Journal of Biomedical Science and Engineering
Vol.08 No.10(2015), Article ID:60283,10 pages
10.4236/jbise.2015.810065
Sequence Motif-Based One-Class Classifiers Can Achieve Comparable Accuracy to Two-Class Learners for Plant microRNA Detection
Malik Yousef1,2*, Jens Allmer3,4, Waleed Khalifa1,2
1The Institute of Applied Research, The Galilee Society, Shefa Amr, Israel
2Computer Science, The College of Sakhnin, Sakhnin, Israel
3Molecular Biology and Genetics, Izmir Institute of Technology, Izmir, Turkey
4Bionia Incorporated, IZTEKGEB A8, Izmir, Turkey
Email: *malik.yousef@gmail.com
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 2 September 2015; accepted 11 October 2015; published 14 October 2015
ABSTRACT
microRNAs (miRNAs) are short nucleotide sequences expressed by a genome that are involved in post transcriptional modulation of gene expression. Since miRNAs need to be co-expressed with their target mRNA to observe an effect and since miRNAs and target interactions can be cooperative, it is currently not possible to develop a comprehensive experimental atlas of miRNAs and their targets. To overcome this limitation, machine learning has been applied to miRNA detection. In general binary learning (two-class) approaches are applied to miRNA discovery. These learners consider both positive (miRNA) and negative (non-miRNA) examples during the training process. One-class classifiers, on the other hand, use only the information for the target class (miRNA). The one-class approach in machine learning is gradually receiving more attention particularly for solving problems where the negative class is not well defined. This is especially true for miRNAs where the positive class can be experimentally confirmed relatively easy, but where it is not currently possible to call any part of a genome a non-miRNA. To do that, it should be co-expressed with all other possible transcripts of the genome, which currently is a futile endeavor. For machine learning, miRNAs need to be transformed into a feature vector and some currently used features like minimum free energy vary widely in the case of plant miRNAs. In this study it was our aim to analyze different methods applying one-class approaches and the effectiveness of motif- based features for prediction of plant miRNA genes. We show that the application of these one- class classifiers is promising and useful for this kind of problem which relies only on sequence- based features such as k-mers and motifs comparing to the results from two-class classification. In some cases the results of one-class are, to our surprise, more accurate than results from two-class classifiers.
Keywords:
microRNA, One-Class, Plant, Machine Learning

1. Introduction
1.1. microRNAs
microRNAs (miRNAs) are short RNA sequences that form a hairpin structures which harbor one or more mature miRNAs of about 21 nucleotides in length [1] . Mature miRNAs, when incorporated into RISC, provide a template sequence for the recognition of their target mRNAs which are then either degraded or their translation efficiency is affected [2] . Since their discovery by Lee and colleagues [3] , they have received increasing attention and it is now clear that in cases of animals they are also involved in many diseases [4] and in cases of plants play essential roles in regulation, development, response to cold stress, and nutrient deprivation [5] . microRNAs are found in multicellular organisms ranging from sponges [6] to human, but the plant miRNA pathway may have evolved distinctly from the animal one [7] .
1.2. Computational Detection of Pre-microRNAs
Since experimental detection of novel miRNAs is difficult and since it seems futile to aim to discover all miRNAs and their targets of an organism, computational prediction of miRNAs and their targets has become a research focus. Different approaches to computational miRNA detection have been applied, but most approaches are based on feature extraction followed by machine learning [8] [9] . The so called ab initio miRNA detection methodology is well established in animal models for which abundant learning data is available for example in miRBase [10] . Various computational approaches (apart from machine learning; or in combination with it) have been employed for example based on sequence conservation and/or structural similarity [11] - [15] .
For example, NOVOMIR [16] , using a series of filters and a statistical model reportedly detects pre-miRNA with a sensitivity of 80% at a specificity of 99%. MiRenSVM combines three SVMs for prediction reported a sensitivity of 93% at a specificity of 97% [17] . Xue and colleagues also trained a support vector machine on human data (93% sensitivity at 88% specificity) and achieved high accuracies of up to 90% in other species using the same model [18] . Jiang and colleagues [19] added some featores to Xue and colleagues’ and used Random Forrest as a classifier. They achieved a sensitivity of 95% at a specificity of 98%. Studies that don’t exactly fall into the realm of ab initio miRNA detection exist. For example, Zeller and coworkers employed structure/ sequence conservation, homology to known microRNAs, and phylogenetic footprinting [20] . Others have used homology searches for revealing paralog and orthologmiRNAs [12] [21] - [24] . Additionally, Wang and others [25] developed a method based on sequence and structure alignment for miRNA identification. Finally Hertel and Stadler included multiple sequence alignment for microRNA detection [26] . These approaches employ evolutionary models which may not be always applicable and which can be hard to parameterize.
Therefore, we will focus on ab initio detection of miRNAs using parameterized pre-miRNAs. Such approaches need positive (true miRNAs) and negative (non-miRNAs) data to become functional. Unfortunately, the positive data (usually derived from miRBase) may contain non-miRNAs [27] . Even worse, the negative class cannot be established experimentally and, therefore, most of these methods require the generation of an artificial negative class which may lead to problems [2] . In general, these algorithms need a sufficient number of positive as well as negative examples. Although many miRNA genes seem to be unique in any organism, positive training examples can easily be found whereas negative examples are hard to come by [17] [28] - [30] . Some negative examples that were picked in studies, for example mRNA sequences [31] are dubious since to our current knowledge miRNAs can originate from any part of a pri-miRNA. Thus, defining the negative class is a major challenge in training machine learning algorithms for miRNA discovery. For this reason, one-class machine learning which only needs positive examples has been tried [30] .
