Engineering
Vol.5 No.9A(2013), Article ID:36784,9 pages DOI:10.4236/eng.2013.59A003
Nanocellulose-Based Biosensors: Design, Preparation, and Activity of Peptide-Linked Cotton Cellulose Nanocrystals Having Fluorimetric and Colorimetric Elastase Detection Sensitivity
1Southern Regional Research Center, USDA-ARS, New Orleans, USA
2School of Renewable Natural Resources, Louisiana State University Agricultural Center, Baton Rouge, USA
Email: *vince.edwards@ars.usda.gov
Copyright © 2013 J. Vincent Edwards et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received March 25, 2013; revised April 25, 2013; accepted May 6, 2013
Keywords: cellulose nanocrystal; cotton; human neutrophil elastase; fluorescent; colorimetric; point of care biosensor; peptide-cellulose conjugate
ABSTRACT
Nanocrystalline cellulose is an amphiphilic, high surface area material that can be easily functionalized and is biocompatible and eco-friendly. It has been used singularly and in combination with other nanomaterials to optimize biosensor design. The attachment of peptides and proteins to nanocrystalline cellulose and their proven retention of activity provide a route to bioactive conjugates useful in designs for point of care biosensors. Elastase is a biomarker for a number of inflammatory diseases including chronic wounds, and its rapid sensitive detection with a facile approach to sensing is of interest. An increased interest in the use of elastase sensors for point of care diagnosis is resulting in a variety of approaches to elsastase sensors utilizing different detection technologies. Here elastase substrate peptide-celluose conjugates synthesized as colorimetric and fluorescent sensors on cotton cellulose nanocrystals are compared. The structure of the sensor peptide-nanocellulose crystals when modeled with computational crystal structure parameters demonstrates the spatio-stoichiometric features of the nanocrystalline surface that allows ligand to active site protease interacttion. An understanding of the structure/function relations of enzyme and conjugate substrate of the peptides covalently attached to nancellulose has implications for enhancing the biomolecular transducer. The potential applications of both fluorescent and colorimetric detection to markers like elastase using peptide cotton cellulose nanocrystals as a transducer surface to model point of care biosensors for protease detection are discussed.
1. Introduction
Advances in biosensor technology hold promise to revolutionize healthcare and diagnosis through assessment of health status and disease onset and progression using non-invasive methods [1,2]. This is especially the case in the area of sensor and imaging for wound healing, where approaches for biochemical and cellular markers are emerging [3].
Biosensor mechanism design may be defined as the interface of a molecular or receptor-recognition property with a cellular or biochemical activity, triggering a “biomolecular switch” that in turn is connected with a detector signal. Biosensors can generally be divided into electrical, optical, and mechanical, which include a wide range of detector technologies: colorimetric, fluorescent, bioluminescent, electrochemical, piezoelectric, quartz microbalance, acoustic and conductometric signals [4]. The “biomolecular switch” may be defined by a host of bioactive molecules or biological changes ranging from pH to enzyme or cellular sources. Detectors have been adapted to matrices that function at varying levels of sensitivity based on wavelength selectivity and sensitivity of resolution within the electromagnetic spectrum. Although the sophistication and sensitivity of detectors underlie much of the progress in the rapidly growing field of biosensors, the composition of the transducer material and its interaction at the bio-molecular interface is of equal value. Detector interface with the biological environment can complicate and dampen the sensitivity of detection. Thus, biosensor materials must ideally be compatible with the complexity of the bio-system by being resistant to nonselective adsorption by lipids, proteins, polysaccharides, cellular debris and breakdown of the transducer surface.
Nanomaterials with high surface area and biocompatible properties are good transducer surfaces. Nanocellulose in particular provides noteworthy advantages, but has received less attention than other nanomaterials for sensor applications [5]. As a highly crystalline biopolymer with a hydrophilic, high surface area, it possesses reactive hydroxyls that can be derivatized to covalently append a wide range of biologically active molecules. An image of freeze dried cotton cellulose nanocrystals displayed in Figure 1(a) reveals a translucent crystalline appearance with an x-ray diffraction pattern signature of cellulose I [6]. Prior to freeze drying cotton nanowhiskers display the morphology shown in the TEM image of Figure 1(b). Cellulose nanocrystals have also been reported to have chiral neumatic structures arising from unique self-assembly yielding inherent optical properties that are environmentally sensitive [7].
Cellulose does not tend to irreversibly adsorb proteins as do more hydrophobic material surfaces making it promise in the form of cellulose-fibrin nanocomposites
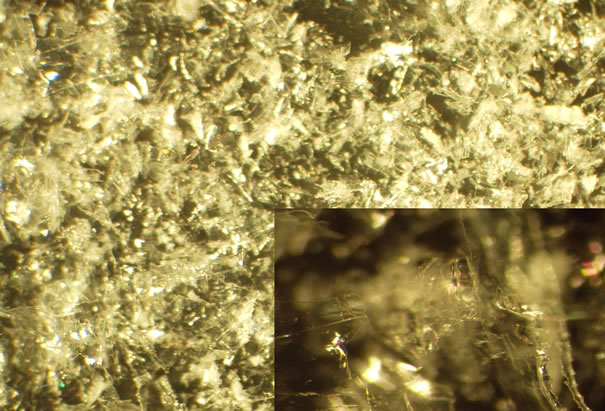

Figure 1. a(top) Untreated freeze-dried cotton nanowhiskers magnified 15X and 60X(inset). b(bottom) TEM of cotton nanowhiskers.
more biocompatible. Recent studies have shown that its use in artificial vascular graft applications is a good example of its biocompatibility [8]. Cellulose in a nanocrystalline state provides a very high surface area matrix for functionalizing and hosting selectively active biomolecules, while abrogating nonspecific protein binding. Thus nanocellulose has the potential to increase sensitivity to the bioactive response of interest. Depending on its functionalized structure, nanocellulose may be interfaced with sensitive spectrosocopic, conductive, colorimetric, fluorescent, and quartz impedance detector technologies.
2. Applications of Nanocellulose to Biosensors
Varying forms of nanocellulose have been characterized and discussed for their morphological properties in previous reviews, and self-assembled nanocrystalline materials include cellulose nanowhiskers, microfibrils, and microfibrillated cellulose [9,10]. These forms of nanocellulose have dimensions and morphology as shown in Figure 1(b) that give rise to useful properties applicable to biosensors [5]. Nanocellulosic materials as nancompomsites, gels, microdialysis membranes, and surface grafted molds have been shown to function at the aqueous bio-interface of a probe.
