Paper Menu >>
Journal Menu >>
 Chinese Medicine, 2010, 1, 31-38 doi:10.4236/cm.2010.12006 Published Online September 2010 (http://www.SciRP.org/journal/cm) Copyright © 2010 SciRes. CM Antimicrobial Activity of Polyalthia longifolia (Sonn.) Thw. var. Pendula Leaf Extracts Against 91 Clinically Important Pathogenic Microbial Strains Sumitra Chanda*, Rathish Nair Phytochemical, Pharmacological and Microbiological Laboratory Department of Biosciences, Saurashtra University, Rajkot, Gujarat, India E-mail: svchanda@gmail.com Received May 21, 2010; revised July 21, 2010; accepted July 26, 2010 Abstract The methanol, acetone and 1,4-dioxan fractions of leaves of Polyalthia longifolia (Sonn.) Thw. were evalu- ated for antibacterial and antifungal activity. 91 clinically important strains were used for the study which were both clinical isolates as well as identified strains. Piperacillin and gentamicin were used as standards for antibacterial assay, while nystatin and flucanazole were used as standards for antifungal assay. The antibac- terial activity was more pronounced against gram positive bacterial and fungal strains. Poor activity was shown against gram negative bacterial strains studied. Keywords: Antibacterial, Antifungal, Polyalthia longifolia, Clinical Isolates, Organic Solvent Extracts 1. Introduction Due to the increasing development of drug resistance in human pathogens as well as the appearance of undesir- able effect on certain antimicrobial agents, there is a need to search for new agents. The world health organi- zation in 1997 suggested that effective locally available plants be used as substitutes for drugs. Research work on medicinal plants be intensified and information on these plants be exchanged. This thought will go a long way in the scientific exploration of medicinal plants for the benefit of man and is likely to decrease the dependence on importance of drugs [1]. Polyalthia longifolia (An- nonaceae) is a tree, which is widely distributed in Bang- ladesh, Srilanka and throughout the hotter parts of India [2]. In India, the seeds of this plant were used as febri- fuge [3]. Literature survey revealed that most of the plants of annonaceae family contain antitumor and anticancer principles [4,5]. The bark is also used as a febrifuge in the Balasore district of Orissa [6]. The ex- tract of stem bark and the alkaloids isolated from this were found to demonstrate a good antibacterial and antifungal activities [7]. In the present study, antim- icrobial potentiality of the P. longifolia leaves was in- vestigated against a few clinically isolated as well as standard microbial cultures. 2. Materials and Methods 2.1. Plant Material Polyalthia longifolia (Sonn.) Thw. (Annonaceae) leaves were collected in May, 2004 from Rajkot in the State of Gujarat (Western India) and identified by comparison with specimens (PSN 4) available at the Herbarium of the Department of Biosciences, Saurashtra University, Rajkot, Gujarat, India. 