Advances in Bioscience and Biotechnology
Vol.4 No.5A(2013), Article ID:31991,8 pages DOI:10.4236/abb.2013.45A003
Identification of regulatory sequence signatures in microRNA precursors implicated in neurological disorders
![]()
DBT Centre for Bioinformatics, Presidency University, Kolkata, India
Email: *sayakbif@yahoo.com
Copyright © 2013 Sayak Ganguli et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 26 March 2013; revised 3 May 2013; accepted 21 May 2013
Keywords: Neurological Diseases; Regulatory Motif Signatures; Parkinson’s disease; Alzheimer’s disease; Huntington Disease
ABSTRACT
MicroRNAs have emerged as one of the major classes of non-coding RNAs. Recent reports have placed them in high abundance in the nervous system, having key roles in development. Neurological disorders such as Parkinson’s disease, Alzheimer’s disease as well as Huntington disease have also been studied and several microRNAs associated with diseases pathogenesis have been identified. Various such findings indicate differential expression levels of many of these microRNAs. Such changes in the expression levels not only indicate towards a control of the biogenesis of these microRNAs but also indicate towards critical yet unelucidated roles of regulatory proteins, which probably act in concert to control the production or maturation of these molecules. In this work, a collection of overrepresented regulatory motif signatures were identified in the DNA and RNA sequences of the precursor microRNAs. The identification of such regulatory sequence signatures promises to provide new insights into many facets of microRNA regulation and neurological disorders.
1. INTRODUCTION
Along with the steady rise in human life expectancy, there has been an increase in the prevalence of neurodegenerative disorders. Limited knowledge about the actual molecular processes involved in disease occurrence and progression has proved to be major hurdles in the identification of suitable drug design experiments and other therapeutic strategies.
Since their discovery in C. elegans [1], microRNAs have emerged as one of the key cellular regulators which possess sequence specific inhibitory functions [2] and regulate a large number of cellular processes. Recent studies have indicated the presence of a large number of microRNAs in the brain and spinal cord [3,4]. A large number of these molecules have been reported to exhibit brain specific expression patterns. Many works have also indicated the expression of a large number of microRNAs in the neuron, astrocytes and oligodendritic cells [5-15]. Experimental data from such reports reveal the preferential expression levels of the various microRNAs. MicroRNA expression profiling has been the standard procedure for exploration of differentially expressed microRNAs in neurodegenerative diseases. Table 1 summarizes the information on various miRNAs and their disease associations.
The most common form of dementia is Alzheimer’s disease [AD]. This is characterized by a progressive degenerative neurological disorder and is generally sporadic in nature. Short β-amyloid [Aβ] peptides form plaques which progressively produce the disease condition [16-18]. Increased BACE1 expression is the most dominant risk factor in the brain as BACE1/β-secretase cleavage of APP is the rate-limiting step for Aβ peptide production [19,20]. Many microRNAs have been observed to exhibit altered expression levels under AD progression, and most of them regulate BACE1[19]. Findings further suggest that loss of specific microRNAs miR-107, miR-29a and miR-29b-1 contributes to increased BACE1 and Aβ levels in sporadic AD. In contrast to above, miR-9, miR-125b, and miR-128 have been found to increase their expression levels in the brain under Alzheimer’s disease progression [21]. Other studies shed light on the control of brain inflammatory responses by miR-146a which regulates complement factor H indica-
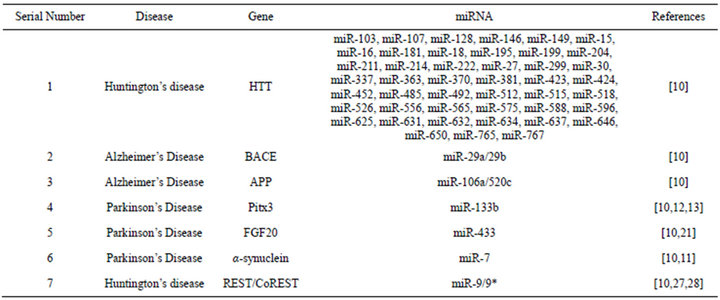
Table 1. Summary of identified microRNAs implicated in the three major neurological disorders.
ting towards a possible role of miRNA control of the neuroinflammatory process associated with deposition of the Aβ peptide. Conglomerative evidence suggests that microRNA regulation may play a role in AD pathogenesis.