2.1. Biosensors with Polymer, Biopolymer and Enzyme Composites of Cellulose
Materials including carbon nanotubes, mesoporous silica, gold, quantum dots, dendrimers, biopolymers and block copolymers when combined with nanocellulose have been shown to be useful in optimizing biosensor design [11-17].
Cellulose derivatives in the form of block copolymers have been proposed as stimuli responsive mediators that can be prepared, regioselectively modified through dissolution of cellulose in ionic liquids, and modified through radical polymerization [11]. Regioselectively modified cellulose as a polyethylene glycol derivative was also synthesized and employed to give rise to honeycomb patterned films that are formed based on amphiphilic cellulosic properties [12]. Fluorescent molecules and quantum dots were attached to the films to demonstrate the potential for site specific functionality suitable for biosensors.
Enzymes can be formulated with nanocellulose as covalent linkages, salt bridges, and with physical inclusion complexes [13-21]. The hydrophilic surface properties provide a biocompatible matrix for selective activity to occur. Glucose oxidase immobilized on nanocrystalline cellulose with gold nanoparticles attached through thiols of polyethylenimine derivatives gave varying levels of activity as a function of the thiol linker [13]. Dendrimers as linkers attached to antigens were also linked to nanocellulose and combined with zeolite to improve specific antibody recognition [14]. Glucose oxidase, which is of interest in glucose monitoring for diabetes control has its origin as well in the first reported biosensor [15]. Glucose oxidase has also been immobilized on bacterial cellulose-filtered carbon nanotube composites deposited on electrodes. The sensor demonstrated direct electron transfer between glucose and glucose oxidase [16]. Quarternized cellulose nanoparticles were combined with acetylene black and used for physical inclusion of glucose oxidase on the surface of electrodes giving both good glucose and hydrogen peroxide sensitivity [17].
Edwards et al. reported very high lysozyme activity when the hydrolase was attached to cotton cellulose nanocrystals [18], which suggests the activity of anti-microbial enzymes can be bolstered when immobilized on cotton cellulose nanocrystals, and this approach may have further implications for use to detect as well as inhibit formation of microbial biofilms [19]. Finally it is noteworthy that cellulases have been used in an approach that demonstrates how enzymatic digestion of cellulose in combination with soft lithography can be used to obtain patterned surfaces [20].
2.2. Detection of Cellular Markers
Detection of cellular markers has shown to be facilitated through a nanocelluose matrix. Electrospun cellulose nitrate nanofibers when combined with conductive magnetic nanoparticles were shown to be useful in the sensitive detection of E coli bacteria [21]. A sensor that detects urate, a metabolite useful as a diagnostic marker of S. aureus and P. aeruginosa, utilized a carbon fiber sensor laminate in combination with cellulose acetate, which filtered out large proteins, and was partially successful when used with blood and blister fluid [22].
2.3. Cellulose-Based Fluorescent Detection
Bioactive, fluorescent nanocrystalline cellulose (NCC) has recently been shown to provide numerous interesting applications [23] which includes quantification and localization of nanoparticles within cells [24] using positively charged fluorescent NCC for bio-imaging, and the attachment of peptide and enzyme fluorophores to NCC for biosensing. For example fluorescent cellulose analogs derivatized with rhodamine B or fluorescein-5- isothiocyanate [25] have been assessed for cellular imaging. Whereas the fluorescent NCC analog 1-pyrenebutyryl-3- aminopropyl-silanized cellulose and terpyridine-modified pyrylene cellulose, a self assembled supramolecular complex with high affinity for transition metal [26], both have potential for biosensing and imaging applications. Fluorescent coumarin and anthracene analogs of TEMPO oxidized, propargylamino-cellulose were also prepared through “Click” chemistry [27].
Fluorescent Peptide Analogs
Fluorescent tryptophan-containing peptide conjugates of TEMPO oxidized nanocellulose were prepared and retained their fluorescent properties [28]. A higher yield general route to peptide conjugation on cellulose surfaces has also been recently disclosed using a xyloglucanpeptide conjugate for activation [29]. Intramolecularly quenched fluorogenic substrates of neutrophil serine proteases that distinguish human neutrophil elastase, proteinase 3, and chathepsin G activities at free and membrane bound subnanomolar concentrations of HNE have been reported [30] as sensitive biosensors for HNE. Here we examine the use of HNE elastase substrates that are fluorescent peptide.
2.4. Cotton Cellulose Nanocrystals as Sensors
Shown in Figure 2 is a diagram of the process for the preparation of cotton cellulose nanocrystals. Using this approach as described by Wu [6] nanocrystals with rodlike geometrical shapes (159-nm length and 15-nm average diameter) as shown in Figure 1(b) have been prepared and characterized with TEM and X-ray crystallography. When calculated with a value of 1.5 g∙cm−3 for the density of crystalline cellulose the specific surface area of cotton cellulose nanowhiskers is 186.2 m2∙g−1 and for dimensions closer to microfibrillated cellulose (700 - 900 or 1000 nm) 512 nm in length and 10 - 40 nm in diameter), surface area is approximately 131 m2∙g−1 which is consistent with the TEM images.
Figure 3 shows the synthesis of the peptide-cellulose conjugate. Peptide incorporation on cotton cellulose nanocrystals was characterized with DS levels ranging from 0.02 - 0.06 using the approach described for colorimetric elastase sensors [31]. The relationship of the specific surface area to the DS levels for these types of conjugates has been characterized in terms of derivatization of available hyroxymethyls on the surface of the

Figure 2. Schematic flowchart of the preparation of cotton cellulose nanocrystals.
