. 2.5. Microbiological Analysis of Milk Samples A total of 30 raw milk samples collected from the vari- ous farms and from the control study were microbiologi- cally analyzed following the procedure of the bacterio- logical analytical manual [21]. The samples were tested for the presence of Staphylococcus aureus, E. coli, Lis- teria monocytogenes, Listeria innocua, total aerobic count and total coliforms. In the procedure, for the enumeration of E. coli and the total aerobic microorg anisms, a 10 ml portion of each milk sample was aseptically diluted with 90 ml of sterilized peptone-water (PW)1. Serial dilutions from 10−1 to 10−3 were prepared and an aliquot of 0.1 ml of the homogenate was spread on Plate Count Agar1 plates for the total aerobic count while Rapid E. coli (REC 2/agar)1 agar was used by the pour plate technique for the enumeration of E. coli. Plates were then incubated for 48 hours at 37˚C. All colonies on PCA and REC were enumerated; purple-pink colonies were identified as E. coli, whereas blue colonies as coliforms. For S. aureus, a 25 ml portion of each sample was treated in an equiva- lent manner, but diluted with 225 ml PW and plated on Rapid Staphylococcus (R. Staph)1 agar plates. Selected black colonies with a white ring were enumerated and tested for coagulase and catalase activity using the Pas- torex (Staph plus/Latex)1 test as a biochemical confirma- tion test [21]. Finally, the presence of L. monocytogenes was assessed by taking an additional 25 ml portion of each sample and diluted with sterile Listeria Enrichment Broth (Fraser 1 broth)1 supplemented with Listeria En- richment Broth Supplement (Fraser 12 broth supple- ment)1 and spread on plates of Palcam agar (Palcam)1 supplemented with Palcam supplement1. Black colonies were identified as presumptive L. monocytogenes, whereas white colonies were presumptive L. innocua. Then, Fra- zer One supplement (Frazer 1 broth supplement), (0.1 ml/1ml) was added to the homogenate to further enrich the remaining Listeria species for complete detection and were incubated for another 24 hours at 30˚C. 0.1 ml of serial dilutions from 10−1 to 10−3 were then plated on Palcam agar plates in duplicate. The suspected colonies of L. monocytogenes, L. innocua, S. aureus, E. coli and coliforms were then isolated and frozen stocks of each isolate were prepared in sterile microtubes with 1 ml of 30% glycerol and stored at −20˚C to be used in the sub- sequent susceptible tests. 2.6. Antimicrobial Susceptibility Tests Bacterial isolates from the milk samples were tested for antimicrobial susceptibility using the disc agar diffusion method according to the procedures recommended by the Clinical and Laboratory Standards Institute (CLSI) [22]. Gentamicin and Streptomycin, the most commonly used antibiotics, were chosen to assess the resistance of the isolated pathogens. Bacterial isolates from the above mi- crobiological analyses were grown on PCA plates. Strep- tomycin and Gentamicin (10 μg, BIORAD) antibiotic discs and empty discs as control were tested on duplicate plates. The plates were incubated for 24 hours at 37˚C and the zones of inhibition were measured with a metric ruler and interpreted as resistant or sensitive according to the CLSI guidelines. Predisposed strains did not grow in the area around the disc; whereas, resistant strains en- dured the antibiotic. 2.7. Statistical Analyses Data subjected to statistical analysis were analyzed using the software SPSS 15.0 for Windows Evaluation Version. Gentamicin and Streptomycin residue levels were com- 1BIO-RAD; boulevard Raymond-Poincare 92430, Marnes-La-Coquette- France. Copyright © 2013 SciRes. FNS  Quantification of Antibiotic Residues and Determination of Antimicrobial Resistance Profiles of Microorganisms Isolated from Bovine Milk in Lebanon 4 pared by means of 1-way ANOVA at P < 0.05 among the milk samples from the selected farms to determine if antibiotic usage among farmers were similar. Concerning the antimicrobial resistance patterns in milk, statistical analyses (Chi-square analysis at P < 0.05) were con- ducted to compare resistance patterns to Gentamicin with resistance patterns to Streptomycin. To verify if a rela- tion between Streptomycin or Gentamicin residue levels and microorganism levels in the milk samples exists, regression analysis was conducted at P < 0.05. 