On the other hand Parkinson’s disease [PD] is characterized by the progressive neurodegeneration of dopaminergic neurons in the substantia nigra. Several evidences have accumulated elucidating the role of miR-133b and its regulation of Pitx3—a paired like homeodomain transcription factor. A disruption of this negative feedback mechanism may be instrumental in causing the multitude of symptoms associated with PD [22-24]. MiR-433 binding site disruption have been reported to result in increased expression of fibroblast growth factor 20 [FGF20] which has been earlier identified to be an important risk factor for PD and correlated with overexpression of α-synuclein [25]. miR-7 has been reported to have a neuron specific expression and has also been shown to reduce α-synuclein protein levels [11].
The third most common neurological disorder is Huntington disease [HD] which is a dominant inherited neurodegenerative disorder. This is primarily caused by a trinucleotide repeat expansion of the gene encoding Huntingtin [Htt]. In non-affected individuals, the transcriptional repressor protein REST is found in the cytoplasm through interaction with Htt. The trinucleotide repeat expansion renders, REST incapable of binding Htt and thus accumulates in the nucleus. In the nucleus, REST recruits CoREST, and inactivates neuron-specific genes [26]. Recent studies have revealed miR-124a and miR-132 to be dysregulated under effects of REST repression. In vitro studies have also shown that miR-9/ miR-9* have the ability to target REST and CoREST, respectively [18]. The above background suggests the presence of a complex regulatory circuit of microRNAs and their subsequent expression leading to a large number of neurological disorders. The main aim of the work is to identify possible regulatory signatures in the microRNA precursor sequences which could possibly provide insight into the regulatory control of mature microRNA production.
2. MATERIALS AND METHOD
Information regarding the microRNAs associated with neurological disorders was identified from existing literature and was validated with the Phenomir database. The common miRNAs were then selected and their precursor sequences were obtained from miRBase. Once the precursor sequences were obtained they were converted to their complimentary DNA form using a very basic PERL script and the sets of sequences were divided in two parts—one was the complimentary DNA data set and the other was miRNA precursor dataset [RNA form]. The total dataset [Set 1 + Set 2] comprised of 400 sequences belonging to both human and mouse. Now to test the presence of regulatory motifs, a large number of position specific weight matrices were obtained from the established transcription factor databases such as JASPAR [27] and DFTBS [28]. And for the analyses of RNA regulatory elements, position specific weight matrices were obtained from RegRNA [29]. These weight matrices were compared to the weight matrices obtained through the method described by Wasserman and Sandelin (2004) [30]and Sandelin et al. (2004) [31]. Matching results were then validated using the MELINA server [32] for DNA motif identification. Following the identification of the motifs a multiple sequence alignment was constructed using the UGENE tool [33] and the motifs were mapped to the conserved positions. Figure 1
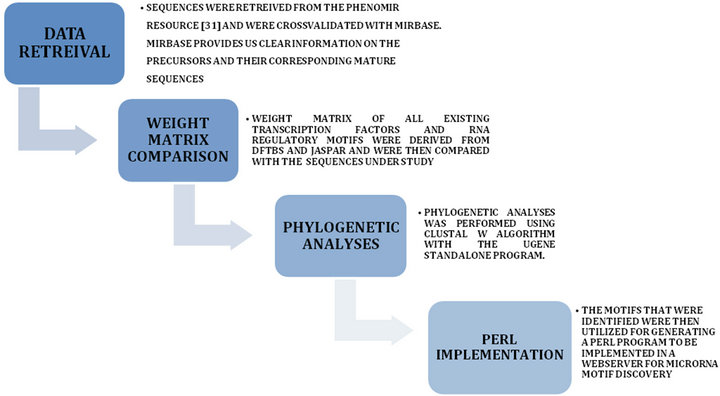
Figure 1. The Analyses Pipeline.
represents the workflow for the analyses.