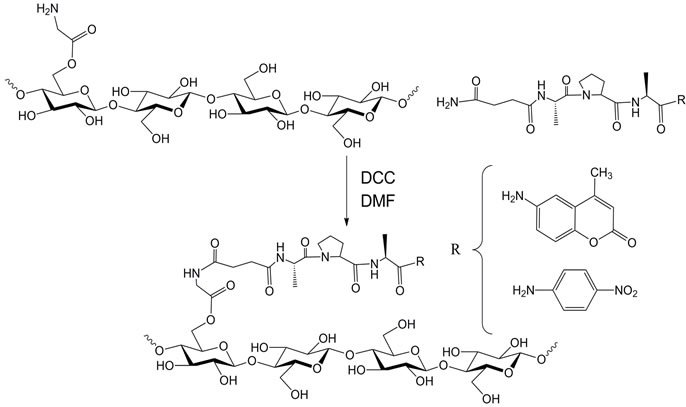
Figure 3. Synthesis of peptide-cellulose conjugate. DCC = dicyclohexylcarbodiimide. DMF = dimethylformamide
nanocrystal. Other work on cellulose nanocrystals has examined the limits of specific surface area coverage in functionalization reactions, and demonstrated in some cases that maximum reaction of hydroxymethyls is possible on the surface [32]. When peptide nanocellulose conjugates of cotton were compared with paper the cellulose nanocrystaline surface was found to have a 8000 times greater surface area. The activity of the peptidecellulose conjugate on nanocrystalline cellulose is greater than what was found on paper. Thus specific surface area plays a role in activity, yielding higher sensitivity when colorimetrically assessed than paper in the detection of elastase activity.
3. Human Neutrophil Elastase (HNE) in Chronic Diseases
Human neutrophil elastase (HNE) is a serine protease that is often released in higher than normal concentrations from immune cells e.g. neutrophils under inflamematory disease conditions. Numerous inflammatorybased disease states secrete HNE and matrix metalloproteases which in turn have prolonged deleterious effects stemming from excessive degradation of extracellular matrix proteins [33-35], fibronectin [36,37], and growth factors [38]. In some cases these effects are central to the pathology of inflammatory-based diseases as rheumatoid arthritis [39] chronic obstructive pulmonary disease [40], adult respiratory distress syndrome [41], and glomerulonephritis [42]. Some of these diseases have therapeutic protocols that either have suggested or required treatment with HNE inhibitors [43,44].
HNE has been shown to be a specific marker for psoriasis [45], an early prognostic marker of acute pancreatitis [46] and of cardiovascular events in patients with angina pectoris [47]. HNE has also been documented as a biomarker in neonatal sepsis [48] and septic shock [49], and has been suggested as a biomarker for newborns with cystic fibrosis transmembrane conductance regulator related metabolic syndrome (CRMS), a form of cystic fibrosis (CF) that does not meet the diagnostic criteria for clinically diagnosed CF [50]. Thus, the prevalence of HNE in some chronic inflammatory diseases suggests its use as an analyte for diagnosis and treatment.
4. HNE Biosensors
Some of the types of sensor approaches to HNE detection are outlined in Table 1" target="_self"> Table 1. The molecular basis of some of these sensor motifs include peptide cross-linked dextran hydrogels with a tunable quartz crystal microbalance detection [51], chromogenic peptide substrates tethered to collagen, polyamide polyesters, silica gel [52], ethoxyacrylate resin [53], and cellulose [54], and DNA-based aptasensors [55]. A variety of sensor design motifs have also been employed, and these include: 1) a microchip integrated with reagent-release capillaries is reported as a “drop-and-sip” technique, which utilizes a single microliter droplet of HNE-containing solution with fluorescence image analysis of the hydrolyzed substrate product [56]; 2) fluorometric detection of HNE activity with synthetic supramolecular pore sensors [57,58] the covalent immobilization of HNE on biosensor chips having surface plasma resonance capability has been employed for analysis of HNE inhibitors [59]; 3) a microdialysis sampling assay of HNE activity where the substrate is delivered through the microdialysis probe to external solutions containing HNE, and the product, pNA, is recovered back into the probe [60].