3. Results The main recurrent antibiotics used among the inter- viewed farmers were Gentamicin, Canamycin, Penicillin, Oxytetracyclin, Tetracycline and Streptomycin with Gen- tamicin and Streptomycin being the most frequently used (88% and 92% respectively). All farmers reported treat- ing their cattle only during illness in doses specified ei- ther by the veterinarian or according to the manufac- turer’s instructions on the label. They also claimed to give the antibiotic treatment for as long as specified and withhold as directed. 3.1. Analysis of Antibiotic Residues 3.1.1. Controlled Study This aimed to determine from a controlled study of a known antibiotic dosage, the residual levels of antibiotics during withholding days to compare with the experimen- tal samples and validate farmers’ claims. Both cows that were treated with Gentamicin had similar antibiotic resi- due levels in their milk samples throughout the 10-day interval of the study. On average, an initial residue level of 107 ng/ml of Gentamicin was noticed in the milk sam- ples on day one followed by a peak residue level of 132 ng/ml on day 4 and a decrease to 57 ng/ml on the 10th day (Figure 1). However, on the 7th day, the final day of withholding period according to the directions, Gen- tamicin residue levels reached a mean concentration of 71.6 ng/ml (Figure 1). Similarly, the initial Streptomycin residue level was 108 ng/ml on day one followed by a peak of 120 ng/ml on day 2 and a decrease to 74 ng/ml of Streptomycin re- sidue levels on the 10th day. At the 6th day, the final day of withholding period as directed, Streptomycin residue levels reached a mean of 107.91 ng/ml (Figure 2). 3.1.2. Experimental Samples The random experimental milk samples were also ana- lyzed for Gentamicin and Streptomycin. The levels of Gentamicin and Streptomycin residues in all the tested samples were less than the recommended maximum residue limit as set by the FAO/WHO; MRL = 200 μg/L (Table 1). This result in milk indicates that the a = Gentamicin residue level at day 7, the international specified day for withholding after treatment of cattle with Gentamicin; Sample detection limit = 2.5 ng/ml; Number of samples = 20 (10 from each cow). Figure 1. Mean Gentamicin residue levels in milk samples collected for 10 days after treating 2 control cows with Gen- tamicin. Streptomycin residue levels in milk after cow treatment with st re pt om ycin a y = -0. 3689x 2 - 0.9245x + 116.46 R 2 = 0. 7926 0.00 20.00 40.00 60.00 80.00 100.00 120.00 140.00 0246810 Days Streptomycin residue levels ng/ml 12 a = Streptomycin residue level at day 7, the international specified day for withholding after treatment of cattle with Streptomycin; Sample detection limit = 2.5 ng/ml; Number of samples = 20; 10 from each cow. Figure 2. Mean Streptomycin residue levels in milk samples collected for 10 days after treatment of 2 cows with Strep- tomycin. application doses by Lebanese farmers may not be ex- ceeding the recommendations. In addition, the milk sam- ples collected from the selected farms had their Gen- tamicin and Streptomycin residue levels equivalent to those obtained from the milk samples in the controlled study post the required withholding period (~71 and 107 ng/ml, respectively). 3.2. Microbiological Analysis of Milk Samples 3.2.1. Controlled study All tested samples were contaminated with S. aureus (103 to 104 CFU/g) and L. monocytogenes (106 to 107 CFU/g); while 62.5% of the milk samples had their total aerobic counts ranging between 103 and 104 CFU/g. None of the samples had E. coli; whereas, 12.5% of samples had total coliform levels between 104 and 105 CFU/g. Finally, L. innocua was detected in 62.5% of the tested samples. 