2.2. Extraction Leaves of P. longifolia were collected, air dried and then powdered in a homogenizer and 10 gm was used for dif- ferent solvent extractions (Methanol, Acetone, N, N-dimethylformamide); the sample was extracted in sol- vent kept on a rotary shaker overnight, and then the fil- trate was collected and centrifuged at 5000 rpm. The solvent was then evaporated to dryness under reduced pressure and the extracted compound left was used for the antimicrobial assay. The percentage yield of 1, 4-dioxan, methanol and acetone extracts were 20.56, 29.30 and 13.52 respectively. Microorganisms Studied 91 clinically important mi- crobial strains which included 23 gram positive, 56 gram 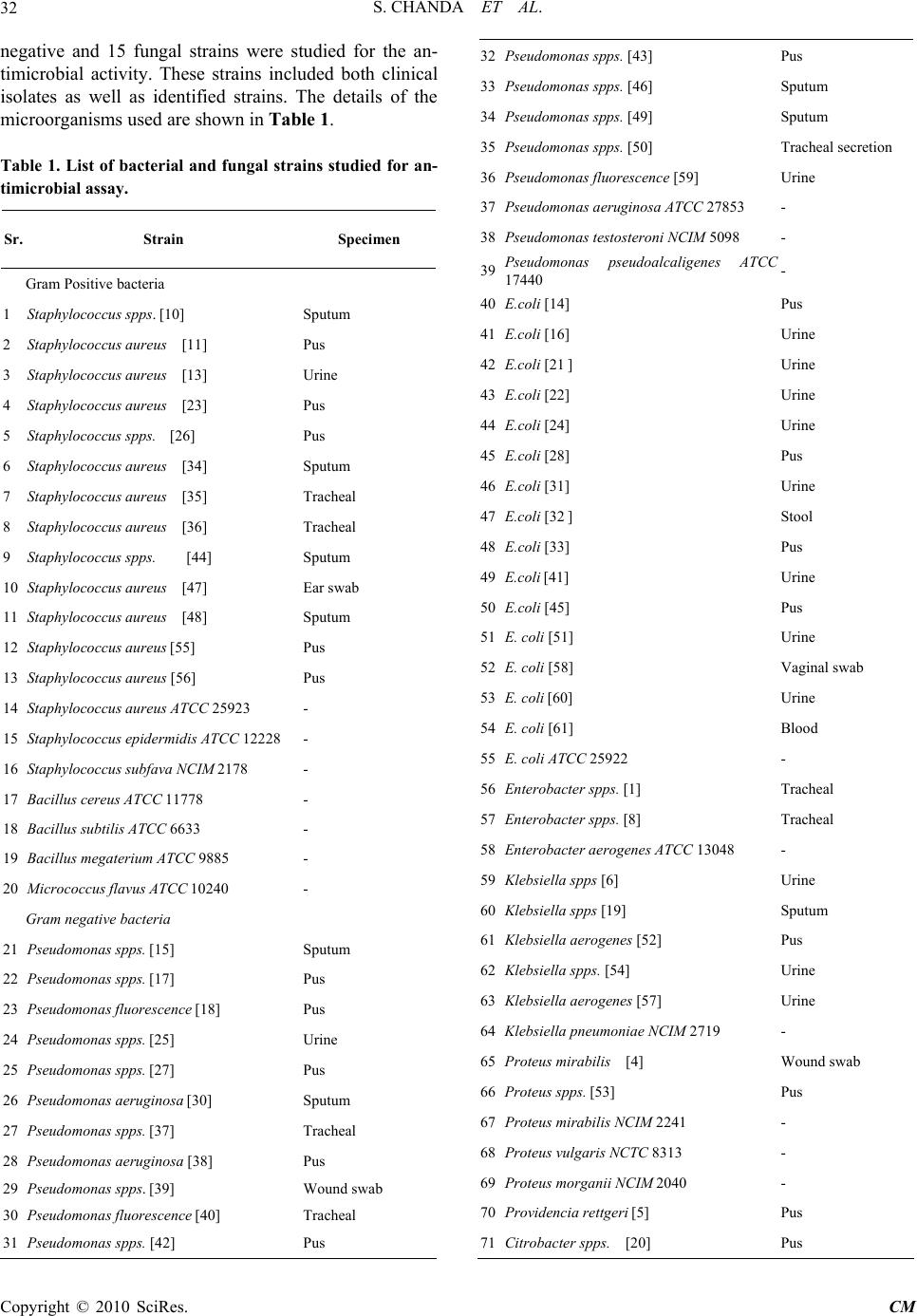 S. CHANDA ET AL. Copyright © 2010 SciRes. CM 32 negative and 15 fungal strains were studied for the an- timicrobial activity. These strains included both clinical isolates as well as identified strains. The details of the microorganisms used are shown in Table 1. Table 1. List of bacterial and fungal strains studied for an- timicrobial assay. Sr. Strain Specimen Gram Positive bacteria 1 Staphylococcus spps. [10] Sputum 2 Staphylococcus aureus [11] Pus 3 Staphylococcus aureus [13] Urine 4 Staphylococcus aureus [23] Pus 5 Staphylococcus spps. [26] Pus 6 Staphylococcus aureus [34] Sputum 7 Staphylococcus aureus [35] Tracheal 8 Staphylococcus aureus [36] Tracheal 9 Staphylococcus spps. [44] Sputum 10 Staphylococcus aureus [47] Ear swab 11 Staphylococcus aureus [48] Sputum 12 Staphylococcus aureus [55] Pus 13 Staphylococcus aureus [56] Pus 14 Staphylococcus aureus ATCC 25923 - 15 Staphylococcus epidermidis ATCC 12228 - 16 Staphylococcus subfava NCIM 2178 - 17 Bacillus cereus ATCC 11778 - 18 Bacillus subtilis ATCC 6633 - 19 Bacillus megaterium ATCC 9885 - 20 Micrococcus flavus ATCC 10240 - Gram negative bacteria 21 Pseudomonas spps. [15] Sputum 22 Pseudomonas spps. [17] Pus 23 Pseudomonas fluorescence [18] Pus 24 Pseudomonas spps. [25] Urine 25 Pseudomonas spps. [27] Pus 26 Pseudomonas aeruginosa [30] Sputum 27 Pseudomonas spps. [37] Tracheal 28 Pseudomonas aeruginosa [38] Pus 29 Pseudomonas spps. [39] Wound swab 30 Pseudomonas fluorescence [40] Tracheal 31 Pseudomonas spps. [42] Pus 32 Pseudomonas spps. [43] Pus 33 Pseudomonas spps. [46] Sputum 34 Pseudomonas spps. [49] Sputum 35 Pseudomonas spps. [50] Tracheal secretion 36 Pseudomonas fluorescence [59] Urine 37 Pseudomonas aeruginosa ATCC 27853 - 38 Pseudomonas testosteroni NCIM 5098 - 39 Pseudomonas pseudoalcaligenes ATCC 17440 - 40 E.coli [14] Pus 41 E.coli [16] Urine 42 E.coli [21 ] Urine 43 E.coli [22] Urine 44 E.coli [24] Urine 45 E.coli [28] Pus 46 E.coli [31] Urine 47 E.coli [32 ] Stool 48 E.coli [33] Pus 49 E.coli [41] Urine 50 E.coli [45] Pus 51 E. coli [51] Urine 52 E. coli [58] Vaginal swab 53 E. coli [60] Urine 54 E. coli [61] Blood 55 E. coli ATCC 25922 - 56 Enterobacter spps. [1] Tracheal 57 Enterobacter spps. [8] Tracheal 58 Enterobacter aerogenes ATCC 13048 - 59 Klebsiella spps [6] Urine 60 Klebsiella spps [19] Sputum 61 Klebsiella aerogenes [52] Pus 62 Klebsiella spps. [54] Urine 63 Klebsiella aerogenes [57] Urine 64 Klebsiella pneumoniae NCIM 2719 - 65 Proteus mirabilis [4] Wound swab 66 Proteus spps. [53] Pus 67 Proteus mirabilis NCIM 2241 - 68 Proteus vulgaris