3. RESULTS AND DISCUSSION
The results of the analyses clearly indicate the presence of regulatory element signatures which are cis acting in nature in the complimentary DNA sequences of the precursors and when RNA sequence regulatory elements were analyzed a large number of such elements were identified in those precursor sequences as well. This leads us to comment that our understanding of the myriad mechanisms controlling the biogenesis of microRNAs is still in its nascent stage as each microRNA family may possess specific modular regulatory elements in both the DNA and RNA levels indicative of a failsafe loop which makes the biogenesis specific and reduces the error rate in the production of the mature functional microRNAs. We have earlier identified secondary structural motifs in the precursors of microRNAs [34-39] and have correlated them with their minimum free energy variations.
4. DNA REGULATORY MOTIFS
It is interesting to note that the occurrence of the sequence signatures are not restricted to any specific type of secondary structural motif apart from the ComA motif which is observed to occur predominantly in loop regions (Figure 2). A large number of the splicing regulatory motifs were identified in the RNA sequence motif discovery pipeline (Table 2). Transcription factor binding sites [TFBS] are generally defined as binding sites on DNA which interact with specific transcription factors and regulate nearby genes. In our study we identified three conserved motifs in all the sequences of the dataset (Figure 3). The presence of most of these motifs are significant as FNR protein is a fumarate and nitrate reductase protein in the prokaryotic system while it shares homology with the eukaryotic cytochrome p450s. HrcA is a repressor protein which controls gene expression as a part of the heat shock response [28] and shares its helix loop helix DNA binding motif with eukaryotic proteins while the GlnR protein which shares homology with numerous eukaryotic Helix turn helix proteins. It is to be noted further that the presence of these regulatory motifs were conserved in miRNAs which were differentially regulated under disease conditions (Phenomir DB; Figure 3).
5. RNA REGULATORY MOTIFS
Several workers have reported that alternative splicing is a conserved regulatory phenomenon leading to neural protein production. Licatalosi and Darnell (2006) [40] have stated that the phenomenon of alternative splicing enables “the cell to fine tune its protein composition” which helps it to adapt to the changes associated with different stimuli. Mutations in these splicing regulatory motifs can result in neurologic disorders such as phakomatoses as well as muscular dystrophies. Thus the presence of such motifs indicates that there are possibilities
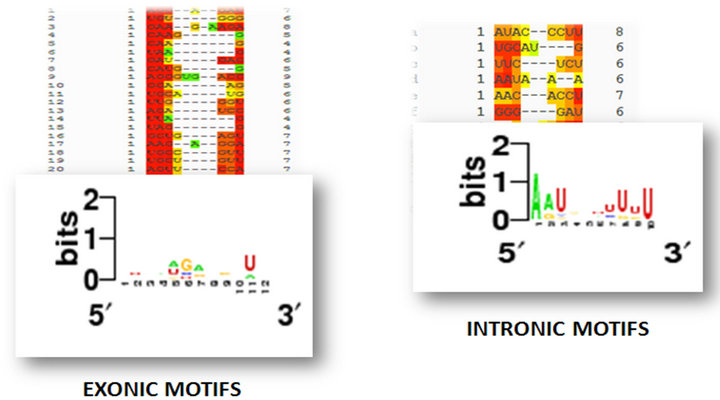
Figure 2. Intronic and Exonic regulatory motifs and their consensus motif structure which indicate phylogenetic conservedness.
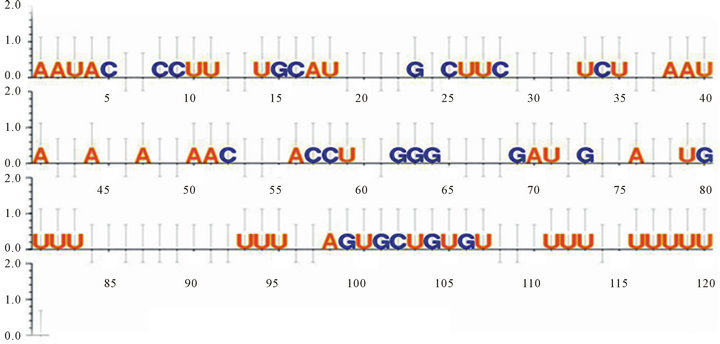
Figure 3. Intronic and Exonic regulatory motifs and their phylogenetically conserved positions as identified through multiple sequence alignment using the UGENE tool.