1.3. One Class Machine Learning and Sequence Motifs for Pre-miRNA Detection
In general machine learning uses positive and negative training and testing examples to learn a model describing the data. One class machine learning is trained only using examples of one class during training and during testing known examples of that class are paired with unknown examples which may be of the class used in training or not. The trained model must then decide whether the examples belong to the class used for training or whether they are unknown. Here we provided parameterized pre-miRNAs as the target class and during testing added unseen positive as well as negative examples. Similar to the one-class method, MiRank [18] , is based on a random walk through a graph consisting of known miRNA examples and unknown candidate sequences and, therefore, is also independent of negative examples.
Plant miRNAs may have evolved distinct from animal ones and thus the approaches for miRNA detection introduced so far may need to be adapted when applied to plant miRNA detection. It has been found that plant miRNAs are more variable in size and very heterogeneous, but usually larger than animal miRNAs. Also their base pairing propensity (bonds in the stem) seems to be more extensive and their length is close to 21 nucleotides [32] . Billoud and colleagues predicted miRNAs in brown algae, which are different from both land plants and animals using a set of normalized features like Shannon entropy that have previously been used for detection of miRNAs in plants and animals [33] . Other studies also use tools developed for miRNA detection in animals for studies in plants [16] [34] [35] . PlantMiRNAPred reported an accuracy of more than 90% when used with multiple plant species [35] . One study showed that generalized training using multiple plant data as input for training a decision tree leads to sensitivity of 84% at a specificity of 99% [36] . This may be due to their concurrent usage of structural features and targeting parameters for miRNA prediction which is beneficial for the accuracy of miRNA prediction [37] . In Arabidopsis thaliana, one approach searched for all complementary pairs of sequences within its transcriptome of the expected size of a miRNA-mRNA duplex and then filtered the results according to divergence patterns [38] .
Most of the studies that consider using motifs to predict microRNAs are based on structure motifs and not on precursor sequence motifs as we suggest in this study and in our previous one [39] . Liu, He et al. 2012 [40] represent pre-miRNA and non-pre-miRNA hairpins as string sequence-structure motifs (ss-motifs). Using only ss-motifs as features in a support vector machine (SVM) for pre-miRNA identification they achieved 99.2% specificity and 97.6% sensitivity on human test data.
The main idea of our method is based on that the microRNAs are clustered into families which are likely to share similar sequence motifs and that sequence motifs can distinguish between positive and negative class. Obviously, there is no single motif which could lead to proper discrimination into classes and therefore we suggest that combination of motifs could be used as a signature that can distinguish the positive class from the negative class.
The one-class method employed in this study, was more accurate (on average ~91%) than Triplet-SVM (~74% on average), but slightly less accurate than our previous study (~92% on average), PlantMiRNAPred (~96%), but equally successful as microPred (~91%). However, training and testing data can potentially be contaminated and two-class classifiers may be more affected by this than one-class ones. Oppositely, one-class performance is unfairly negatively influenced by false negative examples since they are not used in training. Strikingly in this study, one-class classifiers are similar in performance to the best two-class ones and, therefore, we suggest that the use of one-class predictors should be further investigated.
2. Materials and Methods
2.1. Data
We downloaded all available microRNAs from selected plant species from miRBase (Releases 20 and 21). The selected species were: Glycine max (gma), Zeamayis (zma), Medicagotruncatula (mtr), Sorghum bicolor (sbi), Physcomitrella patens (ppt), Arabidopsis thaliana (ath), Populustrichocarpa (ptc), and Oryza sativa (osa). We considered Brassicaceae with 699 pre-miRNAs, that consists of Arabidopsis lyrata (205 precursors), Arabidopsis thaliana (298 precursors), Brassica napus (90 precursors), Brassica oleracea (10 precursors), and Brassica rapa (96 precursors). We also included the data published on the web server PlantMiRNAPred [35] whose training dataset consist of 980 real pre-miRNAs and 980 pseudo pre-miRNAs (we refer to this data as PlantMiRNAPred data in the following). Our negative data pool of the 980 pseudo pre-miRNAs consists of the PlantMiRNAPred dataset.