NCTC 8313 - 69 Proteus morganii NCIM 2040 - 70 Providencia rettgeri [5] Pus 71 Citrobacter spps. [20] Pus  S. CHANDA ET AL. Copyright © 2010 SciRes. CM 33 72 Citrobacter freundii [29] Pus 73 Citrobacter freundii ATCC 10787 - 74 Alcaligenes fecalis ATCC 8750 - 75 Salmonella typhimurium ATCC 23564 - Fungus 76 Candida albicans [1] Urine 77 Candida albicans [2] Sputum 78 Candida spps. [3] Sputum 79 Candida spps. [4] Sputum 80 Candida spps. [5] Urine 81 Candida albicans ATCC 2091 - 82 Candida albicans ATCC 18804 - 83 Candida glabrata NCIM 3448 - 84 Candida tropicalis ATCC 4563 - 85 Candida apicola NCIM 3367 - 86 Cryptococcus neoformans ATCC 34664 - 87 Cryptococcus luteolus ATCC 32044 - 88 Trichosporan beigelii NCIM 3404 - 89 Aspergillus flavus NCIM 538 - 90 Aspergillus candidus NCIM 883 - 91 Aspergillus niger ATCC 6275 - 2.3. Preparation of Samples Methanol, acetone and 1,4-dioxan extracts were dis- solved in 100% DMSO at a concentration of 25 mg/ml and 12.5 mg/ml and were used as working stocks respec- tively. Sterile discs (Hi-media Labs) were impregnated with 20 µl of the stock solution. Gentamicin (10 µg/disc) and Piperacillin (100 µg/disc) for bacteria; nystatin (100 units/disc) and flucanazole (10 µg/disc) (Himedia Labs) for fungus were used as positive control and pure DMSO was used as a negative control. 2.4. Antimicrobial Study Antimicrobial activity was performed by agar disc diffu- sion method [8,9]. The bacterial strains were grown in nutrient broth while fungal strains were grown in MGYP (Malt glucose yeast peptone) broth. Mueller Hinton agar no. 2 was the media used to study the antibacterial sus- ceptibility while Sabroaud agar was used to study the antifungal susceptibility test. The cultures were grown for 24 hours, and the turbidity of the culture was main- tained according to the 0.5 MacFarland standards. The inoculum’s size was 1 × 108 cells/ml. 2.5. Agar Disc Diffusion The media (Mueller Hinton Agar No.2 and MRS media) and the test bacterial cultures were poured into Petri dishes (Hi-Media). The test strain (200 µl) was inocu- lated into the media (inoculums size 108 cells/ml) when the temperature reached 40-42°C. The test compound (20 µl) was impregnated in to sterile discs (7 mm) (Hi-Media) and was then allowed to dry. The disc was then intro- duced into medium with the bacteria. The plates were incubated overnight at 37°C for bacterial strains and 28°C for fungal strains. The experiment was performed under strictly aseptic conditions. Microbial growth was determined by measuring the diameter of the zone of inhibition. The experiment was performed in triplicates and the mean values of the result are shown in Table 2. 