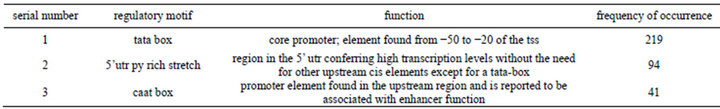
Table 2. Identified DNA regulatory elements that were overrepresented.
that alternative splicing may occur in presursors as well which might explain their ability to produce multiple mature microRNAs (Table 3).
6. PHYLOGENETIC CONSERVEDNESS
Phylogenetically conserved positions (Figures 3-5) identified through multiple sequence alignment and the fact that these conserved positions form intricate part of the regulatory elements further signify the assumption that the regulatory modules are indeed functional and hence have retained their position specificity throughout the events of divergence and occurrence of this particular
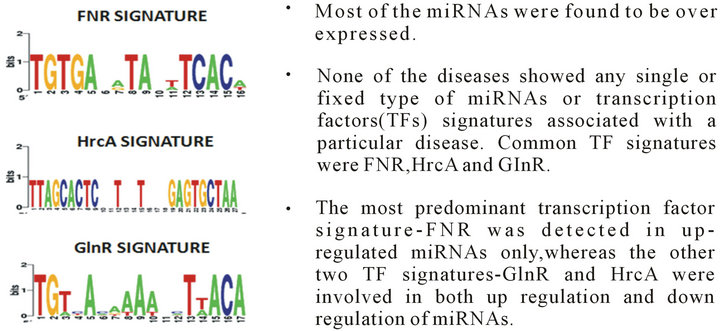
Figure 4. Conserved ubiquitous TF signatures identified in microRNAs implicated in various neurological disorders.
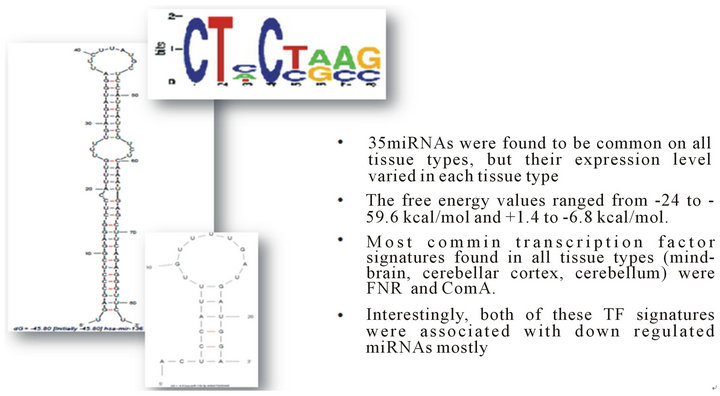
Figure 5. Conserved ubiquitous ComA signatures identified in microRNAs implicated in various neurological disorders and its predominant loop specific occurrence.
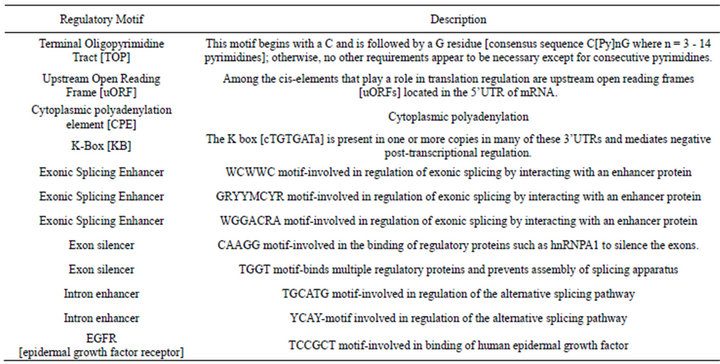
Table 3. Identified RNA regulatory elements that were overrepresented.
family of micro RNA.