Recently the discussion of a clinical pathway that includes a point of care protease test as part of the decision making process in managing and treating non-healing wounds has been introduced, and the numerous potential clinical advantages of using a point of care protease test in guiding therapy have been cited [3]. Although all chronic wounds have not been found to have elevated HNE and proteases the advantages of detecting pathological levels of HNE at an early stage has been well documented over the years [3,61,62]. Thus, the in situ detection of HNE and other proteases through dressing design motif is of potential diagnostic value in providing improved healing resolution in chronic wounds.
5. Cotton Cellulose Nanocrystals as HNE Sensors
Figure 4 shows a model of a peptide-conjugated nanocrystal constructed from a “minimal size” cellulose nanocrystal containing 180 anhydroglucose units, and constructed to reflect the proximal DS level on the nanocrystalline surface observed (0.06) as recently reported [31]. The molecular model portrays the putative accessibility of the peptide substrate on the surface of the nanocrystal by serine protease elastase. The glycinesuccinyl-linker that is esterified to cellulose places the
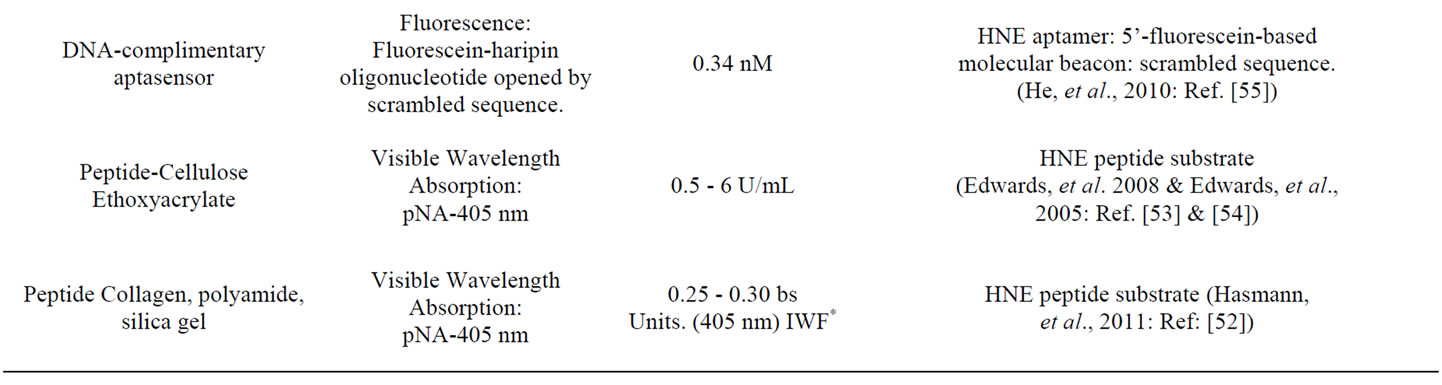
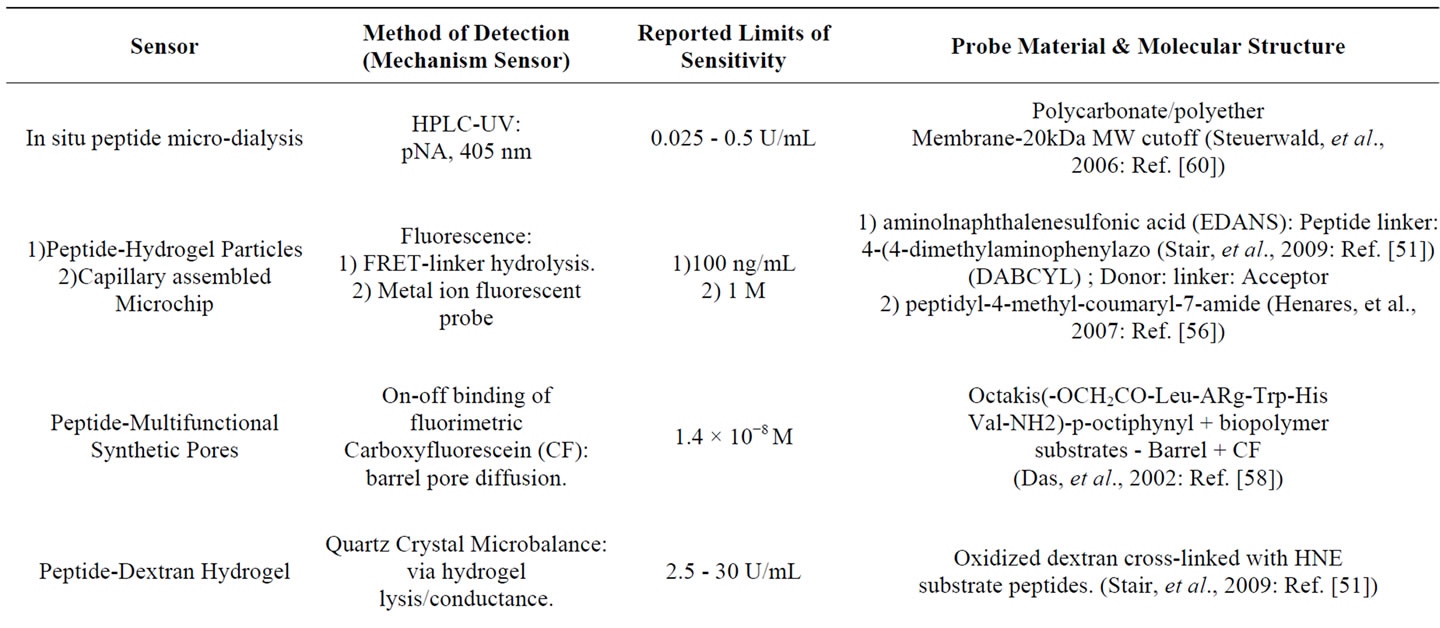
Table 1. Design and mode of detection for several human neutrophil elastase (HNE) sensors.
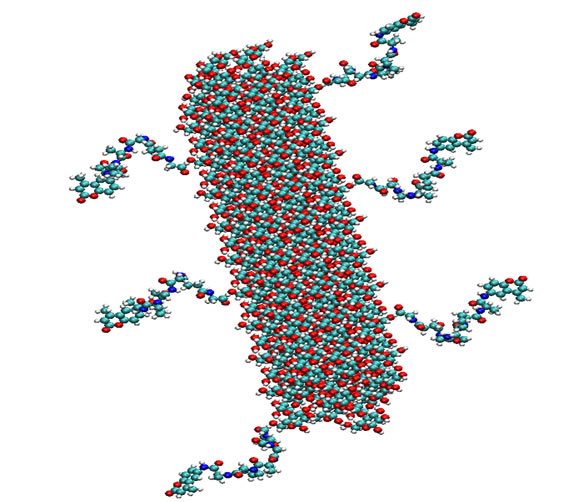
Figure 4. Model of peptide-conjugated cellulose nanocrystal. Six tetrapeptides (Succinyl-Ala-Ala-Pro-Val-CMA) with amino-terminal of peptide attached to glycine-cellulose (see Figure 3).
peptide at a distance from the surface of the crystal. However, it can be seen how elastase may dock the COOH-terminal subsites of enzyme recognition without apparent steric interference from the crystal surface. On the other hand, on an extended sheet-like cellulose crystalline surface, elastase may bind to the surface and hydrolyze available peptide substrates releasing the chromophore or fluorophore.
The affinity of elastase for the tripeptide sequence is enhanced when it is conjugated to nanocellulose as reflected in Michaelis Menten kinetic Km values for both the freely soluble peptide and the peptide-nanocellulose conjugate. The substrate binding affinity is increased 30-fold when the substrate is annealed to the cellulose whisker (chemical structure shown in Figure 3). The increased affinity of elastase for the cellulose nanocrystalline bound substrate may be due to enhanced binding of the cationic surface of elastase to the negatively charged cellulose nanocrystalline surface. It is noteworthy in this regard that the cellulose nanocrystalline surface is negatively charged with up to 10% of the surface hydroxyls being sulfated due to the sulfuric acid step of the nanocrystalline preparation (Figure 2).
The role of the COOH-terminal peptide leaving group in sensor detection merits some comment. Both paranitroaniline (pNA) and coumadinamide (CMA) have been used to assess elastase substrate activity through colorimetric and fluorescence detection [63]. Although the pNA substrate is highly specific for elastase the AMC substrate is much more sensitive. Fluorescent elastase substrates attached to cotton cellulose nanocrystals have recently been prepared similar to a colorimetric HNE sensor reported [31] and shown to be sensitive to low levels of elastase concentrations. Figure 5" target="_self"> Figure 5 shows the fluorescent response of 2 milligrams of a tetrapeptidenanocellulose analog prepared from cotton cellulose nanocrystals upon incubation with human neutrophil elastase. The analogous colorimetric responses for elastase are shown in Figure 6" target="_self"> Figure 6 based on single time point determinations elastase units.