2.2. Parameters for Machine Learning
2.2.1. Motif Parameters
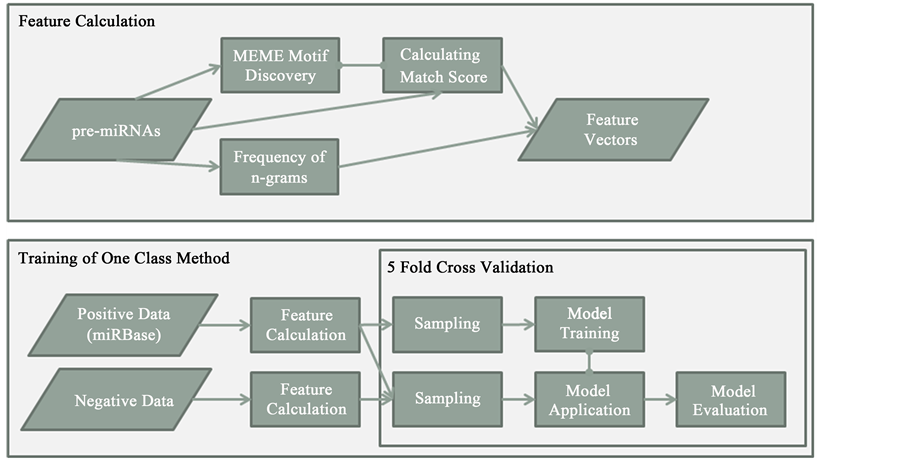
Here a sequence motif is a short stretch of nucleotides that is widespread among plant pre-miRNAs. Motif discovery in turn is the process of finding such short sequences within a larger pool of sequences; here in plant hairpins. The MEME (Multiple EM for Motif Elicitation) [41] suite web server is used for motif discovery in our study (Figure 1).
The algorithm is based on [42] which works by repeatedly searching for ungapped sequence motifs that occur in input sequences. MEME provides the results as regular expressions (Table 1, Motif row).
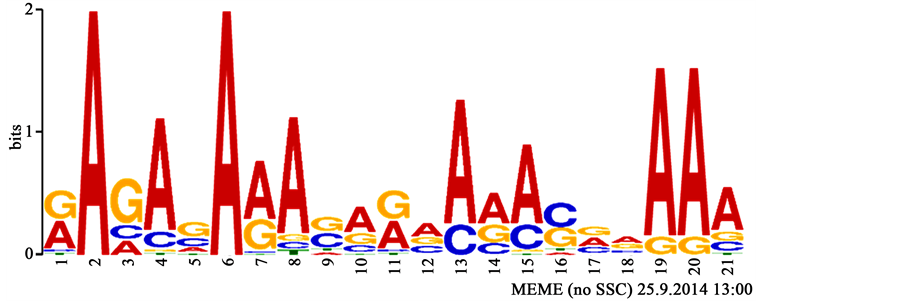
Nucleotides within brackets represent alternatives at the given position in the sequence; without brackets only the given nucleotide occurs abundantly within all collected sequences representing the motif. More visual representations of such motifs are sequence logos (Figure 2). MEME was instructed to generate 20 motifs, each of which must appear in at least 10 hairpins to be an acceptable motif.
Figure 1. Workflow for feature calculation and one-class method training and evaluation. Pre-miRNAs are used to discover motifs with MEME and they are then scored against the discovered candidates. N-gram frequency is added to the calculation and thus a feature vectors describing the hairpins are created. These feature vectors are calculated for positive and negative data and the positive examples are sampled and used for model training. Remaining positive examples and negative examples are then used for model evaluation.
Figure 2. The sequence logo corresponding to one of the motifs discovered in this study. Size of letters in stacks represents their frequencies while the height of the stack represents the information content. Not all options in the profile may be incorporated into its corresponding regular expression (see Table 1).
Table 1. Example of match score between a motif and a part of a sequence. The number of matches is 6. For the assessment the score is normalized by the length of the motif. The final match score is 6/19 = 0.31.
2.2.2. Sequence-Based and Motif Features for Plant Pre-miRNA Detection
Simple sequence-based features have been described and used for ab initio pre-miRNA detection in numerous studies (see Hairpin Feature Calculation). These simple features, also called words, k-mers, or n-grams, describe a short sequence of nucleotides. For example a 1-gram over the relevant alphabet [39] an produce the words A, U, C, and G; while a 2-gram over {A, U, C, G} can generate: AA, AC, AG, AU, CA, CC, CG, CU, GA, GC, GG, GU, UA, UC, UG, and UU. Higher n have also been used [37] and here we chose 1, 2, and 3-grams as features.
Motif features are different from n-grams in that they are not exact and allow some degree of error tolerance. In this study motifs are represented as regular expressions (see above). Regular expressions are widespread in approximate pattern matching and many programs allow searching with regular expressions (e.g.: most Linux tools such as grep). Here we use PatMatch [43] to analyze whether a pattern is within a hairpin (1) or not (0). The hairpin is analyzed using the following algorithm:
Let w be the length of the given motif
Let max be 0
For i: 0 to len(sequence)-w + 1
Align w sized window with ith position of sequence
Let ls be the calculated match score (normalized to the length of the motif)
updateMax(ls,max)
Report max as Match Score
As a summary the features vector consists of 84 words and n motifs (n is specified in Table 2 for some datasets):
a, c, g, t, aa, ac, ag, at, ca, cc, cg, ct, ga, gc, gg, gt, ta, tc, tg, tt, aaa, aac, aag, aat, aca, acc, acg, act, aga, agc, agg, agt, ata, atc, atg, att, caa, cac, cag, cat, cca, ccc, ccg, cct, cga, cgc, cgg, cgt, cta, ctc, ctg, ctt, gaa, gac, gag, gat, gca, gcc, gcg, gct, gga, ggc, ggg, ggt, gta, gtc, gtg, gtt, taa, tac, tag, tat, tcg, tct, tga, tgc, tgg, tgt, tta, ttc, ttg, ttt, Motif_1, Motif_2, Motif_3, …, Motif_n
Words are transformed into features by using their frequency and motifs by using their match score.