3.2.2. Experimental Samples Microbiological results of the raw milk samples collected from the various selected farms showed a mean value of Copyright © 2013 SciRes. FNS 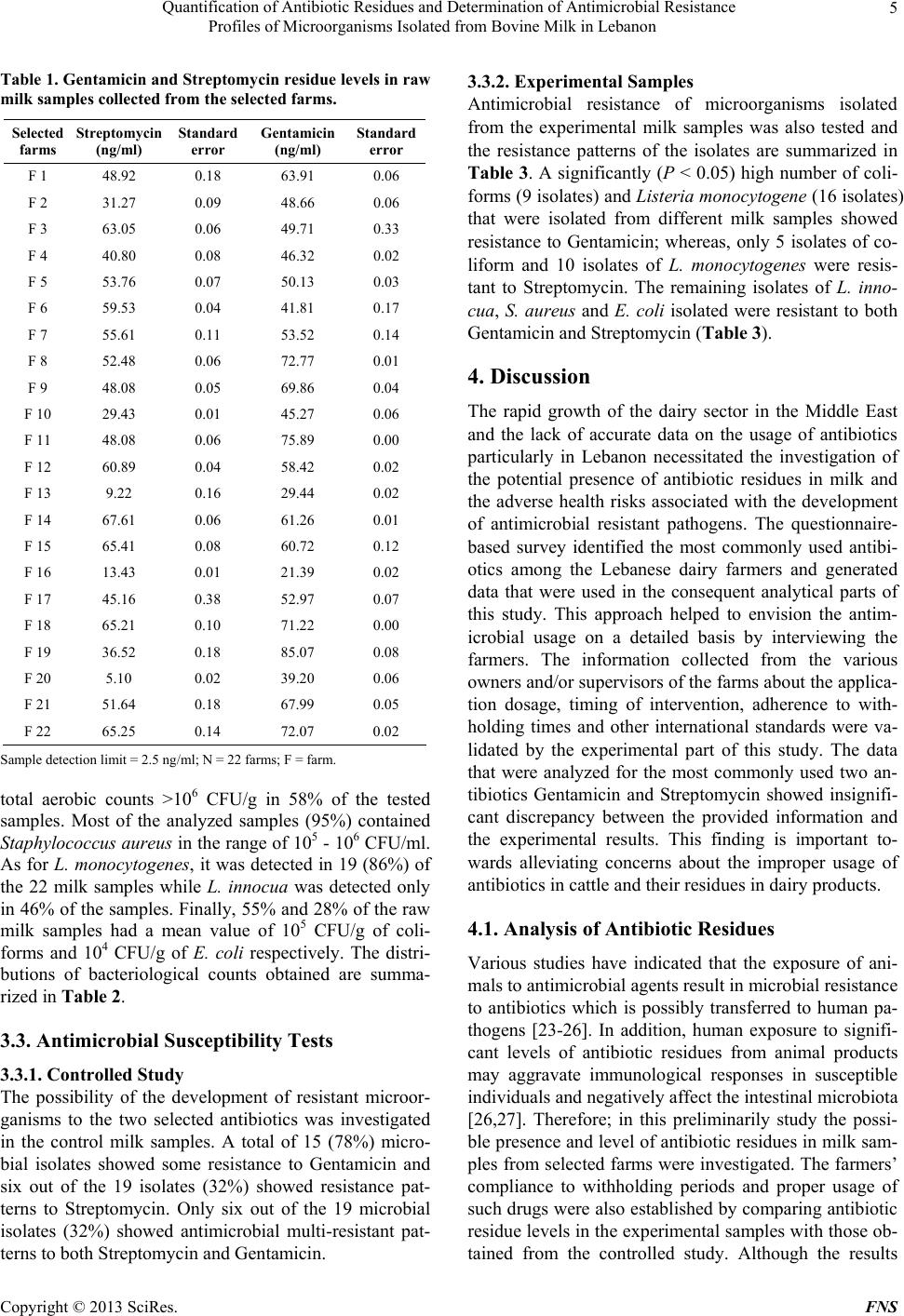 Quantification of Antibiotic Residues and Determination of Antimicrobial Resistance Profiles of Microorganisms Isolated from Bovine Milk in Lebanon 5 Table 1. Gentamicin and Streptomycin residue levels in raw milk samples collected from the selected farms. Selected farms Streptomycin (ng/ml) Standard error Gentamicin (ng/ml) Standard error F 1 48.92 0.18 63.91 0.06 F 2 31.27 0.09 48.66 0.06 F 3 63.05 0.06 49.71 0.33 F 4 40.80 0.08 46.32 0.02 F 5 53.76 0.07 50.13 0.03 F 6 59.53 0.04 41.81 0.17 F 7 55.61 0.11 53.52 0.14 F 8 52.48 0.06 72.77 0.01 F 9 48.08 0.05 69.86 0.04 F 10 29.43 0.01 45.27 0.06 F 11 48.08 0.06 75.89 0.00 F 12 60.89 0.04 58.42 0.02 F 13 9.22 0.16 29.44 0.02 F 14 67.61 0.06 61.26 0.01 F 15 65.41 0.08 60.72 0.12 F 16 13.43 0.01 21.39 0.02 F 17 45.16 0.38 52.97 0.07 F 18 65.21 0.10 71.22 0.00 F 19 36.52 0.18 85.07 0.08 F 20 5.10 0.02 39.20 0.06 F 21 51.64 0.18 67.99 0.05 F 22 65.25 0.14 72.07 0.02 Sample detection limit = 2.5 ng/ml; N = 22 farms; F = farm. total aerobic counts >106 CFU/g in 58% of the tested samples. Most of the analyzed samples (95%) contained Staphylococcus aureus in the range of 105 - 106 CFU/ml. As for L. monocytogenes, it was detected in 19 (86%) of the 22 milk samples while L. innocua was detected only in 46% of the samples. Finally, 55% and 28% of the raw milk samples had a mean value of 105 CFU/g of coli- forms and 104 CFU/g of E. coli respectively. The distri- butions of bacteriological counts obtained are summa- rized in Table 2. 3.3. Antimicrobial Susceptibility Tests 3.3.1. Controlled Study The possibility of the development of resistant microor- ganisms to the two selected antibiotics was investigated in the control milk samples. A total of 15 (78%) micro- bial isolates showed some resistance to Gentamicin and six out of the 19 isolates (32%) showed resistance pat- terns to Streptomycin. Only six out of the 19 microbial isolates (32%) showed antimicrobial multi-resistant pat- terns to both Streptomycin and Gentamicin. 3.3.2. Experimental Samples Antimicrobial resistance of microorganisms isolated from the experimental milk samples was also tested and the resistance patterns of the isolates are summarized in Table 3. A significantly (P < 0.05) high number of coli- forms (9 isolates) and Listeria monocytogene (16 isolates) that were isolated from different milk samples showed resistance to Gentamicin; whereas, only 5 isolates of co- liform and 10 isolates of L. monocytogenes were resis- tant to Streptomycin. The remaining isolates of L. inno- cua, S. aureus and E. coli isolated were resistant to both Gentamicin and Streptomycin (Table 3). 4. Discussion The rapid growth of the dairy sector in the Middle East and the lack of accurate data on the usage of antibiotics particularly in Lebanon necessitated the investigation of the potential presence of antibiotic residues in milk and the adverse health risks associated with the development of antimicrobial resistant pathogens. The questionnaire- based survey identified the most commonly used antibi- otics among the Lebanese dairy farmers and generated data that were used in the consequent analytical parts of this study. This approach helped to envision the antim- icrobial usage on a detailed basis by interviewing the farmers. The information collected from the various owners and/or supervisors of the farms about the applica- tion dosage, timing of intervention, adherence to with- holding times and other international standards were va- lidated by the experimental part of this study. The data that were analyzed for the most commonly used two an- tibiotics Gentamicin and Streptomycin showed insignifi- cant discrepancy between the provided information and the experimental results. This finding is important to- wards alleviating concerns about the improper usage of antibiotics in cattle and their residues in dairy products. 