3. Results and Discussion Herbal medicine in developing countries is commonly used for the traditional treatment of health problems [10]. In recent years multiple drug resistance in human patho- genic microorganisms has developed due to the indis- criminate use of commercial antimicrobial drugs com- monly used in the treatment of infectious diseases, mak- ing it a global growing problem [11-13]. In addition to this problem antibiotics are sometimes associated with adverse effects on host including hypersensitivity, im- mune suppression and allergic reactions [14]. Therefore there is a need to develop alternative antimicrobial drugs for the treatment of infections obtained from various sources such as medicinal plants [15,16]. In the present study, P. longifolia leaf extracts extracted in 1, 4-dioxan (PDE), methanol (PME) and acetone extracts (PAE) were investigated at two different concentrations for their antimicrobial potentiality against 91 clinically important microbial strains. All the three extracts (PDE, PME and PAE at 500 µg/disc concentration) were active against 95% of the total gram positive bacterial strains studied. PDE was active against 18.18% of the total gram nega- tive bacterial strains studied (active against 21% of Pseudomonas spps., 33.3% of Enterobacter spps., 16% of Klebsiella spps., 33.3% of Proteus spps. and 66.6% of Citrobacter spps.). PME and PAE were active against 12.72% of the total gram negative bacterial strains stud- ied. P. aeruginosa is most common pathogen of im- muno-compromised individuals [17]. Infections caused by Pseudomonas spps. are among the most difficult to treat with conventional antibiotics. Both PME and PAE were active against 5.26% of the Pseudomonas spps. and 66.6% of Enterobacter spps. PME was active against 33.3% of Klebsiella spps. and Proteus spps., while PAE was active against 66.6% of Klebsiella spps. and Proteus spps. studied. Salmonellosis is an important public 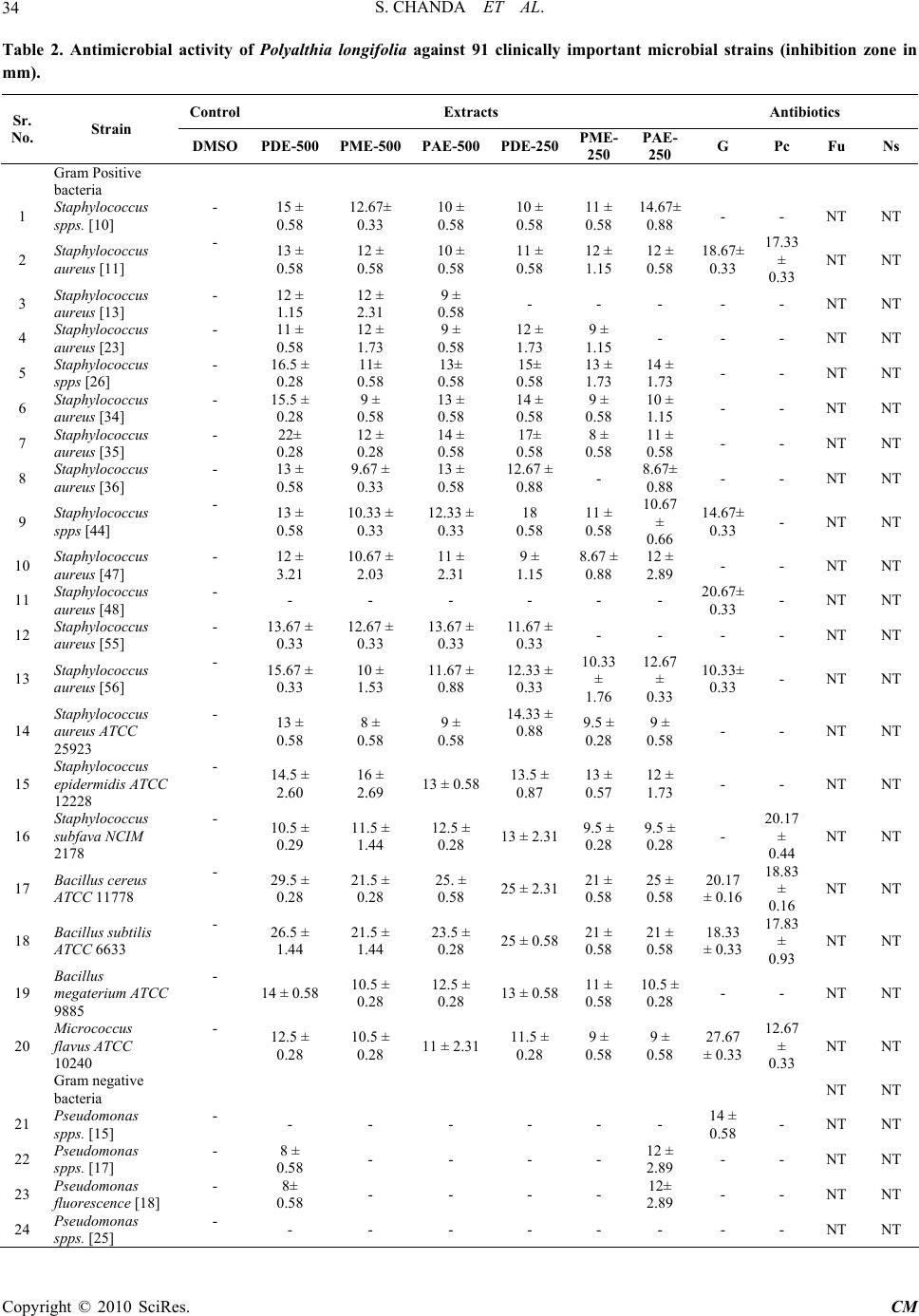 S. CHANDA ET AL. Copyright © 2010 SciRes. CM 34 Table 2. Antimicrobial activity of Polyalthia longifolia against 91 clinically important microbial strains (inhibition zone in mm). Control Extracts Antibiotics Sr. No. Strain DMSOPDE-500 PME-500PAE-500 PDE-250 PME- 250 PAE- 250 G Pc Fu Ns Gram Positive bacteria 1 Staphylococcus spps. [10] - 15 ± 0.58 12.67± 0.33 10 ± 0.58 10 ± 0.58 11 ± 0.58 14.67± 0.88 - - NT NT 2 Staphylococcus aureus [11] - 13 ± 0.58 12 ± 0.58 10 ± 0.58 11 ± 0.58 12 ± 1.15 12 ± 0.58 18.67± 0.33 17.33 ± 0.33 NT NT 3 Staphylococcus aureus [13] - 12 ± 1.15 12 ± 2.31 9 ± 0.58 - - - - - NT NT 4 Staphylococcus aureus [23] - 11 ± 0.58 12 ± 1.73 9 ± 0.58 12 ± 1.73 9 ± 1.15 - - - NT NT 5 Staphylococcus spps [26] - 16.5 ± 0.28 11± 0.58 13± 0.58 15± 0.58 13 ± 1.73 14 ± 1.73 - - NT NT 6 Staphylococcus aureus [34] - 15.5 ± 0.28 9 ± 0.58 13 ± 0.58 14 ± 0.58 9 ± 0.58 10 ± 1.15 - - NT NT 7 Staphylococcus aureus [35] - 22± 0.28 12 ± 0.28 14 ± 0.58 17± 0.58 8 ± 0.58 11 ± 0.58 - - NT NT 8 Staphylococcus aureus [36] - 13 ± 0.58 9.67 ± 0.33 13 ± 0.58 12.67 ± 0.88 - 8.67± 0.88 - - NT NT 9 Staphylococcus spps [44] - 13 ± 0.58 10.33 ± 0.33 12.33 ± 0.33 18 0.58 11 ± 0.58 10.67 ± 0.66 14.67± 0.33 - NT NT 10 Staphylococcus aureus [47] - 12 ± 3.21 10.67 ± 2.03 11 ± 2.31 9 ± 1.15 8.67 ± 0.88 12 ± 2.89 - - NT NT 11 Staphylococcus aureus [48] - - - - - - - 20.67± 0.33 - NT NT 12 Staphylococcus aureus [55] - 13.67 ± 0.33 12.67 ± 0.33 13.67 ± 0.33 11.67 ± 0.33 - - - - NT NT 13 Staphylococcus aureus [56] - 15.67 ± 0.33 10 ± 1.53 11.67 ± 0.88 12.33 ± 0.33 10.33 ± 1.76 12.67 ± 0.33 10.33± 0.33 - NT NT 14 Staphylococcus aureus ATCC 25923 - 13 ± 0.58 8 ± 0.58 9 ± 0.58 14.33 ± 0.88 9.5 ± 0.28 9 ± 0.58 - - NT NT 15 Staphylococcus epidermidis ATCC 12228 - 14.5 ± 2.60 16 ± 2.69 13 ± 0.5813.5 ± 0.87 13 ± 0.57 12 ± 1.73 - - NT NT 16 Staphylococcus subfava NCIM 2178 - 10.5 ± 0.29 11.5 ± 1.44 12.5 ± 0.28 13 ± 2.319.5 ± 0.28 9.5 ± 0.28 - 20.17 ± 0.44 NT NT 17 Bacillus cereus ATCC 11778 - 29.5 ± 0.28 21.5 ± 0.28 25. ± 0.58 25 ± 2.3121 ± 0.58 25 ± 0.58 20.17 ± 0.16 18.83 ± 0.16 NT NT 18 Bacillus subtilis ATCC 6633 - 26.5 ± 1.44 21.5 ± 1.44 23.5 ± 0.28 25 ± 0.5821 ± 0.58 21 ± 0.58 18.33 ± 0.33 17.83 ± 0.93 NT NT 19 Bacillus megaterium ATCC 9885 - 14 ± 0.58 10.5 ± 0.28 12.5 ± 0.28 13 ± 0.5811 ± 0.58 10.5 ± 0.28 - - NT NT 20 Micrococcus flavus ATCC 10240 - 12.5 ± 0.28 10.5 ± 0.28 11 ± 2.3111.5 ± 0.28 9 ± 0.58 9 ± 0.58 27.67 ± 0.33 12.67 ± 0.33 NT NT Gram negative bacteria NT NT 21 Pseudomonas spps. [15] - - - - - - - 14 ± 0.58 - NT NT 22 Pseudomonas spps. [17] - 8 ± 0.58 - - - - 12 ± 2.89 - - NT NT 23 Pseudomonas fluorescence [18] - 8± 0.58 - - - - 12± 2.89 - - NT NT 24 Pseudomonas spps. [25] - - - - - - - - - NT NT 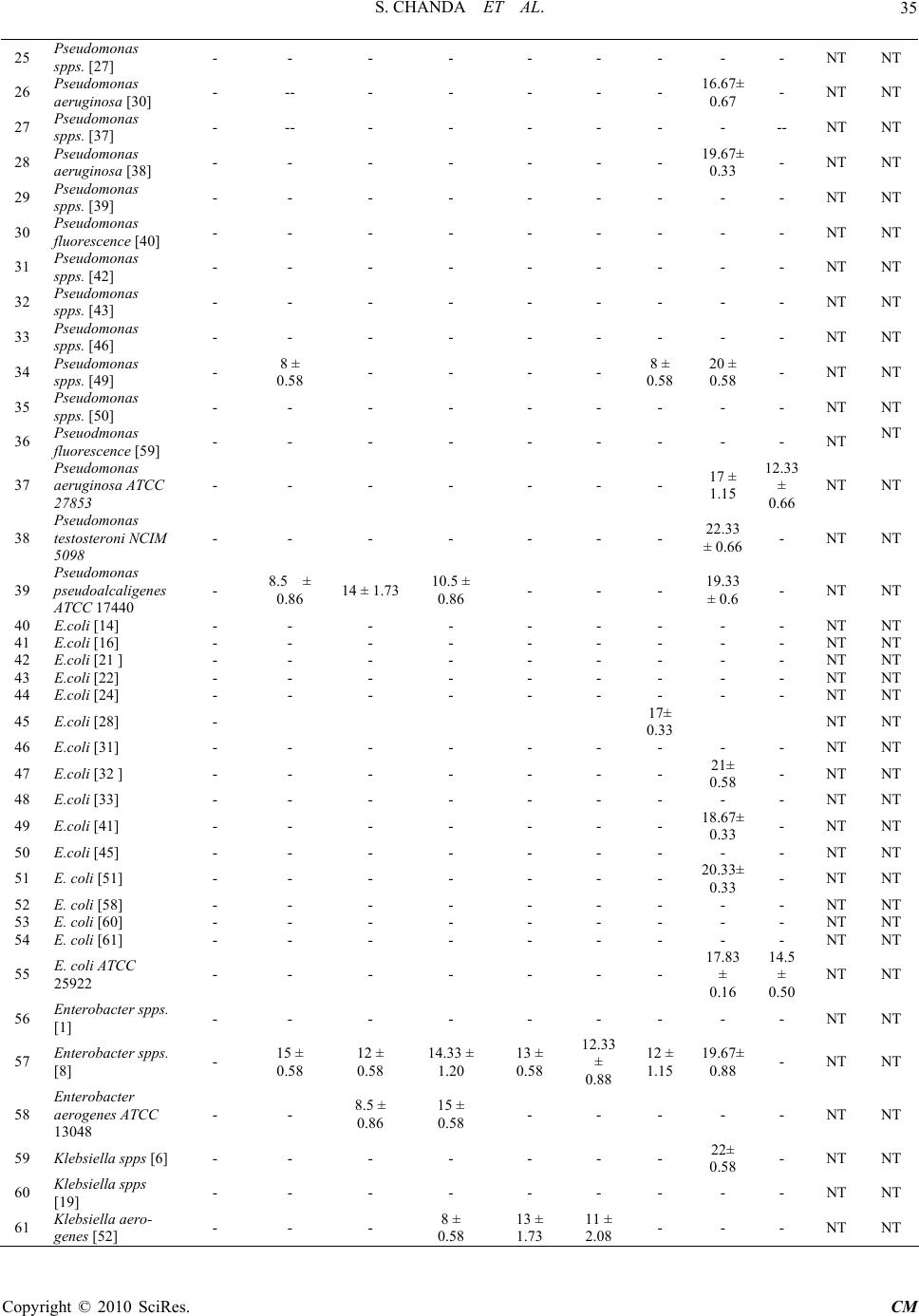 S. CHANDA ET AL. Copyright © 2010 SciRes. CM 35 25 Pseudomonas spps. [27] - - - - - - - - - NT NT 26 Pseudomonas aeruginosa [30] - -- - - - - - 16.67± 0.67 - NT NT 27 Pseudomonas spps. [37] - -- - - - - - - -- NT NT 28 Pseudomonas aeruginosa [38] - - - - - - - 19.67± 0.33 - NT NT 29 Pseudomonas spps. [39] - - - - - - - - - NT NT 30 Pseudomonas fluorescence [40] - - - - - - - - - NT NT 31 Pseudomonas spps. [42] - - - - - - - - - NT NT 32 Pseudomonas spps. [43] - - - - - - - - - NT NT 33 Pseudomonas spps. [46] - - - - - - - - - NT NT 34 Pseudomonas spps. [49] - 8 ± 0.58 - - - - 8 ± 0.58 20 ± 0.58 - NT NT 35 Pseudomonas spps. [50] - - - - - - - - - NT NT 36 Pseuodmonas fluorescence [59] - - - - - - - - - NT NT 37 Pseudomonas aeruginosa ATCC 27853 - - - - - - - 17 ± 1.15 12.33 ± 0.66 NT NT 38 Pseudomonas testosteroni NCIM 5098 - - - - - - - 22.33 ± 0.66 - NT NT 39 Pseudomonas pseudoalcaligenes ATCC 17440 - 8.5 ± 0.86 14 ± 1.7310.5 ± 0.86 - - - 19.33 ± 0.6 - NT NT 40 E.coli [14] - - - - - - - - - NT NT 41 E.coli [16] - - - - - - - - - NT NT 42 E.coli [21 ] - - - - - - - - - NT NT 43 E.coli [22] - - - - - - - - - NT NT 44 E.coli [24] - - - - - - - - - NT NT 45 E.coli [28] - 17± 0.33 NT NT 46 E.coli [31] - - - - - - - - - NT NT 47 E.coli [32 ] - - - - - - - 21± 0.58 - NT NT 48 E.coli [33] - - - - - - - - - NT NT 49 E.coli [41] - - - - - - - 18.67± 0.33 - NT NT 50 E.coli [45] - - - - - - - - - NT NT 51 E. coli [51] - - - - - - - 20.33± 0.33 - NT NT 52 E. coli [58] - - - - - - - - - NT NT 53 E. coli [60] - - - - - - - - - NT NT 54 E. coli [61] - - - - - - - - - NT NT 55 E. coli ATCC 25922 - - - - - - - 17.83 ± 0.16 14.5 ± 0.50 NT NT 56 Enterobacter spps. [1] - - - - - - - - - NT NT 57 Enterobacter spps. [8] - 15 ± 0.58 12 ± 0.58 14.33 ± 1.20 13 ± 0.58 12.33 ± 0.88 12 ± 1.15 19.67± 0.88 - NT NT 58 Enterobacter aerogenes ATCC 13048 - - 8.5 ± 0.86 15 ± 0.58 - - - - - NT NT 59 Klebsiella spps [6] - - - - - - - 22± 0.58 - NT NT 60 Klebsiella spps [19] - - - - - - - - - NT NT 61 Klebsiella aero- genes [52] - - - 8 ± 0.58 13 ± 1.73 11 ± 2.08 - - - NT NT  S. CHANDA ET AL. Copyright © 2010 SciRes. CM 36 62 Klebsiella spps. [54] - - - - - - - - - NT NT 63 Klebsiella aero- genes [57] - - - - - - - - - NT NT 64 Klebsiella pneu- moniae NCIM 2719 - 12 ± 0.58 12 ± 0.58 10.5 ± 0.28 10.5 ± 0.86 10.5 ± 0.86 11 ± 0.58 - 24.67 ± 0.33 NT NT 65 Proteus mirabilis [4] - - - - - - - - 14± 0.58 NT NT 66 Proteus spps. [53] - - - - - -- - - - NT NT 67 Proteus mirabilis NCIM 2241 - 10.5 ± 0.86 10.5 ± 0.28 9.5 ± 0.86 - - - 18.67 ± 0.33 - NT NT 68 Proteus vulgaris NCTC 8313 - - 9 ± 1.15 - - - - 18 ± 1.00 - NT NT 69 Proteus morganii NCIM 2040 - 9 ± 0.58 - - 8 ± 0.58- - - - NT NT 70 Providencia rett- geri [5] - - - - - - - - - NT NT 71 Citrobactor spps [20] - 8 ± 0.58 8 ± 0.58 8 ± 0.58 - - - - - NT NT 72 Citrobactor freundii [29] - - - - - - - 12.33± 0.33 - NT NT 73 Citrobactor freundii ATCC 10787 - 11 ± 0.58 - - 11.5 ± 0.28 10 ± 0.58 9.5 ± 0.28 - - NT NT 74 Alcaligenes fecalis ATCC 8750 - - - - - - - 18.33 ± 0.66 - NT NT 75 Salmonella ty- phimurium ATCC 23564 - - - - - - - 18.5 ± 0.28 - NT NT Fungus 76 Candida albicans [1] - 7.5 ± 0.29 8 ± 0.58 - 7.5 ± 0.29 10.5 ± 0.29 10 ± 0.58 NT NT - 11.33 ± 0.33 77 Candida albicans [2] - - - 10 ± 0.58 13.33 ± 0.88 9 ± 0.58 - NT NT - 18 ± 0.58 78 Candida spps. [3] - - - 9.5 ± 0.29 14.33 ± 0.66 12.5 ± 0.86 8 ± 0.58 NT NT - 14 ± 0.58 79 Candida spps. [4] - 11 ± 2.13 10.5 ± 2.02 11.5 ± 2.06 8 ± 0.58 8.5 ± 0.29 12.5 ± 0.86 NT NT - 14 ± 0.58 80 Candida spps. [5] - 7.5 ± 0.29 8.5 ± 0.29 9.5 ± 0.29 7.5 ± 0.29 - - NT NT - 10 ± 0.58 81 Candida albicans ATCC 2091 - 11.5 ± 2.60 11 ± 2.31 8 ± 0.58 7.5 ± 0.29 7.5 ± 0.29 10.5 ± 2.02 NT NT 17.67 ± 0.33 13 ± 0.58 82 Candida albicans ATCC 18804 - 10.5 ± 0.29 8 ± 0.58 - - 11 ± 0.58 15 ± 1.15 NT NT - 14.33 ± 0.33 83 Candida glabrata NCIM 3448 - - - - - - - NT NT 39.67 ± 0.88 22 ± 0.58 84 Candida tropicalis ATCC 4563 - - - 7.5 ± 0.29 11 ± 0.58 12 ± 0.58 9.5 ± 0.29 NT NT - 8.33 ± 0.33 85 Candida apicola NCIM 3367 - 23 ± 3.60 26 ± 0.58 28 ± 1.15 25.33 ± 0.88 24 ± 0.58 21.66 ± 0.33 NT NT - 21.33 ± 0.88 86 Cryptococcus neoformans ATCC 34664 - 7.5 ± 0.29 8 ± 0.58 - - - 9.5 ± 1.4 NT NT 21.33 ± 0.33 17 ± 0.58 87 Cryptococcus luteolus ATCC 32044 - 14 ± 0.58 11.5 ± 0.86 11 ± 1.15 9.5 ± 1.44 8.5 ± 0.86 8.5 ± 0.88 NT NT 23.66 ± 0.88 17.66 ± 0.88 88 Trichosporan beigelii NCIM 3404 - 12 ± 0.58 13 ± 1.73 10.5 ± 2.02 - - - NT NT - - 89 Aspergillus flavus NCIM 538 - - - - 14.67 ± 4.34 22 ± 0.58 10.33 ± 2.02NT NT - -  S. CHANDA ET AL. Copyright © 2010 SciRes. CM 37 90 Aspergillus can- didus NCIM 883 - 10.5 ± 0.29 9 ± 1.15 11 ± 0.58 - - - NT NT - - 91 Aspergillus niger ATCC 6275 - - - - 11 ± 2.31 - - NT NT - - Mean ± SEM, n = 3, zone includes disc diameter 7 mm; G – Gentamicin (10 µg/disc), Pc – Piperacillin (100 µg/disc), Ns – Nystatin (100 units/disc), Fu – Fluconazole (10 µg/disc); PME – Methanol extract, PAE – Acetone extract, PDE – Dioxan extract, DMSO – Dimethylsulphoxide. health problem worldwide. Salmonella infection is pri- marily associated with gastroenteritis. This illness poses a more serious