2.3. One-Class Methods
In general a binary learning (two-class) approach to miRNA discovery considers both positive (miRNA) and negative (non-miRNA) classes by providing examples for the two-classes to a learning algorithm in order to build a classifier that will attempt to discriminate between them. One-class classification uses only the information for the target class (positive class; miRNA) building a classifier which is able to recognize the examples belonging to its target and rejecting others as outliers. The one-class approach in machine learning is receiving increaing attention particularly for solving problems where the negative class is not well defined [30] [44] - [50] ; moreover, the one-class approach has been successfully applied in various fields including text mining [48] [50] , and miRNA gene and target discovery [49] . We used the dd-tools package [51] for the implementation of the one-class models.
As previously described in Yousef et al. 2010, we used multiple one-class classifiers [49] . One of them (OC-SVM) is based on a SVM learner, another on k-means clustering (OC-Kmeans), one on a Gaussian model (OC-Gauss), another on principal component analysis (OC-PCA) and the last one on neural networks (OC-kNN). Additionally, we calculated the majority vote among one class classifiers (OC-MV).
Table 2. Dataset description and number of generated motifs per dataset.
2.4. Evaluation Methods
Previously published algorithms, based on two-class classification, have been evaluated using sensitivity, specificity, and accuracy as measures for their predictive power. We have previously shown that positive data derived from miRBase contains contaminating non-miRNAs [27] . Additionally, it is clear that the negative class cannot be established experimentally and that the proposed negative datasets are likely to contain miRNAs. Due to the unknown quality of the training and testing data used, it is questionable whether using sensitivity, specificity, and accuracy is a valid approach. Unfortunately, in the absence of a better comparison measure, we have to acknowledge this drawback and are forced to use these statistics to compare among trained models. This may lead to a lower accuracy of the one-class method’s results since examples assigned to the negative class may in reality be positive ones and vice versa. This is less so for two-class classifiers since they are trained on both classes and will, therefore, “correctly” return the wrong classification since it was part of their negative training set. The one-class method is trained on positive data only and, therefore, will call data from the negative class miRNA which may in fact be miRNAs but will be scored as non-miRNAs since they are part of the negative dataset.
The statistics are calculated as follows:
Sensitivity (SE) = TP/(TP + FN)
Specificity (SP) = TN/(TN + FP)
Accuracy (ACC) = (TP + TN)/(TP + TN + FP + FN)
Where TP is true positive, FP is false positive, TN is true negative and FN is false negative.
3. Results and Discussion
Comparing one-class classifiers with two-class ones, is not without danger. For example, erroneous example labels in training and testing data affect the results differently. In our comparison, on the two-class classifier features selection was performed. Feature selection for one-class classifiers, however, is an active area of research and no suitable standards have been established, yet. Thus the results of the one-class classifier are expected to be not as good as the two-class classifier’s for these two problems.
The PlantMiRNAPred data was divided into two parts, PlantMiRNAPred-p1 data consisting of 450 pre-miRNAs (positive data) and 450 pseudo pre-miRNAs (negative data) and PlantMiRNAPred-p2 data composed of 530 pre-miRNAs and 530 pseudo pre-miRNAs. The Brassicaceae data was also divided into two parts, first part consists of one third of the data (233 sequences; named Brassicaceae-p1) the remaining two third (named Brassicaceae-p2) contain 466 sequences. MEME software was used to discover motifs in the dataset as described in the Materials and Methods Section, and several motifs were found in all datasets as seen in Table 2. Additionally MEME was used to discover motifs in one part of the divided dataset (p1) and the same motifs were used for representation of the remainder of the data (p2) to ensure that the extracted motifs are meaningful and not dataset dependent.
For the two-class model (Two-Class SVM) the selected motifs and the n-grams, were used to train a support vector machine model for which the accuracy and other performance measures were established (Table 3). Table 3 presents the average performance of our previous SVM classifier―MotifmiRNAPred [39] ―using five- fold cross validation while for the one-class methods we applied 100 iterations during training.
For the motifs extracted from PlantMiRNAPred-p1 and applied to PlantMiRNAPred-p2 we see a decrease in performance of the two-class SVM model by about 12% which indicates that there is some data dependency of
Table 3. The result of one-class methods compared to two-class MotifmiRNAPred applied to different plant miRNA data. Two-class reference is highlighted in gray and best value per column is bolded (except if it is OC-MV). OC-MV shows the majority vote result using the four one-class classifiers. OC: one-class, MV: majority vote, ACC: accuracy, SE: sensitivity, SP: specificity.
the motifs in this case while in the one-class considering the OC-Kmeans and OC-Gauss results we see almost same performance (94.0%, 95.7% and 90.2%, −91.2%, respectively). The K-means and OC-Gauss performance for PlantMiRNAPred-p2 data is much higher (~14% and ~10%) than for the two-class classifier. For Brassicaceae and for one-class and two-class there was no significant difference between the datasets p1 and p2 which shows that in this case stable motifs were generated that are not affected by differences in the tested datasets. For Brassicaceae-p2 comparing the two-class SVM with one-class we see that the one-class is achieving similar or even slightly better results (1% - 2%) more than the two-class in the case of OC-Kmeans and OC-Gaussian while using majority vote of the one-class classifiers we achieve even 2.4% better results than the two-class classifier. Overall, the one-class classifiers achieved the highest accuracy in all datasets. The average accuracies between OC-Gauss and the two-class SVM differ by less than 0.5% while OC-Kmeans is better than the two- class by about 4%. This is a striking result since in our experience and due to the problems pointed out initially, we wouldn’t expect the two methods to perform so similar.