4.1. Analysis of Antibiotic Residues Various studies have indicated that the exposure of ani- mals to antimicrobial agents result in microbial resistance to antibiotics which is possibly transferred to human pa- thogens [23-26]. In addition, human exposure to signifi- cant levels of antibiotic residues from animal products may aggravate immunological responses in susceptible individuals and negatively affect the intestinal microbiota [26,27]. Therefore; in this preliminarily study the possi- ble presence and level of antibiotic residues in milk sam- ples from selected farms were investigated. The farmers’ compliance to withholding periods and proper usage of such drugs were also established by comparing antibiotic residue levels in the experimental samples with those ob- tained from the controlled study. Although the results Copyright © 2013 SciRes. FNS  Quantification of Antibiotic Residues and Determination of Antimicrobial Resistance Profiles of Microorganisms Isolated from Bovine Milk in Lebanon Copyright © 2013 SciRes. FNS 6 Table 2. Percentage of samples tested positive for L. monocytogenes, L. innocua, S. aureus, E. c oli, coliforms and total aerobic counts found in raw milk samples collecte d from 22 r a ndomly selec ted c a ttle far ms in Lebanon. Type of bacteria 0 ND* (%) <103 CFU/g (%) 103 to 104 CFU/g (%) 104 to 105 CFU/g (%) 105 to 106 CFU/g (%) 106 to 107 CFU/g (%) Total (%)**** TAC** 0 9*** 14 14 5 58 22 (100) S. aureus 14 27 9 41 4 21 (95) Coliform 45 18 18 19 0 0 12 (55) E. coli 72 23 5 0 0 0 6 (28) L. mono 14 23 9 27 18 9 19 (86) L. innocua 54 5 14 27 0 0 10 (46) *ND not detected in 25 g of sample tested; **Total aerobic count; ***Percentage of samples having a mean count in the specified range; ****Total number of samples tested positive—number of samples tested negative for the microorganism; N: number of milk samples = 22. Table 3. Resistance, multiresistance and intermediate resis- tance patterns of microorganisms in the milk samples to Gentamicin and Streptomycin. **N (%) isolates resistant to Microorganism *Number of isolates GM SM S. aureus 21 21(100)a 20 (95)a E. coli 10 10 (100)a 6 (60)a Coliforms 9 9 (100)a 5 (55)b L. monocytogenes 17 16 (94)a 10 (58)b L. inocua 7 7 (100)a 6 (85)a Percentages in same row with different superscripts are significantly differ- ent at P < 0.05; GM = Gentamicin; SM = Streptomycin. *Number of isolates isolated from the milk samples for each microorganism. **Number and percentage of isolates of each microorganism resistant to Gentamicin and Streptomycin. obtained in this study show a widespread usage of Strep- tomycin and Gentamicin in Lebanon, a general compli- ance with international regulations for their uses and proper withdrawal times were evident. However, since all the samples collected from the visited farms contained a certain level of antibiotic residues, this may imply a repeated and prolonged treatment of dairy cattle with these antibiotics. Hence, despite the fact that the usage in general is in compliance with regulations with respect to dosages and withholding periods, the excessive and ex- tended applications among farmers may result in the de- velopment of antimicrobial resistant microorganisms in raw milk. This hypothesis was further investigated in this study for the main pathogenic microorganisms isolated from the various milk samples. In other studies, for in- stance, S. aureus has been reported to frequently show multiple antimicrobial resistance patterns [28-31]. 