Figure 6 portrays the color change of the nanocrystals upon amplification of the colorimetric response when para-nitroaniline is released from the peptide and reacts with a color amplifying reagent. As shown recently for the reaction of elastase with the peptide-nanocellulose conjugates there is a strong color response associated with the amplification reaction of para-nitroaniline with amplifying reagents [31]. The images of the nanocrystalline sensor following incubation with elastase demonstrate the visual color response positive for the presence of elastase activity. Treatment of the elastase sensor with HNE concentrations commensurate with wound fluid activity results in a visible color within minutes. The color response is a result of the reaction of paranitroaniline, which is hydrolyzed from the nanocellulose
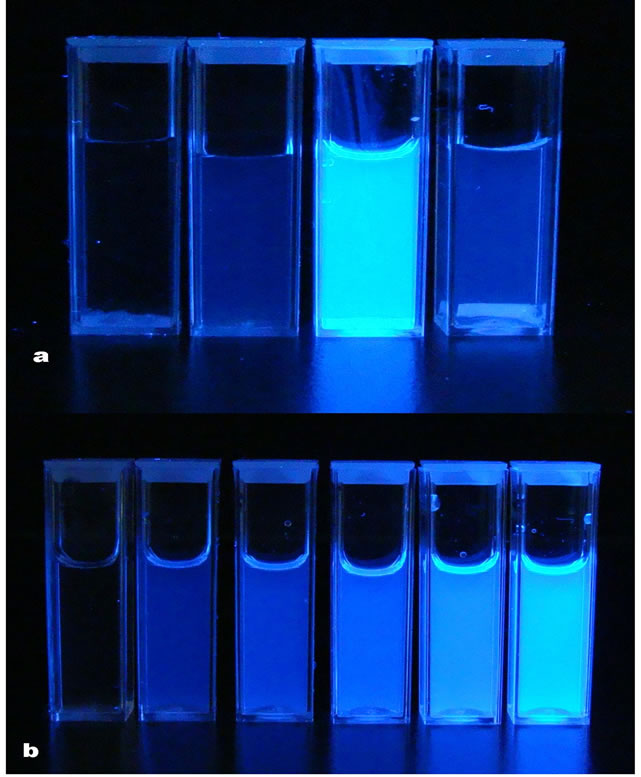
Figure 5. a(top, l. to r.) Fluorescent response of untreated cotton nanocrystals, tetrapeptide-nanocellulose, tetrapeptidenanocellulose with 2 U/mL of HNE and tetrapeptide-filter paper with 2 U/mL of HNE. b(bottom) fluorescent response of the tetrapeptide in solution at different concentrations ranging (l. to r.) 0 to 0.25 µmole/mL with 2 U/mL of HNE.
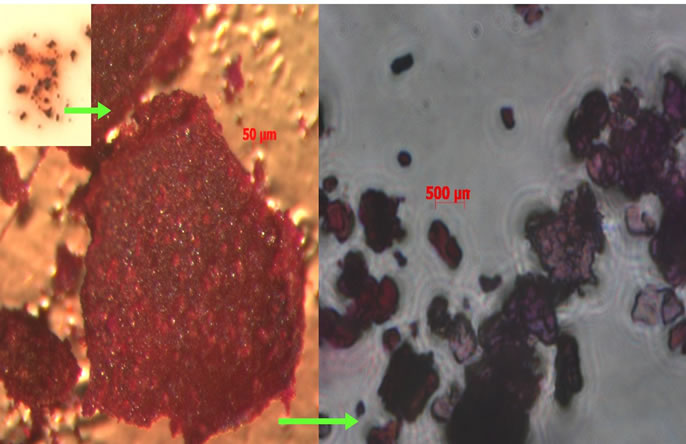
Figure 6. Cellulose nanocrystals after released pNA has reacted with the color amplifying reagent.
peptide conjugate, and is diazotized or reacts with a cinnamonaldehyde analog. In a similar manner fluorescent CMA analogs gives a strong reponse when visualized on a fluorescent lamp (Figure 5). These types of adducts may have potential as point of care diagnostics where incubation times are minimized [3]. This type of sensor approach has also been aligned with dialysis cellulose membranes to demonstrate the potential for in situ wound dressing use [31].
6. Conclusion
The versatility and sensitivity that cellulose nanocrystals afford are playing a role in providing a biocompatible, high surface area material that can be combined with biologically active molecules as peptides and proteins in biosensor design. The cotton cellulose nanocrystals shown here provide an example of how bioactive conjugates with high enzyme affinity can be used to prepare a sensitive biosensor for a clinically relevant protease.
REFERENCES
- C. Gouvea, “Biosensors for Health Applications,” In: P. A. Serra, Ed., Biosensors for Health, Environment and Biosecurity, InTech, 2011, Chapter 3. http://dx.doi.org/10.5772/17103
- B. Bohunicky and S. A. Mousa, “Biosensors: The New Wave in Cancer Diagnosis,” Nanotechnology Science and Applications, Vol. 4, No. 1, 2011, pp. 1-10.
- T. R. Dargaville, B. L. Farrugia, J. A. Broadbent, S. Pace and Z. Upton, “Sensors and Imaging for Wound Healing: a Review,” Biosensors and Bioelectronics, Vol. 41, 2013, pp. 30-42. doi:10.1016/j.bios.2012.09.029
- J. Tamayo, P. M. Kosaka, J. J. Ruz, A. S. Paulo and M. Calleja, “Biosensors Based on Nanomechanical Systems,” Chemical Society Review, Vol. 42, No. 3, 2013, pp. 1287-1311. doi:10.1039/c2cs35293a
- V. Incani, C. Danumah and Y. Boluk, “Nanocomposites of Nanocrystalline Cellulose for Biosensor Applications,” Proceedings of the 2012 NSTI Nanotechnology Conference and Expo, Santa Clara, 18-21 June 2012, pp. 13-16.