When comparing the results on PlantMiRNAPred-p1 with the results achieved by PlantMiRNAPred [35] it can be seen that our methodology achieves a similar performance (Table 4). PlantMiRNAPred achieves accuracies between 92% and 100% when the data is separated into species with a trend to be more successful for smaller datasets. It needs to be noted, that PlantMiRNAPred was trained and tested on some of the data used here.
In Table 4 we considered the data from PlantMiRNAPred web server [35] to perform a comparison performance with the classification results of PlantMiRNAPred, TripletSVM [18] , and microPred [52] . The data was represented by 174 features consisting of 84 n-grams and 90 motifs. For the two-class MotifmiRNAPred the top 60 selected features by SVM-RFE, feature selection method available in WEKA [53] , were considered and the performance resulting from a 5-fold cross validation are presented while for the one-class all features are used with 100 iteration (Table 3).
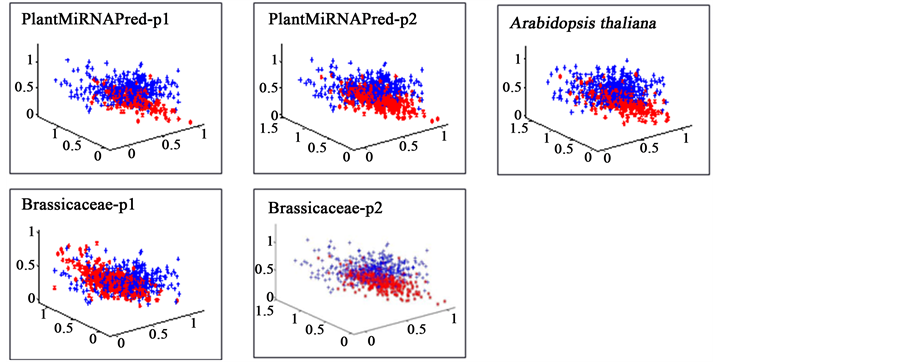
This introduces a slight bias and it would be better to perform feature selection for the features of the one-class classifiers as well. Figure 3 shows how the data distributes when mapped into two dimensional space. Projection into this low dimensional space suggests that separation in higher dimensional space will be possible. It also suggests that not many dimensions seem to be necessary and that feature selection should be performed. Unfortunately, features selection for one-class classifiers is an open problem.
The comparison in Table 4 shows that using motifs for miRNA detection is comparably to using traditional features while at times even slightly more successful. It is clear that OC-Kmeans is achieving better results compared to the two-class method Triplet-SVM considering the average (Table 4, last row). Triplet-SVM achieved 73.6% accuracy while OC-Kmeans achieved 91.4% which represents a 17.8% increase. Our previous method, using two-class classification (MotifMiRNAPred) performs about the same as OC-Kmeans and similar to PlantMiRNAPred and microPred. As pointed out above, comparison between these methods is not entirely
Figure 3. Mapping of separation data into two dimensional space. The separation appears difficult in two dimensions, but in higher dimensions it becomes possible. Blue dots represent outlier class, while red dots depict the target class.
Table 4. Comparison of the best one-class method (OC-Kmeans; highlighted in gray) with MotifmiRNAPred and other two- class methods. The first 4 columns taken from the PlantMiRNAPred paper while the MotifmiRNAPred are taken from the paper [39] . Best results per row are bolded. ACC: accuracy, SE: sensitivity, SP: specificity.
fair and biases towards two-class methods. Neither the positive data nor the negative data are of pure class. The result is therefore biased towards two-class methods since two-classes are also used for accuracy evaluation [39] . Despite this, one-class classifiers developed in this study perform similarly if not better than existing methods using two-class classification (Table 4) and the difference in average performance over datasets is less than 4% compared to the best performing two-class classifier on these datasets.
4. Conclusions
An abundance of features describing miRNA hairpins have been proposed which are mostly based on structural, statistical and thermodynamic features [54] . Here we show that for plant miRNA detection, motif based features are useful and they by themselves lead to a good recognition of pre-miRNAs as we present using different methods applying one-class approaches for prediction of plant miRNA genes. We show that the application of one-class classification is promising and useful for this kind of problem that rely only on features from the sequence such as k-mers and motifs comparing to the results from two-class classification.
More importantly, we show that the accuracy of OC-Gauss and OC-Kmeans, based on a biased assessment using an artificial negative class is still comparable to two-class classifiers that are trained on the biased dataset.