4.2. Microbiological Analysis of Milk Samples The unnecessary use of therapeutic doses of antibiotics or as growth promoters in producing animals may be a main cause for the selection of multiple resistant strains of bacterial pathogens which can result in serious human and animal infections [32,33]. The microbiological anal- yses of both the experimental and control raw milk sam- ples in this study allowed for the selection of the com- monly present microorganisms in milk to be tested for antibiotic resistance against Gentamicin and Streptomy- cin. Interestingly, when comparing the microbiological levels in the samples to the microbiological criteria as set by the Commission Regulation [34], total aerobic counts, L. monocytogenes and S. aureus exceeded the limit al- lowed by the legislation for raw milk samples. As for E. coli and coliforms, although present in the milk samples, their levels were acceptable. These findings although not the main scope of this study, indicate a possible health risk because S. aureus may produce a heat stable toxin in raw milk [35,36]. Furthermore, S. aureus has been known to be the most prevalent pathogen to cause intramam- mary infections in dairy ruminants leading to major eco- nomic losses [37,38]. However, the S. aureus isolated from the above samples were resistant to both Gentami- cin and Streptomycin (Table 3) and this result is quite alarming because if the drugs were or are to be used to treat and control the condition, regular doses may no longer be effective; thus, promoting a high health and residual levels risk on the animals and humans. Another serious pathogen, L. monocytogenes, was also isolated from all collected milk samples. L. monocytogenes has been linked with numerous outbreaks associated with milk and milk products [39-41]. This pathogen may have contaminated the milk samples through inadequate sani- tation during milking, storage and transport or from in- fected cows on the farms [42]. Thus, proper animal mo- nitoring and handling techniques for milk should be ap- plied on Lebanese farms. Furthermore, poor environ- mental sanitation noticed during the farm visits may be the cause for the elevated levels of total coliforms and E. coli in the analyzed milk samples. Studies have shown that E. coli, a normal habitat of human and animal intes- tines, when constantly gets exposed to antibiotics; it de-  Quantification of Antibiotic Residues and Determination of Antimicrobial Resistance Profiles of Microorganisms Isolated from Bovine Milk in Lebanon 7 velops resistance in order to survive. When these resis- tant isolates are excreted to the environment by feces, they tend to spread resistance genes by vertical gene transfer to pathogens [43-45]. Thus, this will result in re- sistance to antimicrobial drugs used in treating infec- tious diseases leading to serious health implications in both humans and animals [46-48]. The above risks are reflected in the results that showed all isolated microorganisms from the experimental and the controlled study samples resistant to Gentamicin (Ta- ble 3). With respect to Streptomycin, the findings were as alarming because all microorganisms, except for E. coli and coliforms isolated from the controlled milk sam- ples, showed various resistance patterns. This may indi- cate that a high percentage of the milk supply in the Le- banese market may contain resistant strains of major pa- thogens against the two drugs, Gentamicin and Strepto- mycin, commonly used in therapeutic treatments; thus, incurring a major public health concern. The excessive use of Gentamicin and Streptomycin among the farms may be a main cause for the emerging of antimicrobial- resistant bacterial pathogens [49]. Furthermore, accord- ing to Prescott and Baggot [16] resistance of animal pa- thogens to aminoglycosides, particularly Streptomycin is very common. 4.3. Conclusion The findings of this study preliminarily validated the claims of dairy Lebanese farmers about the proper usages of antibiotics with respect to doses and withholding pe- riods. However, the results highlighted a potential public health problem reflected in the development of multi- resistant pathogenic bacteria to both Gentamicin and Strep- tomycin. A nationwide study is necessary in the near future with more in depth analyses of the isolated resis- tant pathogens perhaps at the genotypic levels. 