- Y. Yue, C. Zhou, A. D. French, G. Xia, G. Han, A. Wang and Q. Wu, “Comparative Properties of Cellulose NanoCrystals from Native and Mercerized Cotton Fibers,” Cellulose, Vol. 19, No. 4, 2012, pp. 1173-1187. doi:10.1007/s10570-012-9714-4
- K. E. Shopsowitz, H. Qi, W. Y. Hamad and M. J. MacLachlan, “Free Standing Mesoporous Silica Films with Tunable Chiral Nematic Structures,” Nature, Vol. 468, No. 7322, 2010, pp. 422-425. doi:10.1038/nature09540
- E. E. Brown, D. Hu, N. Abu Lail and X. Zhang, “Potential of Nanocrystalline Cellulose-Fibrin Nanocomposites for Artificial Graft Applications,” Biomacromolecules, Vol. 14, No. 4, 2013, pp. 1063-1071 doi:10.1021/bm3019467
- D. Klemm, F. Kramer, S. Moritz, T. Lindstrom, M. Ankerfors, D. Gray and A. Dorris, “Nanocelluloses: A New Family of Nature-Based Materials,” Angewandte Chemie International Edition, Vol. 50, No. 24, 2011, pp. 5438-5466. doi:10.1002/anie.201001273
- Y. Habibi, L. A. Lucia and O. J. Rojas, “Cellulose Nanocrystals: Chemistry, Self-Assembly, and Applications,” Chemical Reviews, Vol. 110, 2010, pp. 3479-3500. doi:10.1021/cr900339w
- H. Kang, R. Liu and Y. Huang, “Cellulose Derivatives and Graft Copolymers as Blocks for Functional Materials,” Polymer International, Vol. 62, No. 3, 2013, pp. 338-344. doi:10.1002/pi.4455
- W. Z. Xu and J. F. Kadla, “Honeycomb Films of Cellulose Azide: Molecular Structure and Formation of Porous Films,” Langmuir, Vol. 29, No. 2, 2012, pp. 727-733. doi:10.1021/la303835e
- V. Incani, C. Danumah and Y. Boluk, “Nanocomposites of Nanocrystalline Cellulose for Enzyme Immobilization,” Cellulose, Vol. 20, No. 1, 2013, pp. 191-200. doi:10.1007/s10570-012-9805-2
- A. J. Ruiz-Sanchez, M. I. Montanez, C. Mayorga, M. J. Torres, N. S. Kehr, Y. Vida, D. Collado, F. Najera, L. De Cola and E. Perez-Inestrosa, “Dendrimer-Modified Solid Supports: Nanostructured Materials With Potential Drug Allergy Diagnostic Applications,” Current Medicinal Chemistry, Vol. 19, No. 29, 2012, pp. 4942-4954. doi:10.2174/0929867311209024942
- L. C. Clark and C. Lyons, “Electrode Systems for Continuous Monitoring in Cardiovascular Surgery,” Annals of the New York Academy of Sciences, Vol. 102, No. 1, 1962, pp. 29-45. doi:10.1111/j.1749-6632.1962.tb13623.x
- Y.-H. Kim, S. Park, K. Won, H. J. Kim and S. H. Lee, “Bacterial Cellulose—Carbon Nanotube Composite as a Biocompatible Electrode for the Direct Electron Transfer of Glucose Oxidase,” Journal of Chemical Technology & Biotechnology, Vol. 88, No. 6, 2013, pp. 1067-1070. doi:10.1002/jctb.3939
- M. Li, Y. Qi, Y. Ding, Q. Zhao, J. Fei and J. Zhou, “Electrochemical Sensing Platform Based on the Quaternized Cellulose Nanoparticles/Acetylene Black/Enzymes Composite Film,” Sensors and Actuators B: Chemical, Vol. 168, 2012, pp. 329-335. doi:10.1016/j.snb.2012.04.030
- J. V. Edwards, N. Prevost, B. Condon, A. French and Q. Wu, “Immobilization of Lysozyme-Cellulose AmideLinked Conjugates on Cellulose I and Ii Cotton Nanocrystalline Preparations,” Cellulose, Vol. 19, No. 2, 2012, pp. 495-506. doi:10.1007/s10570-011-9637-5
- B. Thallinger, E. N. Prasetyo, G. S. Nyanhongo and G. M. Guebitz, “Antimicrobial Enzymes: An Emerging Strategy to Fight Microbes and Microbial Biofilms,” Biotechnology Journal, Vol. 8, No. 1, 2013, pp. 97-109. doi:10.1002/biot.201200313
- R. Kargl, T. Mohan, S. Köstler, S. Spirk, A. Doliška, K. Stana-Kleinschek and V. Ribitsch, “Functional Patterning of Biopolymer Thin Films Using Enzymes and Lithographic Methods,” Advanced Functional Materials, Vol. 23, No. 3, 2013, pp. 308-315. doi:10.1002/adfm.201200607
- L. Yilun, S. Nartker, M. Wiederoder, H. Miller, D. Hochhalter, L. T. Drzal and E. C. Alocilja, “Novel Biosensor Based on Electrospun Nanofiber and Magnetic Nanoparticles for the Detection of E. Coli O157:H7,” IEEE Transactions on Nanotechnology, Vol. 11, No. 4, 2012, pp. 676-681. doi:10.1109/TNANO.2011.2174801
- D. Sharp, S. Forsythe and J. Davis, “Carbon Fibre Composites: Integrated Electrochemical Sensors for Wound Management,” Journal of Biochemistry, Vol. 144, No. 1, 2008, pp. 87-93. doi:10.1093/jb/mvn045
- E. Lam, K. B. Male, J. H. Chong, A. C. W. Leung and J. H. T. Luong, “Applications of Functionalized and Nanoparticle-Modified Nanocrystalline Cellulose,” Trends in Biotechnology, Vol. 30, No. 5, 2012, pp. 283-290. doi:10.1016/j.tibtech.2012.02.001
- K. A. Mahmoud, J. A. Mena, K. B. Male, S. Hrapovic, A. Kamen and J. H. T. Luong, “Effect of Surface Charge on the Cellular Uptake and Cytotoxicity of Fluorescent Labeled Cellulose Nanocrystals,” ACS Applied Materials & Interfaces, Vol. 2, No. 10, 2010, pp. 2924-2932. doi:10.1021/am1006222
- Q. Yang and X. Pan, “A Facile Approach for Fabricating Fluorescent Cellulose,” Journal of Applied Polymer Science, Vol. 117, No. 6, 2010, pp. 3639-3644.
- M. L. Hassan, C. M. Moorefield, H. S. Elbatal, G. R. Newkome, D. A. Modarelli and N. C. Romano, “Fluorescent Cellulose Nanocrystals Via Supramolecular Assembly of Terpyridine-Modified Cellulose Nanocrystals and Terpyridine-Modified Perylene,” Materials Science and Engineering: B, Vol. 177, No. 4, 2012, pp. 350-358. doi:10.1016/j.mseb.2011.12.043
- I. Filpponen, H. Sadehgifar and D. A. Argyropoulos, “Photoresponsive Cellulose Nanocrystals,” Nanomaterials and Nanotechnology, Vol. 1, No. 1, 2011, pp. 34-43.