In the future, we plan to create a new negative dataset and aim to add additional features to the one-class model developed here. We believe that in the absence of experimentally proven negative data, one-class classification needs to be further developed since two-class classifiers are more strongly affected by wrong examples among the negative examples. One-class classification is only affected during the evaluation phase whereas two- class classification is affected in the training phase by wrongly assigned negative examples. We believe the latter is quite dangerous and urge for further development of one-class classification in the field of miRNA detection. We should emphasizes again that the performance of the one-class is based on all the features while the two-class is with selected features that clearly improve the results even in some cases dramatically and still we are achieving similar performance. In the future, we will investigate methods for feature selection for one-class classification.
Acknowledgements
The work was supported by the Scientific and Technological Research Council of Turkey [grant number 113E326] to JA. Conflict of interest: none declared.
Cite this paper
MalikYousef,JensAllmer,WaleedKhalifa,11, (2015) Sequence Motif-Based One-Class Classifiers Can Achieve Comparable Accuracy to Two-Class Learners for Plant microRNA Detection. Journal of Biomedical Science and Engineering,08,684-694. doi: 10.4236/jbise.2015.810065
References
- 1. Erson-Bensan, A.E. (2014) Introduction to microRNAs in Biological Systems. Methods in Molecular Biology, 1107, 1-14.
http://dx.doi.org/10.1007/978-1-62703-748-8_1 - 2. Allmer, J. and Yousef, M. (2012) Computational Methods for ab Initio Detection of microRNAs. Front in Genet, 3, 209.
http://dx.doi.org/10.3389/fgene.2012.00209 - 3. Lee, R.C., Feinbaum, R.L. and Ambros, V. (1993) The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell, 75, 843-854.
http://dx.doi.org/10.1016/0092-8674(93)90529-Y - 4. Tüfekci, K.U., Oner, M.G., Meuwissen, R.L.J. and Genc, S. (2014) The Role of microRNAs in Human Diseases. Methods in Molecular Biology, 1107, 33-50.
http://dx.doi.org/10.1007/978-1-62703-748-8_3 - 5. Zhang, Z., Yu, J., Li, D., et al. (2010) PMRD: Plant microRNA Database. Nucleic Acids Research, 38, D806-D813.
http://dx.doi.org/10.1093/nar/gkp818 - 6. Kim, V.N., Han, J. and Siomi, M.C. (2009) Biogenesis of Small RNAs in Animals. Nature Reviews Molecular Cell Biology, 10, 126-139.
http://dx.doi.org/10.1038/nrm2632 - 7. Chapman, E.J. and Carrington, J.C. (2007) Specialization and Evolution of Endogenous Small RNA Pathways. Nature Reviews Genetics, 8, 884-896.
http://dx.doi.org/10.1038/nrg2179 - 8. Sacar, M.D. and Allmer, J. (2013) Comparison of Four ab Initio microRNA Prediction Tools. International Conference on Bioinformatics Models, Methods and Algorithms, SciTePress, Science and Technology Publications, Barcelona, 190-195.
- 9. Lopes, I.D.O.N., Schliep, A. and de Carvalho, A.C.P.D.L.F. (2014) The Discriminant Power of RNA Features for Pre-miRNA Recognition. BMC Bioinformatics, 15, 124.
http://dx.doi.org/10.1186/1471-2105-15-124 - 10. Kozomara, A. and Griffiths-Jones, S. (2011) miRBase: Integrating microRNA Annotation and Deep-Sequencing Data. Nucleic Acids Research, 39, D152-D157.
http://dx.doi.org/10.1093/nar/gkq1027 - 11. Lim, L.P., Glasner, M.E., Yekta, S., Burge, C.B. and Bartel, D.P. (2003) Vertebrate MicroRNA Genes. Science, 299, 1540.
http://dx.doi.org/10.1126/science.1080372 - 12. Weber, M.J. (2005) New Human and Mouse MicroRNA Genes Found by Homology Search. FEBS Journal, 272, 59- 73.
http://dx.doi.org/10.1111/j.1432-1033.2004.04389.x - 13. Lim, L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., et al. (2003) The MicroRNAs of Caenorhabditis elegans. Genes & Development, 17, 991-1008.
http://dx.doi.org/10.1101/gad.1074403 - 14. Lai, E.C., Tomancak, P., Williams, R.W. and Rubin, G.M. (2003) Computational Identification of Drosophila MicroRNA Genes. Genome Biology, 4, R42.
http://dx.doi.org/10.1186/gb-2003-4-7-r42 - 15. Grad, Y., Aach, J., Hayes, G.D., Reinhart, B.J., Church, G.M., Ruvkun, G. and Kim, J. (2003) Computational and Experimental Identification of C. elegans MicroRNAs. Molecular Cell, 11, 1253-1263.
http://dx.doi.org/10.1016/S1097-2765(03)00153-9 - 16. Teune, J.-H. and Steger, G. (2010) NOVOMIR: De Novo Prediction of MicroRNA-Coding Regions in a Single Plant-Genome. Journal of Nucleic Acids, 2010, Article ID: 495904.
http://dx.doi.org/10.4061/2010/495904 - 17. Ding, J.D., Zhou, S.G. and Guan, J.H. (2010) MiRenSVM: Towards Better Prediction of MicroRNA Precursors Using an Ensemble SVM Classifier with Multi-Loop Features. BMC Bioinformatics, 11, S11.