5. Acknowledgements The researchers would like to thank the University Re- search Board at the American University of Beirut for funding this study, the Agricultural Research and Educa- tion Center (AREC), Faculty of Agricultural and Food Sciences of the American University of Beirut for mak- ing the controlled study possible, farmers and our labo- ratory technician for helping in the analysis. REFERENCES [1] S. Joshi, “HPLC Separation of Antibiotics Present in Formulated and Unformulated Samples,” Journal of Phar- maceutical and Biomedical Analysis, Vol. 28, No. 5, 2002, pp. 795-809. doi:10.1016/S0731-7085(01)00706-3 [2] M. Choueiri, “Emerging Contaminants in Lebanon: Anti- biotic Residues in Cow Manure and Soil,” MS Thesis, American University of Beirut, Beirut, 2008. [3] W. C. John and E. H. Fred, “Antibiotics: III; Mechanism of Action of Antimicrobial and Antitumor Agents,” Sprin- ger-Verlag, New York, 1975. [4] A. A. Bergwerff and J. Schloesser, “Antibiotics and Drugs Residue Determination,” Encyclopaedia of Food Science, Food Technology and Nutrition, 2003, p. 254. [5] M. Dubois, D. Fluchard, E. Sior and P. H. Delahaut, “Identification and Quantification of Five Macrolide An- tibiotics in Several Tissues, Eggs and Milk by Liquid Chromatography-Electrospray Tandem Mass Spectrome- try,” Journal of Chromatography B: Biomedical Sciences and Applications, Vol. 753, No. 2, 2001, pp. 189-202. doi:10.1016/S0378-4347(00)00542-9 [6] The Council of the European Communities, “Council Re- gulation (EEC) No.2377/90,” Official Journal of the Eu- ropean Communities, No. L 224/P, 1990, pp. 1-8. [7] Food and Drug Administration, “Tolerances for Residues of New Animal Drugs in Food,” 2008. http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/C FRSearch.cfm?fr=530.41 [8] K. N. Woodward, “Antibiotics and Drugs Uses in Food Production,” Encyclopedia of Food Science, Food Tech- nology and Nutrition, 1993, p. 249. [9] F. M. Aarestrup, “Antimicrobial Resistance in Bacteria of Animal Origin,” ASM Press, Washington DC, 2006. [10] L. Guardabassi, S. Schwarz and D. H. Lloyd, “Pet Ani- mals as Reservoirs of Antimicrobial-Resistant Bacteria,” Journal of Antimicrobial Chemotherapy, Vol. 54, No. 2, 2004, pp. 321-332. doi:10.1093/jac/dkh332 [11] I. Phillips, M. Casewell, T. Cox, B. De Groot, C. Friis, R. Jones, C. Nightingale, R. Preston and J. Waddell, “Does the Use of Antibiotics in Food Animals Pose a Risk to Human Health? A Critical Review of Published Data,” Journal of Antimicrobial Chemotherapy, Vol. 53, No. 1, 2004, pp. 28-52. doi:10.1093/jac/dkg483 [12] D. L. Smith, J. Dushoff and J. G. Morris, “Agricultural Antibiotics and Human Health,” PLoS Medicine, Vol. 2, No. 8, 2005, p. 232. doi:10.1371/journal.pmed.0020232 [13] E. J. Threlfall, “Antimicrobial Drug Resistance in Salmo- nella: Problems and Perspectives in Food- and Water- Borne Infections,” FEMS Microbiology Reviews, Vol. 26, No. 2, 2002, pp. 141-148. [14] A. Casadevall, “Crisis in Infectious Diseases: Time for a New Paradigm?” Clinical Infectious Diseases, Vol. 23, No. 4, 1996, pp. 790-794. doi:10.1093/clinids/23.4.790 [15] T. C. Eickhoff, “Antibiotics and Nosocomial Infections,” In: J. V. Bennett and P. S. Brachman, Eds., Hospital In- fections, 3rd Edition, Little, Brown and Company, Boston, 1992, pp. 245-264. [16] J. F. Prescott and J. D. Baggot, “Antimicrobial Therapy in Veterinary Medicine,” Iowa State University Press, Ames, 1993. [17] F. Peles, M. Wagner, L. Varga, I. Hein, P. Rieck, K. Gut- ser, P. Keresztúri, G. Kardos, I. Turcsányi, B. Béri and A. Szabó, “Characterization of Staphylococcus aureus Strains Copyright © 2013 SciRes. FNS  Quantification of Antibiotic Residues and Determination of Antimicrobial Resistance Profiles of Microorganisms Isolated from Bovine Milk in Lebanon 8 Isolated from Bovine Milk in Hungary,” International Journal of Food Microbiology, Vol. 118, No. 2, 2007, pp. 186-193. doi:10.1016/j.ijfoodmicro.2007.07.010 [18] P. M. Griffin and A. V. Tauxe, “The Epidemiology of Infections Caused by Escherichia coli O157:H7, Other Enterohemorrhagic E. coli and the Associated Hemolytic Uremic Syndrome,” Epidemiologic Reviews, Vol. 13, 1991, pp. 60-98. [19] H. P. R. Seeliger and D. Jones, “Genus Listeria,” In: J. G. Holt, Ed., Bergey’s Manual of Systematic Bacteriology, Vol. 2, 8th Edition, Williams and Wilkins, Baltimore, 1986, pp. 1235-1245. [20] B.