7. POSSIBLE SIGNIFICANCE OF MOTIF OCCURRENCE
It is clearly known that upstream DNA regulatory elements enable the proper transcription of the genes and exhibit a modular control; however, the question that remains is why would they remain in clusters in phylogenetically conserved positions in an RNA molecule that is already a product of transcription and has passed through a primary processing step? Still to date information is scanty regarding the actual temporal regulation of many microRNA products and as to how their expression levels are controlled; thus it may be possible that these regulatory motifs serve as binding sites of cellular regulatory factors and enable the regulation of the production of mature microRNAs from their precursors. Our observations are in accordance with those of Piriyapongsa et al. (2011) [41], who have identified numerous transcription factor binding sites in the pre-miRNAs. Song Gao et al. (2010) [42] have also reported the presence of atypical promoter elements in microRNA precursor fragments. Many proteins have been identified which bind to the precursor microRNA and regulate its function viz—the NF90—NF45 complex which binds directly to the stem loop region and may also interact via NF90 to DGCR8. Other proteins such as R-Smads, KSRP, hnRNPA1 and LIN28 have also been observed to bind specific pre miR stem or loop regions. The identification of such conserved elements indicate that there may be many more protein factors or factors with multiple binding ability which may act in unison to regulate the expression of the microRNAs associated with neurological disorders.
8. CONCLUSION
From the obtained results we can safely conclude that a large number of regulatory elements and Transcription factor binding site signatures exist in the precursor sequences of microRNAs implicated in neurological disorders such as Alzheimer’s disease, Parkinson’s disease and Huntington disease. Whether these regulatory sites are cryptic it is difficult to predict computationally however, many cellular proteins do exist which have high affinity towards such conserved signature motifs. Differential expression levels of the various microRNAs implicated in the disorders provide ample evidence that these small regulators are themselves controlled by regulatory interactions much of which still stands unelucidated.
9. ACKNOWLEDGEMENTS
The authors acknowledge the fund provided by the Department of Biotechnology, Government of India for maintenance of the facility.
REFERENCES
- Lee, R.C. and Ambros, V. (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science, 294, 862-864. doi:10.1126/science.1065329
- Hutvagner, G., Simard, M.J., Mello, C.C. and Zamore, P.D. (2004) Sequence-specific inhibition of small RNA function. PloS Biology, 2, E98. doi:10.1371/journal.pbio.0020098
- Bak, M., Silahtaroglu, A., Moller, M., Christensen, M., Rath, M.F., Skryabin, B., Tommerup, N. and Kauppinen, S. (2008) MicroRNA expression in the adult mouse central nervous system. RNA, 14, 432-444. doi:10.1261/rna.783108
- Christensen, M. and Schratt, G.M. (2009) MicroRNA involvement in developmental and functional aspects of the nervous system and in neurological diseases. Neuroscience Letters, 466, 55-62. doi:10.1016/j.neulet.2009.04.043
- Hebert, S.S., Papadopoulou, A.S., Smith, P., Galas, M.C., Planel, E., Silahtaroglu, A.N., Sergeant, N., Buee, L. and De Strooper, B. (2010) Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Human Molecular Genetics, 19, 3959-3969. doi:10.1093/hmg/ddq311
- Hebert, S.S., Horre, K., Nicolai, L., Papadopoulou, A.S., Mandemakers, W., Silahtaroglu, A.N., et al. (2008) Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimers disease correlates with increased BACE1/beta-secretase expression. Proceedings of the National Academy of Sciences of the United States of America, 105, 6415- 6420. doi:10.1073/pnas.0710263105
- Hebert, S.S., Horre, K., Nicolai, L., Bergmans, B., Papadopoulou, A.S., Delacourte, A. and De Strooper, B. (2009) MicroRNA regulation of Alzheimer’s amyloid precursor protein expression. Neurobiology of Disease, 33, 422-428. doi:10.1016/j.nbd.2008.11.009
- Hebert, S.S. and De Strooper, B. (2009) Alterations of the microRNA network cause neurodegenerative disease. Trends in Neurosciences, 32, 199-206. doi:10.1016/j.tins.2008.12.003