health risk to sensitive populations in the community such as the elderly, young and the immuno- compromised, where hospitalization may be required. All the three extracts were inactive against E. coli, A. fecalis and S. typhimurium. Several antimycotic drugs are available at present, its use is limited by a number of factors such as low potency, poor solubility, emergence of resistance strains and drug toxicity. Therefore there is distinct need for the discovery of new, safer and more effective antifungal agents. Candida species have be- come a common cause of hospital acquired infections and a large number of patients die as a result of invasive Candidal infections [18]. All the three extracts were ac- tive against 62.5% of the total fungal strains studied. The three extracts were active against A. candidus while it was inactive against the remaining two moulds (A. flavus and A. niger) studied. The details of the results are given elaborately in Table 2. From the results obtained, it seems that the antibacterial action of the extracts is more pronounced on gram positive than on gram negative bacteria and these findings correlate with the observa- tions of previous screenings of medicinal plants for an- timicrobial activity, where most of the active plants showed activity against gram positive strains only [19- 21]. This difference in susceptibility is because of the difference in cell wall structure of gram positive and gram negative organisms. The lipopolysaccharide con- tent of gram negative bacteria makes them resistant to plant extracts while the peptidoglycan layer of gram positive bacteria is not an effective permeability barrier. 4. Conclusions All the extracts of P. longifolia exhibited the highest rates of antimicrobial activity against gram positive and fungal strains studied. Therefore, it is concluded that P. longifolia extracts should further be studied phyto- chemically to elucidate the active principle in the leaf, which can be used as a leading antibacterial (specific for gram positive) and antifungal agent. 5. Acknowledgements Financial support from Department of Special assistance (DSA) project, New Delhi and supply of clinical isolates by Micro Care and Spandan Diagnostic Laboratories, Rajkot are gratefully acknowledged. 6. References [1] C. K. Amadou, “Promoting Alternative Medicine,” Africa Health Journal, 1998, Vol. 2, pp. 20-25. [2] J. D. Hooker and C. B. Clarke, “Flora of British India, Vol. 1,” L. Reeve and Co. Ltd., London, 1875, pp. 1-741. [3] K. Raghunathan and M. K. Mitra, “Pharmacognosy of Indigenous Drugs, Vol. 1,” Central Council for Research in Ayurveda and Siddha, New Delhi, 1985, pp. 127-139. [4] S. K. Chakrabarti and B. Mukherjee, “Search for Anti- cancer Drug from Indian Medicinal Plants,” Indian Journal of Medical Research, Vol. 56, No. 4, 1968, pp. 445-455. [5] K. Yamaguchi, H. Kinora, S. Natori, Ito, K. Nissbimoio, K. Bando, D. Mizuno and M. Ishignoo, “Screening Tests for Antitumor Activity of Asian Medicinal Herbs I,” Ya- kugaku Zashi, 1964, Vol. 84, pp. 373-377. [6] K. R. Kirtikar and B. D. Basu, “Indian Medicinal Plants,” In: Annonaceae, 2nd