-C. Ye, S. Y. Li, P. Zuo and X.-H. Li, “Simultaneous Detection of Sulfamethazine, Streptomycin, and Tylosin in Milk by Microplate-Array Based SMM-FIA,” Food Chemistry, Vol. 106, No. 2, 2007, pp. 797-803. doi:10.1016/j.foodchem.2007.06.006 [21] H. A. Wallace, G. A. June, P. S. Sherrod, T. S. Hammack and R. M. Amaguana, “Salmonella,” In FDA Bacteriolo- gical Analytical Manual, 8th Edition, Washington DC, 1995. [22] Clinical and Laboratory Standards Institute, “Performance Standards for Antimicrobial Susceptibility Tests,” Na- tional Committee for Clinical Laboratory Standards, Wayne, 2009. [23] C. Chauvin, S. Le Bouquin-Leneveu, A. Hardy, D. Hag- uet, J. P. Orand and P. Sanders, “An Original System for the Continuous Monitoring of Antimicrobial Use in Poul- try Production in France,” Journal of Veterinary Phar- macology and Therapeutics, Vol. 28, No. 6, 2005, pp. 515-523. doi:10.1111/j.1365-2885.2005.00697.x [24] A. L. Aronson, “Potential Impact of the Use of Antim- icrobial Drugs in Farm Animals on Public Health,” Pre- sented at the Meeting on Pharmacology in the Animal Health Sector, Colorado State University, 1975. [25] H. D. Mercer, “Antimicrobial Drugs in Food Producing Animals,” Veterinary Clinics of North America, Vol. 5, 1977, pp. 3-5. [26] S. D. Holmberg, M. T. Osterholm, K. A. Senger and M. L. Cohen, “Drug-Resistant Salmonella from Animals Fed Antimicrobials,” New England Journal of Medicine, Vol. 311, No. 10, 1984, pp. 617-622. doi:10.1056/NEJM198409063111001 [27] P. H. Archimbault, “Persistence in Milk of Active An- timicrobial Intramammary Substances. Veterinary Phar- macology and Toxicology,” MTP Press Ltd., Lancaster, 2005. [28] M. C. Enright, “The Evolution of Resistant Pathogen— The Case of MRSA,” Current Opinion in Pharmacology, Vol. 3, No. 5, 2003, pp. 474-479. doi:10.1016/S1471-4892(03)00109-7 [29] N. H. Kwon, K. T. Park, J. S. Moon, W. K. Jung, S. H. Kim and J. M. Kim, “Staphylococcal Cassette Chromo- some Mec (SCCmec) Characterization and Molecular Ana- lysis for Methicillin-Resistant Staphylococcus aureus and Novel SCCmec Subtype IVg Isolated from Bovine Milk in Korea,” Journal of Antimicrobial Chemotherapy, Vol. 56, No. 4, 2005, pp. 624-632. doi:10.1093/jac/dki306 [30] R. O’Mahony, Y. Abbott, F. C. Leonard, B. K. Markey, P. J. Quinn and P. J. Pollock, “Methicillin-Resistant Staphyl- ococcus aureus (MRSA) Isolated from Animals and Vet- erinary Personnel in Ireland,” Veterinary Microbiology, Vol. 109, No. 3-4, 2005, pp. 285-296. doi:10.1016/j.vetmic.2005.06.003 [31] J. L. Wylie and D. L. Nowicki, “Molecular Epidemiology of Community- and Health Care-Associated Methicillin- Resistant Staphylococcus aureus in Manitoba, Canada,” Journal of Clinical Microbiology, Vol. 43, No. 6, 2005, pp. 2830-2836. doi:10.1128/JCM.43.6.2830-2836.2005 [32] WHO, “Surveillance for the Prevention and Control of Health Hazards Due to Antibiotic Resistant Enterobacte- ria,” Technical Rep ort Series 624, 1978. [33] D. A. Barber, G. Y. Miller and P. E. McNamara, “Models of Antimicrobial Resistance and Food-Borne Illness: Ex- amining Assumptions and Practical Application,” Journal of Food Protection, Vol. 66, No. 4, 2003, pp. 700-709. [34] The Council of the European Communities, “Council Re- gulation (EEC) No. 2073/2005,” Official Journal of the European Communities, No. L 338, 2005, pp. 1-26. [35] G. Normanno, G. La Salandra, A. Dambrosio, N. C. Quaglia, M. Corrente, A. Parisi, G. Santagada, A. Firinu, E. Crisetti and G. V. Celano, “Occurrence, Characteriza- tion and Antimicrobial Resistance of Enterotoxigenic Sta- phylococcus aureus Isolated from Meat and Dairy Prod- ucts,” International Journal of Food Microbiology, Vol. 115, No. 3, 2007, pp. 