- Johnson, R., Zuccato, C., Belyaev, N.D., Guest, D.J., Cattaneo, E. and Buckley, N.J. (2008) A microRNAbased gene dysregulation pathway in Huntington’s disease. Neurobiology of Disease, 29, 438-445. doi:10.1016/j.nbd.2007.11.001
- Junn, E., Lee, K.W., Jeong, B.S., Chan, T.W., Im, J.Y. and Mouradian, M.M. (2009) Repression of alphasynuclein expression and toxicity by microRNA-7. Proceedings of the National Academy of Sciences of the United States of America, 106, 13052-13057. doi:10.1073/pnas.0906277106
- Junn, E., Lee, K.W., Jeong, B.S., Chan, T.W., Im, J.Y. and Mouradian, M.M. (2009) Repression of alphasynuclein expression and toxicity by microRNA-7. Proceedings of the National Academy of Sciences of the United States of America, 106, 13052-13057. doi:10.1073/pnas.0906277106
- Kim, J., Inoue, K., Ishii, J., Vanti, W.B., Voronov, S.V., Murchison, E., Hannon, G. and Abeliovich, A. (2007) A MicroRNA feedback circuit in midbrain dopamine neurons. Science, 317, 1220-1224. doi:10.1126/science.1140481
- Kim, J., Inoue, K., Ishii, J., Vanti, W.B., Voronov, S.V., Murchison, E., et al. (2007) A MicroRNA feedback circuit in midbrain dopamine neurons. Science, 317, 1220- 1224. doi:10.1126/science.1140481
- Kosik, K.S. (2006) The neuronal microRNA system. Nature Reviews Neuroscience, 7, 911-920. doi:10.1038/nrn2037
- Krichevsky, A.M. (2007) MicroRNA profiling: From dark matter to white matter, or identifying new players in neurobiology. Science World Journal, 7, 155-166. doi:10.1100/tsw.2007.201
- Lukiw, W.J. (2007) Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport, 18, 297-300. doi:10.1097/WNR.0b013e3280148e8b
- Lukiw, W.J., Zhao, Y. and Cui, J.G. (2008) An NFkappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. The Journal of Biological Chemistry, 283, 31315-31322. doi:10.1074/jbc.M805371200
- Miska, E.A., Alvarez-Saavedra, E., Townsend, M., Yoshii, A., Sestan, N., Rakic, P., Constantine-Paton, M. and Horvitz, H.R. (2004) Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biology, 5, R68. doi:10.1186/gb-2004-5-9-r68
- Boissonneault, V., Plante, I., Rivest, S. and Provost, P. (2009) MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor proteinconverting enzyme 1. The Journal of Biological Chemistry, 284, 1971-1981. doi:10.1074/jbc.M807530200
- Sethi, P. and Lukiw, W.J. (2009) Micro-RNA abundance and stability in human brain: Specific alterations in Alzheimer’s disease temporal lobe neocortex. Neuroscience Letters, 459, 100-104. doi:10.1016/j.neulet.2009.04.052
- Wang, W.X., Rajeev, B.W., Stromberg, A.J., Ren, N., Tang, G., Huang, Q., Rigoutsos, I. and Nelson, P.T. (2008) The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. The Journal of Neuroscience, 28, 1213-1223. doi:10.1523/JNEUROSCI.5065-07.2008
- Sempere, L.F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E. and Ambros, V. (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biology, 5, R13. doi:10.1186/gb-2004-5-3-r13
- Smith, B., Treadwell, J., Zhang, D., Ly, D., McKinnell, I., Walker, P.R. and Sikorska, M. (2010) Large-scale expression analysis reveals distinct microRNA profiles at different stages of human neurodevelopment. PLoS One, 5, e11109. doi:10.1371/journal.pone.0011109
- Vilardo, E., Barbato, C., Ciotti, M., Cogoni, C. and Ruberti, F. (2010) MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. The Journal of Biological Chemistry, 285, 18344-18351. doi:10.1074/jbc.M110.112664
- Wang, G., van der Walt, J.M., Mayhew, G., Li, Y.J., Zuchner, S., Scott, W.K., et al. (2008) Variation in the miRNA- 433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. The American Journal of Human Genetics, 82, 283-289. doi:10.1016/j.ajhg.2007.09.021
- Packer, A.N., Xing, Y, Harper, S.Q., Jones, L., Davidson, B.L. (2008) The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. The Journal of Neuroscience, 28, 14341-14346. doi:10.1523/JNEUROSCI.2390-08.2008
- Bryne, J.C., Valen, E., Tang, M.H., Marstrand, T., Winther, O., da Piedade, I., Krogh, A., Lenhard, B., Sandelin, A. (2008) JASPAR, the open access database of transcription factor-binding profiles: New content and tools in the 2008 update. Nucleic Acids Research, 36, 102-106.