http://dx.doi.org/10.1186/1471-2105-11-s11-s11 - 18. Xue, C.H., Li, F., He, T., Liu, G.-P., Li, Y.D. and Zhang, X.G. (2005) Classification of Real and Pseudo MicroRNA Precursors Using Local Structure-Sequence Features and Support Vector Machine. BMC Bioinformatics, 6, 310.
http://dx.doi.org/10.1186/1471-2105-6-310 - 19. Jiang, P., Wu, H.N., Wang, W.K., Ma, W., Sun, X. and Lu, Z.H. (2007) MiPred: Classification of Real and Pseudo MicroRNA Precursors Using Random Forest Prediction Model with Combined Features. Nucleic Acids Research, 35, W339-W344.
http://dx.doi.org/10.1093/nar/gkm368 - 20. Keshavan, R., Virata, M., Keshavan, A. and Zeller, R.W. (2010) Computational Identification of Ciona intestinalis MicroRNAs. Zoological Science, 27, 162-170.
http://dx.doi.org/10.2108/zsj.27.162 - 21. Lagos-Quintana, M., Rauhut, R., Lendeckel, W. and Tuschl, T. (2001) Identification of Novel Genes Coding for Small Expressed RNAs. Science, 294, 853-858.
http://dx.doi.org/10.1126/science.1064921 - 22. Lau, N.C., Lim, L.P., Weinstein, E.G. and Bartel, D.P. (2001) An Abundant Class of Tiny RNAs with Probable Regulatory Roles in Caenorhabditis elegans. Science, 294, 858-862.
http://dx.doi.org/10.1126/science.1065062 - 23. Lee, R.C. and Ambros, V. (2001) An Extensive Class of Small RNAs in Caenorhabditis elegans. Science, 294, 862- 864.
http://dx.doi.org/10.1126/science.1065329 - 24. Pasquinelli, A.E., Reinhart, B.J., Slack, F., Martindale, M.Q., Kuroda, M.I., Maller, B., et al. (2000) Conservation of the Sequence and Temporal Expression of Let-7 Heterochronic Regulatory RNA. Nature, 408, 86-89.
http://dx.doi.org/10.1038/35040556 - 25. Wang, X.W., Zhang, J., Li, F., Gu, J., He, T., Zhang, X.G. and Li, Y.D. (2005) MicroRNA Identification Based on Sequence and Structure Alignment. Bioinformatics, 21, 3610-3614.
http://dx.doi.org/10.1093/bioinformatics/bti562 - 26. Hertel, J. and Stadler, P.F. (2006) Hairpins in a Haystack: Recognizing MicroRNA Precursors in Comparative Genomics Data. Bioinformatics, 22, e197-e202.
http://dx.doi.org/10.1093/bioinformatics/btl257 - 27. Sacar, M.D., Hamzeiy, H. and Allmer, J. (2013) Can MiRBase Provide Positive Data for Machine Learning for the Detection of MiRNA Hairpins? Journal of Integrative Bioinformatics, 10, 215.
- 28. Ritchie, W., Gao, D. and Rasko, J.E.J. (2012) Defining and Providing Robust Controls for MicroRNA Prediction. Bioinformatics, 28, 1058-1061.
http://dx.doi.org/10.1093/bioinformatics/bts114 - 29. Wu, Y.G., Wei, B., Liu, H.Z., Li, T.X. and Rayner, S. (2011) MiRPara: A SVM-Based Software Tool for Prediction of Most Probable MicroRNA Coding Regions in Genome Scale Sequences. BMC Bioinformatics, 12, 107.
http://dx.doi.org/10.1186/1471-2105-12-107 - 30. Yousef, M., Jung, S., Showe, L.C. and Showe, M.K. (2008) Learning from Positive Examples When the Negative Class Is Undetermined-MicroRNA Gene Identification. Algorithms for Molecular Biology, 3, 2.
http://dx.doi.org/10.1186/1748-7188-3-2 - 31. Sewer, A., Paul, N., Landgraf, P., Aravin, A., Pfeffer, S., Brownstein, M.J., et al. (2005) Identification of Clustered MicroRNAs Using an ab initio Prediction Method. BMC Bioinformatics, 6, 267.
http://dx.doi.org/10.1186/1471-2105-6-267 - 32. Gomes, C.P.C., Cho, J.-H., Hood, L., Franco, O.L., Pereira, R.W. and Wang, K. (2013) A Review of Computational Tools in MicroRNA Discovery. Frontiers in Genetics, 4, 81.
http://dx.doi.org/10.3389/fgene.2013.00081 - 33. Billoud, B., Nehr, Z., Le Bail, A. and Charrier, B. (2014) Computational Prediction and Experimental Validation of MicroRNAs in the Brown Alga Ectocarpus siliculosus. Nucleic Acids Research, 42, 417-429.
http://dx.doi.org/10.1093/nar/gkt856 - 34. Oliveira, J.S., Mendes, N.D., Carocha, V., Graca, C., Paiva, J.A. and Freitas, A.T. (2013) A Computational Approach for MicroRNA Identification in Plants: Combining Genome-Based Predictions with RNA-Seq Data. Journal of Data Mining in Genomics & Proteomics, 4, 130.