- S. Barazzouk and C. Daneault, “Amino Acid and Peptide Immobilization on Oxidized Nanocellulose: Spectroscopic Characterization,” Nanomaterials, Vol. 2, No. 2, 2012, pp. 187-205. doi:10.3390/nano2020187
- C. Araújo, A. Nakhai, M. Ruda, R. Slättegård, P. Gatenholm and H. Brumer, “A General Route to Xyloglucan-Peptide Conjugates for the Activation of Cellulose Surfaces,” Carbohydrate Research, Vol. 354, 2012, pp. 116-120. doi:10.1016/j.carres.2012.03.038
- B. Korkmaz, S. Attucci, C. Epinette, E. Pitois, M.-L. Jourdan, L. Juliano and F. Gauthier, “Measurement of Neutrophil Elastase, Proteinase 3, and Cathepsin G Activities Using Intramolecularly Quenched Fluorogenic Substrates,” Leucocytes, Vol. 844, 2012, pp. 125-138. doi:10.1007/978-1-61779-527-5_9
- J. V. Edwards, N. Prevost, K. Sethumadhavan, A. Ullah and B. Condon, “Peptide Conjugated Cellulose Nanocrystals with Sensitive Human Neutrophil Elastase Sensor Activity,” Cellulose, Vol. 20, No. 3, 2013, pp. 1223-1235. doi:10.1007/s10570-013-9901-y
- Y. Habibi, H. Chanzy and M. Vignon, “Tempo-Mediated Surface Oxidation of Cellulose Whiskers,” Cellulose, Vol. 13, No. 6, 2006, pp. 679-687. doi:10.1007/s10570-006-9075-y
- G. S. Schultz, G. Ladwig and A. Wysocki, “Extra-cellular Matrix: Review of its Roles in Acute and Chronic Wounds,” 2005. http://www.worldwidewounds.com/2005/august/Schultz/Extrace-Matric-Acute-Chronic-Wounds.html
- S. E. Herrick, P. Sloan, M. McGurk, L. Freak, C. N. McCollum and M. W. Ferguson, “Sequential Changes in Histologic Pattern and Extracellular Matrix Deposition During the Healing of Chronic Venous Ulcers,” The American Journal of Pathology, Vol. 141, No. 5, 1992 pp. 1085-1095.
- S. Herrick, G. Ashcroft, G. Ireland, M. Horan, C. McCollum and M. Ferguson, “Up-Regulation of Elastase in Acute Wounds of Healthy Aged Humans and Chronic Venous Leg Ulcers Are Associate with Matrix Degradation,” Laboratory Investigation, Vol. 77, No. 3, 1997, pp. 281-288.
- F. Grinnell and M. Zhu, “Fibronectin Degradation in Chronic Wounds Depends on the Relative Levels of Elastase, Alpha1-Proteinase Inhibitor and Alpha2-Macroglobulin,” Journal of Investigative Dermatology, Vol. 106, No. 2, 1996, pp. 335-341. doi:10.1111/1523-1747.ep12342990
- F. Grinnell and M. Zhu, “Identification of Neutrophil Elastase as the Proteinase in Burn Wound Fluid Responsible for the Degradation of Fibronectin,” Journal of Investigative Dermatology, Vol. 103, No. 2, 1994, pp. 155- 161.
- D. R. Yager, S. M. Chen, S. I. Ward, O. O. Olutoye, R. F. Diegelmann and I. K. Cohen, “Ability of Chronic Wound Fluids to Degrade Peptide Growth Factors Is Associated with Increased Levels of Elastase Activity and Diminished Levels of Proteinase Inhibitors,” Wound Repair and Regeneration, Vol. 5, No. 1, 1997, pp. 23-32. doi:10.1046/j.1524-475X.1997.50108.x
- L. Ekerot and K. Ohlsson, “Interactions of Granulocyte Proteases with Inhibitors in Rheumatoid Arthritis,” Advances in Experimental Medicine and Biology, Vol. 167, 1984, pp. 335-344. doi:10.1007/978-1-4615-9355-3_28
- S. D. Shapiro, “Proteinases in Chronic Obstructive Pulmonary Disease,” Biochemical Society Transactions, Vol. 30, No. 2, 2002, pp. 98-102. doi:10.1042/BST0300098
- A. B. Groeneveld, P. G. Raijmakers, C. E. Hack and G. T. Thijis, “Interleukin 8-Related Neutrophil Elastase and the Severity of the Adult Respiratory Distress Syndrome,” Cytokine, Vol. 7, No. 7, 1995, pp. 746-752. doi:10.1006/cyto.1995.0089
- T. Oda, O. Hotta, Y. Taguma, H. Kitamura, K. Sudo, I. Horigome, S. Chiba, N. Yoshizawa and H. Nagura, “Involvement of Human Neutrophil Elastase in Crescentic Glomerulonephritis,” Human Pathology, Vol. 28, No. 6, 1997, pp. 720-728. doi:10.1016/S0046-8177(97)90182-9
- P. Sjo, “Neutrophil Elastase Inhibitors: Recent Advances in the Development of Mechanism-Based and Nonelectrophilic Inhibitors,” Future Medicinal Chemistry, Vol. 4, No. 5, 2012, pp. 651-660. doi:10.4155/fmc.12.17
- T. Stevens, K. Ekholm, M. Gränse, M. Lindahl, V. Kozma, C. Jungar, T. Ottosson, H. Falk-Håkansson, A. Churg, J. L. Wright, H. Lal and A. Sanfridson, “AZD 9668: Pharmacological Characterization of a Novel Oral Inhibitor of Neutrophil Elastase,” Journal of Pharmacology and Experimental Therapeutics, Vol. 339, No. 1, 2011, pp. 313-320. doi:10.1124/jpet.111.182139
- A. Orem, O. Deger, G. Cimsit and S. Bahadir, “Plasma Polymorphonuclear Leukocyte Elastase Levels and Its Relation to Disease Activity in Psoriasis,” Clinica Chimica Acta, Vol. 264, No. 1, 1997, pp. 49-56. doi:10.1016/S0009-8981(97)00072-7
- C. Jaffray, J. Yang, G. Carter, C. Mendez and J. Norman, “Pancreatic Elastase Activates Pulmonary Nuclear Factor Kappa B and Inhibitory Kappa B, Mimicking Pancreatitis-Associated Adult Respiratory Distress Syndrome,” Surgery, Vol. 128, No. 2, 2000, pp. 225-231. doi:10.1067/msy.2000.107419