- Sierro, N., Makita, Y., de Hoon, M.J.L. and Nakai, K. (2008) DBTBS: A database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Research, 36, 93-96. doi:10.1093/nar/gkm910
- Huang, Y.H., Chien, C.H., Jen, K.H. and Huang H.D. (2006) RegRNA: A regulatory RNA motifs and elements finder. Nucleic Acids Research, 34, 429-434.
- Wasserman, W.W. and Sandelin, A. (2004) Applied bioinformatics for the identification of regulatory elements. Nature Reviews Genetics, 5, 276-287.
- Sandelin, A. and Wasserman, W.W. (2004) Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. Journal of Molecular Biology, 338, 7-15. doi:10.1016/j.jmb.2004.02.048
- Okumura, T., Makiguchi, H., Makita, Y., Yamashita, R. and Nakai, K. (2007) Melina II: A web tool for comparisons among several predictive algorithms to find potential motifs from promoter regions. Nucleic Acids Research, 35, 227-231.
- Okonechnikov, K., Golosova, O. and Fursov, M. (the UGENE team) (2012) Unipro UGENE: A unified bioinformatics toolkit. doi:10.1093/bioinformatics/bts091
- Adhikary, M., Ganguli, S., Das, G.S. and Datta, A. (2011) Secondary structural analyses of micro RNAs and their precursors in Pan troglodytes. International Journal of Computational Biology, 2, 35-37.
- Das, K.A., Ganguli, S., Gupta, S. and Datta, A. (2011) Secondary structural analysis of microRNA and their precursors in plants. International Journal of Agriculture Sciences, 3, 62-64.
- Ganguli, S., Dey, K.S., Dhar, P., Basu, P., Roy, P. and Datta, A. (2010) Catalytic RNA world relics in Dicer RNAs. International Journal of Genetics, 2, 8-17.
- Ganguli, S., De, M. and Datta, A. (2011) Analyses of Argonaute—MicroRNA interactions in Zea mays. International Journal of Computational Biology, 2, 32-34.
- Ganguli, S. and Datta, A. (2011) RNAi—Interactomics and therapeutics. In: Gupta, V.K., et al., RNAi Technology, Chapter 19, CRC Press, Taylor & Francis Group, Boca Raton, 347-356.
- Ganguli, S., Mitra, S. and Datta, A. (2011) Antagomirbase: A putative antagomir database. Bioinformation, 7, 41-43. doi:10.6026/97320630007041
- Licatalosi, D.D. and Darnell, B.R. (2006) Splicing regulation in neurologic disease. Neuron, 52, 93-101. doi:10.1016/j.neuron.2006.09.017
- Piriyapongsa, J., Jordan, K.I., Conley, B.A., Ronan, T., and Smalheiser, R.N. (2011) Transcription factor binding sites are highly enriched within microRNA precursor sequences. Biology Direct, 6, 61.
- Song Gao, J., Zhang, Y., Li, M., Tucker, L.D., Machan, J.T., Quesenberry, P., Rigoutsos, I. and Ramratnam, B. (2010) Atypical transcription of microRNA gene fragments. Nucleic Acids Research, 38, 2775-2787.
NOTES
*Corresponding author.