Edition, Lalit Mohan Basu, Leader Road, Allahabad, India, Vol. 1, pp. 1993, pp. 72-73. [7] C. M. Hasan, S. N. Islam and M. Ahsan, “Antibacterial Activity of Stem Bark of Polyalthia longifolia,” Dhaka University Studies, Part E, 1988, Vol. 4, pp. 63-66. [8] A. W. Bauer, W. M. M. Kirby, J. C. Sherries and M. Truck, “Antibiotic Susceptibility Testing By Standard Single Disc Diffusion Method,” American Journal of Clinical Pathology, Vol. 45, No. 4, 1966, pp. 426-493. [9] J. Parekh and S. Chanda, “Antibacterial and Phyto- chemical Studies on Twelve Species of Indian Medicinal Plants,” African Journal of Biomedical Research, Vol. 10, No. 2, 2007, pp. 175-181. [10] M. J. Martinez, J. Betancourt, N. Alanso-Gonzalea and A. Jauregui, “Screening of Some Cuban Medicinal Plants for Antimicrobial Activity,” Journal of Ethnopharmacology, Vol. 52, No. 3, 1996, pp. 171-174. [11] J. E. Loper, M. D. Henkels, R. G. Roberts, G. G. Grove, M. J. Willett and T. J. Smith, “Evaluation of Streptomy- cin, Oxytetracycline and Copper Resistance of Erwinia amylavora isolated from pear orchards in Washington State,” Plant Disease, Vol. 75, No. 3, 1991, pp. 287-290. [12] J. Davis, “Inactivation of Antibiotics and Dissemination of Resistance Genes,” Science, Vol. 264, No. 5157, 1994, pp. 375-382. [13] R. F. Service, “Antibiotics That Resist Resistance,” Sci- ence, Vol. 270, No. 5237, 1995, pp. 724-727. [14] I. Ahmad, Z. Mehmood and F. Mohammad, “Screening of Some Indian Medicinal Plants for their Antimicrobial Properties,” Journal of Ethnopharmacology, Vol. 62, No.  S. CHANDA ET AL. Copyright © 2010 SciRes. CM 38 2, 1998, pp. 183-193. [15] A. M. Clark, “Natural Products as Resource of New Drugs,” Pharmaceutical Research, Vol. 13, No. 8, 1996, pp. 1133-1141. [16] G. A. Cordell, “Biodiversity and Drug Discovery a Sym- biotic Relationship,” Phytochemistry, Vol. 55, No. 66, 2000, pp. 463-480. [17] J. R. Zgoda and J. R. Porter, “A Convenient Microdilu- tion Method for Screening Natural Products against Bac- teria and Fungi,” Pharmaceutical Research, Vol. 39, No. 3, 2001, pp. 211-225. [18] T. J. Walsh, J. W. Hathorn, J. D. Sobel, W. G. Merz, V. Sanchez, S. N. Maret, H. R. Buckley, M. A. Pfaller, R. Schaufele, C. Sliva, E. Navarro, J. Lecciones, P. Chandrasekar, J. Lee and P. A. Pizzo, “Detection of Cir- culating Candida enolase by Immunoassay in Patients with Cancer and Invasive Candidiasis,” New England Journal of Medicine, Vol. 324, No. 15, 1991, pp. 1026- 1031. [19] R. M. Herrera, M. Perez, D. A. Martin-Herrera, R. Lopez-Garcia and R. M. Rabanal, “Antimicrobial Activ- ity of Extracts from Plants Endemic to the Canary Is- lands,” Phytotherapy Research, Vol. 10, No. 6, 1996, pp. 364-366. [20] N. A. A. Ali, W. D. Julich, C. Kusnick and U. Lindequist, “Screening of Yemeni Medicinal Plants for Antibacterial and Cytotoxic Activities,” Journal of Ethnopharmaco- logy, Vol. 74, No. 2, 2001, pp. 173-179. [21] R. Nair and S. Chanda, “In vitro Antimicrobial Activity of Psidium guajava L. Leaf Extracts against Clinically Important Pathogenic Microbial Strains,” Brazilian Jour- nal of Microbiology, Vol. 38, No. 3, 2007, pp. 452-458. |