290-296. doi:10.1016/j.ijfoodmicro.2006.10.049 [36] M. L. Marco and M. H. J. Wells-Bennik, “Impact of Bac- terial Genomics on Determining Quality and Safety in the Dairy Production Chain,” International Dairy Journal, Vol. 18, No. 5, 2008, pp. 486-495. [37] O. Akineden, C. Annemüller, A. Hassan, C. Lämmler, W. Wolter and M. Schöck, “Toxin Genes and Other Charac- teristics of Staphylococcus aureus Isolates from Milk of Cows with Mastitis,” Clinical and Diagnostic Laboratory Immunology, Vol. 8, No. 5, 2001, pp. 959-964. [38] A. Pengov, C. V. Flajs, T. Zadnik, J. Marinsek and M. Pogacnik, “Distribution of Chloramphenicol Residues in Lactating Cows Following an External Application,” Analytica Chimica Acta, Vol. 529, No. 1-2, 2005, pp. 347-351. doi:10.1016/j.aca.2004.07.035 [39] D. W. Flemming, S. L. Cochi, K. L. Mackonald, J. Bron- dum, P. S. Hayes and B. D. Plikaytis, “Pasteurised Milk as a Vehicle of Infection in an Outbreak of Listeriosis,” New England Journal of Medicine, Vol. 312, 1985, pp. 404-407. doi:10.1056/NEJM198502143120704 [40] M. J. Linnan, L. Mascola, X. D. Lou, V. Goulet, S. May, C. Salminen, D. W. Hird, M. L. Yonekura, P. Hayes, R. Weaver, A. Audurier, B. D. Plikaytis, S. L. Fannin, A. Kleks and C. V. Broome, “Epidemic Listeriosis Associ- ated with Mexican Style Cheese,” New England Journal of Medicine, Vol. 319, 1988, pp. 823-828. doi:10.1056/NEJM198809293191303 [41] O. Lyytikainen, T. Autio, R. Maijala, P. Ruutu, T. Hon- kanen-Buzalski and M. Miettinen, “An Outbreak of Lis- teria Monocytogenes Serotype 3a Infections from Butter Copyright © 2013 SciRes. FNS  Quantification of Antibiotic Residues and Determination of Antimicrobial Resistance Profiles of Microorganisms Isolated from Bovine Milk in Lebanon Copyright © 2013 SciRes. FNS 9 in Finland,” Journal of Infectious Diseases, Vol. 181, No. 5, 2000, pp. 1838-1841. doi:10.1086/315453 [42] N. Bemrah, M. Sanaa, M. H. Cassin, M. W. Griffiths and O. Cerf, “Quantitative Risk Assessment of Human Liste- riosis from Consumption of Soft Cheese Made from Raw Milk,” Preventive Veterinary Medicine, Vol. 37, No. 1-4, 1998, pp. 129-145. doi:10.1016/S0167-5877(98)00112-3 [43] V. Babak, J. Schlegelova and H. Vlkova, “Interpretation of the Results of Antimicrobial Susceptibility Analysis of Escherichia coli Isolates from Bovine Milk, Meat and Associated Foodstuffs,” Food Microbiology, Vol. 22, No. 4, 2004, pp. 353-358. doi:10.1016/j.fm.2004.08.010 [44] H. Sorum and M. Sunde, “Resistance to Antibiotics in the Normal Flora of Animals,” Veterinary Research, Vol. 32, No. 3-4, 2001, pp. 227-241. doi:10.1051/vetres:2001121 [45] B. Catry, H. Laevens, L. A. Devriese, G. Opsomer and A. de Kruif, “Antimicrobial Resistance in Livestock,” Jour- nal of Veterinary Pharmacology and Therapeutics, Vol. 26, No. 2, 2003, pp. 81-93. doi:10.1046/j.1365-2885.2003.00463.x [46] H. Oppegaard, T. M. Steinum and Y. Wasteson, “Hori- zontal Transfer of a Multi-Drug Resistance Plasmid be- tween Coliform Bacteria of Human and Bovine Origin in a Farm Environment,” Applied and Environmental Mi- crobiology, Vol. 67, No. 8, 2001, pp. 3732-3734. doi:10.1128/AEM.67.8.3732-3734.2001 [47] B. R. Berends, A. E. Van den Bogaard, F. Van Knapen and J. M. Snijders, “Human Health Hazards Associated with the Administration of Antimicrobials to Slaughter Animals. Part II. An Assessment of the Risks of Resistant Bacteria in Pigs and Pork,” Veterinary Quarterly, Vol. 23, No. 1, 2001, pp. 10-21. doi:10.1080/01652176.2001.9695069 [48] J. W. Chow, V. Kak, I. You, S. J. Kao, J. Petrin, D. B. Clewell, S. A. Lerner, G. H. Miller and K. J. Shaw, “Ami- noglycoside Resistance Genes aph(200)-Ib and aac(60)- Im Detected Together in Strains of Both Escherichia coli and Enterococcus faecium,” Antimicrobial Agents and Chemotherapy, Vol. 45, No. 10, 2001, pp. 2691-2694. doi:10.1128/AAC.45.10.2691-2694.2001 [49] S. Magnet and J. S. Blanchard, “Molecular Insights into Aminoglycoside Action and Resistance,” Chemical Re- views, Vol. 105, No. 2, 2005, pp. 477-498. doi:10.1021/cr0301088
|