http://dx.doi.org/10.4172/2153-0602.1000130 - 35. Xuan, P., Guo, M.Z., Liu, X.Y., Huang, Y.C., Li, W.B. and Huang, Y.F. (2011) PlantMiRNAPred: Efficient Classification of Real and Pseudo Plant Pre-miRNAs. Bioinformatics, 27, 1368-1376.
http://dx.doi.org/10.1093/bioinformatics/btr153 - 36. Williams, P.H., Eyles, R. and Weiller, G. (2012) Plant MicroRNA Prediction by Supervised Machine Learning Using C5.0 Decision Trees. Journal of Nucleic Acids, 2012, Article ID: 652979.
http://dx.doi.org/10.1155/2012/652979 - 37. Cakir, M.V. and Allmer, J. (2010) Systematic Computational Analysis of Potential RNAi Regulation in Toxoplasma gondii. 2010 5th International Symposium on Health Informatics and Bioinformatics (HIBIT), Antalya, 20-22 April 2010, 31-38.
http://dx.doi.org/10.1109/HIBIT.2010.5478909 - 38. Adai, A., Johnson, C., Mlotshwa, S., Archer-Evans, S., Manocha, V., Vance, V. and Sundaresan, V. (2005) Computational Prediction of miRNAs in Arabidopsis thaliana. Genome Research, 15, 78-91.
http://dx.doi.org/10.1101/gr.2908205 - 39. Yousef, M., Allmer, J. and Khalifaa, W. (2015) Plant MicroRNA Prediction Employing Sequence Motifs Achieves High Accuracy. (Under Review)
- 40. Liu, X., He, S., Skogerbo, G., Gong, F.Z. and Chen, R.S. (2012) Integrated Sequence-Structure Motifs Suffice to Identify MicroRNA Precursors. PLoS ONE, 7, e32797.
http://dx.doi.org/10.1371/journal.pone.0032797 - 41. Bailey, T.L., Boden, M., Buske, F.A., Frith, M., Grant, C.E., Clementi, L., et al. (2009) MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Research, 37, W202-W208.
http://dx.doi.org/10.1093/nar/gkp335 - 42. Bailey, T.L. and Elkan, C. (1994) Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Biopolymers. Proceedings of the International Conference on Intelligent Systems for Molecular Biology, 2, 28-36.
- 43. Yan, T., Yoo, D., Berardini, T.Z., Mueller, L.A., Weems, D.C., Weng, S., et al. (2005) PatMatch: A Program for Finding Patterns in Peptide and Nucleotide Sequences. Nucleic Acids Research, 33, W262-W266.
http://dx.doi.org/10.1093/nar/gki368 - 44. Kowalczyk, A. and Raskutti, B. (2002) One Class SVM for Yeast Regulation Prediction. SIGKDD Explorations Newsletter, 4, 99-100.
http://dx.doi.org/10.1145/772862.772878 - 45. Chechik, G. (2004) A Needle in a Haystack: Local One-Class Optimization. Proceedings of the 21st International Conference on Machine Learning, Banff, 4-8 July 2004.
- 46. Spinosa, E.J. and Carvalho, A. (2005) Support Vector Machines for Novel Class Detection in Bioinformatics. Genetics and Molecular Research, 4, 608-615.
- 47. Gupta, G. and Ghosh, J. (2005) Robust One-Class Clustering Using Hybrid Global and Local Search. Proceedings of the 22nd International Conference on Machine Learning, Bonn, 7-11 August 2005, 273-280.
http://dx.doi.org/10.1145/1102351.1102386 - 48. Manevitz, L. and Yousef, M. (2007) One-Class Document Classification via Neural Networks. Neurocomputing, 70, 1466-1481.
http://dx.doi.org/10.1016/j.neucom.2006.05.013 - 49. Yousef, M., Najami, N. and Khalifa, W. (2010) A Comparison Study between One-Class and Two-Class Machine Learning for MicroRNA Target Detection. Journal of Biomedical Science and Engineering, 3, 247-252.
http://dx.doi.org/10.4236/jbise.2010.33033 - 50. Manevitz, L.M. and Yousef, M. (2001) One-Class SVMs for Document Classification. Journal of Machine Learning Research, 2, 139-154.
- 51. Tax, D.M.J. (2005) DDtools, the Data Description Toolbox for Matlab.
- 52. Batuwita, R. and Palade, V. (2009) MicroPred: Effective Classification of Pre-miRNAs for Human miRNA Gene Prediction. Bioinformatics, 25, 989-995.
http://dx.doi.org/10.1093/bioinformatics/btp107 - 53. Gewehr, J.E., Szugat, M. and Zimmer, R. (2007) BioWe-ka—Extending the Weka Framework for Bioinformatics. Bioinformatics, 23, 651-653.
http://dx.doi.org/10.1093/bioinformatics/btl671 - 54. Sacar, M.D. and Allmer, J. (2014) Machine Learning Methods for MicroRNA Gene Prediction. In: Malik Yousef, Jens Allmer, Eds., miRNomics: MicroRNA Biology and Computational Analysis, Methods in Molecular Biology, Vol. 1107, Humana Press, New York, 177-187.
http://dx.doi.org/10.1007/978-1-62703-748-8_10
NOTES

*Corresponding author.