- F. B. Smith, F. G. R. Fowkes, A. Rumley, A. J. Lee, G. D. Lowe and C. M. Hau, “Tissue Plasminogen Activator and Leucocyte Elastase as Predictors of Cardiovascular Events in Subjects with Angina Pectoris: Edinburgh Artery Study,” European Heart Journal, Vol. 21, No. 19, 2000, pp. 1607-1613. doi:10.1053/euhj.2000.2127
- R. K. Basu, S. W. Standage, N. Cvijanovich, G. L Allen, N. J. Thomas, R. J. Freishtat, N. Anas, K. Meter, P. A. Checchia, R. Lin, T. P. Shanley, M. T. Bigham, D. S. Wheeler, P. Devarajan, S. L. Goldstein and H. R. Wong, “Identification of Candidate Serum Biomarkers for Severe Septic Shock-Associated Kidney Injury via Microarray,” Critical Care, Vol. 15, No. 6, 2011, p. R273. doi:10.1186/cc10554
- S. F. Kingsmore, N. Kennedy, H. L. Halliday, J. C. Van Velkinburgh, S. Zhong, V. Gabriel, J. Grant, W. D. Beavis, V. T. Tchemev, L. Perlee, S. Lejnine, B. Grimwade, M. Sorette and J. D. Edgar, “Identification of Diagnostic Biomarkers for Infection in Premature Neonates,” Molecular and Cellular Proteomics, Vol. 7, 2008, pp. 1863- 1875. doi:10.1074/mcp.M800175-MCP200
- C. L. Ren, H. Desai, M. Platt and M. Dixon, “Clinical Outcomes in Infants with Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Related Metabolic Syndrome,” Pediatric Pulmonology, Vol. 46, No. 11, 2011, pp. 1079-1084. doi:10.1002/ppul.21475
- J. L. Stair, M. Watkinson and S. Krause, “Sensor Materials for the Detection of Proteases,” Biosensors and Bioelectronics, Vol. 24, No. 7, 2009, pp. 2113-2118. doi:10.1016/j.bios.2008.11.002
- A. Hasmann, U. Gewessler, E. Hulla, K. P. Schneider, B. Binder, A. Francesko, T. Tzanov, M. Schintler, J. Van der Palen, G. M. Guebitz and E. Wehrschuetz-Sigl, “Sensor Materials for the Detection of Human Neutrophil Elastase and Cathepsin G Activity in Wound Fluid,” Experimental Dermatology, Vol. 20, No. 6, 2011, pp. 508-513. doi:10.1111/j.1600-0625.2011.01256.x
- J. V. Edwards, S. Caston-Pierre, A. F. Bopp and W. Goynes, “Detection of Human Neutrophil Elastase with Peptide-Bound Cross-Linked Ethoxylate Acrylate Resin Analogs,” The Journal of Peptide Research, Vol. 66, No. 4, 2005, pp. 160-168. doi:10.1111/j.1399-3011.2005.00284.x
- J. V. Edwards, S. Caston-Pierre, P. Howley, B. Condon and J. Arnold, “A Bio-Sensor for Human Neutrophil Elastase Employs Peptide-Para-Nitroanilide Cellulose Conjugates,” Sensor Letters, Vol. 6, No. 4, 2008, pp. 518- 523. doi:10.1166/sl.2008.418
- J.-L. He, Z.-S. Wu, S.-B. Zhang, G.-L. Shen and R.-Q. Yu, “Fluorescence Aptasensor Based on CompetitiveBinding for Human Neutrophil Elastase Detection,” Talanta, Vol. 80, No. 3, 2010, pp. 1264-1268. doi:10.1016/j.talanta.2009.09.019
- M. Henares, N. Takashi, S. Yoshida, F. Terabe, R. Mizutani, R. Sikizawa and H. Hisamoto, “Integration of Multianayte Sensing Function on a Capillary-Assembled Microchip: Simultaneous Determination of Ion Concentration and Enzymatic Activities by ‘Drop-and-Sip’ Technique,” Analytical Chemistry, Vol. 79, No. 3, 2007, pp. 908-915. doi:10.1021/ac061245i
- N. Sorde, G. Das and S. Matile, “Enzyme Screening with Synthetic Multifunctional Pores: Focus on Biopolymers,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 100, No. 21, 2003, pp. 11964-11969. doi:10.1073/pnas.2132894100
- G. Das, P. Talukdar and S. Matile, “Fluorometric Detection of Enzyme Activity with Synthetic Supramolecular Pores,” Science, Vol. 298, No. 5598, 2002, pp. 1600- 1602. doi:10.1126/science.1077353
- B. Shen, S. Shimmon, M. M. Smith and P. J. Ghosh, “Biosensor Analysis of the Molecular Interactions of Pentosan Polysulfate and of Sulfated Glycosaminoglycans with Immobilized Elastase, Hyaluronidase and Lysozyme Using Surface Plasmon Resonance (SPR) Technology,” Journal of Pharmaceutical and Biomedical Analysis, Vol. 31, No. 1, 2003, pp. 83-93. doi:10.1016/S0731-7085(02)00606-4
- A. J. Steuerwald, J. D. Villeneuve, L. Sun and J. A. Stenken, “In Vitro Characterization of an in Situ Microdialysis Sampling Assay for Elastase Activity Detection,” Journal of Pharmaceutical and Biomedical Analysis, Vol. 40, No. 5, 2006, pp. 1041-1047. doi:10.1016/j.jpba.2005.05.036
- International Consensus, “The Role of Proteases in Wound Diagnostics. An Expert Working Group Review,” Wounds International, London, 2011.
- D. R. Yager and B. C. Nwomeh, “The Proteolytic Environment of Chronic Wounds,” Wound Repair and Regeneration, Vol. 7, No. 6, 1999, pp. 433-441. doi:10.1046/j.1524-475X.1999.00433.x
- M. J. Castillo, K. Nakajima, M. Zimmerman and J. C. Powers, “Sensitive Substrates for Human Leuckocyte and Porcine Pancreatic Elastase: A Study of the Merits of Various Chromophoric and Fluorogenic Leaving Groups in Assays for Serine Proteases,” Analytical Biochemistry, Vol. 99, No. 1, 1979, pp. 53-64. doi:10.1016/0003-2697(79)90043-5
NOTES
*Corresponding author.

