Journal of Diabetes Mellitus
Vol.3 No.4(2013), Article ID:38023,12 pages DOI:10.4236/jdm.2013.34027
Hyperglycemia promotes overexpression of SR-BII isoform of the scavenger receptor class B type I in type 2 diabetes mellitus: A study in Juana Koslay City, San Luis, Argentina
![]()
Laboratory of Diabetes, Department of Biochemistry and Biological Sciences, School of Chemistry, Biochemistry and Pharmacy, National University of San Luis, San Luis, Argentina; *Corresponding Author: msojeda@unsl.edu.ar
Copyright © 2013 Gisela V. Mendoza et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 10 August 2013; revised 10 September 2013; accepted 19 September 2013
Keywords: Type 2 Diabetes Mellitus; Hyperglycemia; Scavenger Receptor Class B Type I and Polymorphism
ABSTRACT
The scavenger receptor class B type I (SR-BI) is a high-density lipoprotein (HDL) receptor involved in reverse cholesterol transport. Some studies reported the association to be stronger in the presence of diabetes. The full length gene encoding SR-BI is comprised in 13 exons that are alternatively spliced to produce two major transcripts: the full length SR-BI and the splice variant SR-BII, in which exon 12 is skipped. Considering that type 2 diabetes status is characterized by changes in the concentration of plasma lipids, modifications in lipoprotein size and composition, which may be important modulators of the SR-BI expression; the aims of the study were to examine the influence of SR-BI polymorphism (rs838895) on lipid profile and SR-BI mRNA expression in a population of diabetic patients living in Juana Koslay City. Blood samples were drawn from controls (n = 40) and Type 2 diabetic patients (n = 66) and DNA and total RNA were obtained. SR-BI mRNA expression was measured by RT-PCR and SR-BI polymorphism was detected by Tetra Primer ARMSPCR. Compared to controls, diabetic patients had higher fasting serum glucose, glycated hemoglobin, triglycerides, total cholesterol, lowdensity lipoprotein cholesterol, and lower highdensity lipoprotein cholesterol. SR-BI mRNA expression was lower in T2DM when compared to controls, suggesting that the hyperglycemia presents in T2DM patients down-regulates SR-BI mRNA expression. Interestingly, we found that decreased SR-BI expression resulted in markedly increased plasma LDL concentrations in T2DM subjects, and the overexpression of SRBII isoform is responsible for the markedly increased plasma LDL-c concentrations. The polymorphism (rs838895) did not modify the mRNA level of SR-BI in leucocytes from control and diabetic patients. This study provides novel evidence suggesting that hyperglycemia may affect reverse cholesterol transport by controlling SRBI expression in diabetic patients. LDL cholesterol levels are associated with low SR-BI mRNA expression in T2DM.
1. INTRODUCTION
Type 2 diabetes mellitus (T2DM) has emerged as one of the most prevalent chronic diseases worldwide. Decreased plasma high-density lipoprotein cholesterol (HDLc) is one of the most common lipid disorders in diabetic subjects [1-3]. Moreover, low HDL-c concentrations have been defined as a key sign of the insulin resistance syndrome, a common metabolic condition linked to a higher cardiovascular risk, which has been estimated to be present in one fourth of all Americans [4,5]. HDL-c is believed to exert its antiatherogenic role mainly through the process of delivering cholesterol from peripheral tissues back to the liver for removal from the body, which is called reverse cholesterol transport [6,7].
Scavenger receptor class B, type I (SR-BI), a major HDL receptor [8,9], plays an important role in reverse cholesterol transport, a major pathway for the clearance of excess cholesterol from the body. In this process, peripheral cholesterol is packaged into HDL from which it is subsequently removed in the liver and excreted into bile. SR-BI mediates the uptake and biliary secretion of HDL-c by the liver [10,11].
Besides its role as a functional HDL receptor involved in reverse cholesterol transport, SR-BI also participates in the metabolism of Apolipoprotein B-containing lipoproteins, including low density lipoprotein (LDL) [12,13] and very low density lipoprotein (VLDL) [14,15].
The full length gene encoding SR-BI (gene symbol SCARB1) is comprised of 13 exons that are alternatively spliced to produce two major transcripts: the full length SR-BI and the splice variant SR-BII, in which exon 12 is skipped. SR-BI and SR-BII splice forms, conserved in both mouse and rat genomes, have different tissue distributions and may influence cellular cholesterol trafficking and homeostasis in different ways [16]. SR-BII is reported to be a minor splice variant in human liver and has shown to be less efficient at reverse cholesterol transport [17].
Studies using animal models have demonstrated that SR-BI has an atheroprotective role. SR-BI overexpression in the liver is associated with reduction of atherosclerosis in LDL receptor-deficient mice under a high fat and cholesterol diet [18]. On the other hand, SR-BI deficiency results in lipid deposition in the aorta and atherosclerosis in mice [19]. In contrast, much less is known about the role of SR-BI in humans.
Genome-wide association studies have uncovered a number of loci associated with type 2 diabetes, but together they account for little of the overall variance and provide little predictive power over traditional risk factors [20]. It is likely that many loci still remain to be discovered. Genes located in chromosomal regions showing linkage to type-2 diabetes in family-based studies are rational candidates for more detailed investigation. SCARB1 lies in a region on chromosome 12q24 that has been linked to type-2 diabetes [21] and more recently to abdominal obesity, a risk factor for type-2 diabetes, in three studies [22-24]. Acton et al. [25] were the first to identify single nucleotide polymorphisms (SNPs) of the SCARB1 in a white European population and associated some of these common variants with plasma lipid levels and body mass index. Genetic variation in the SCARB1 has also been associated with increased risk of coronary artery disease [26], obesity [25], triglycerides [27,28], and HDL-c [29-33], all facets of the metabolic syndrome. Furthermore, there is evidence that diabetes status may modify the SCARB1 association with HDL-c [34].
A poorly studied polymorphism is intron 11 (IVS11) SCARB1 where G is changed by C (rs838896). ChibaFalek et al. [35] showed that in liver tissue of young adult females (but not men) genotypes that carried the mutant allele (C) were associated with generally lower level of SR-BI, a non-significant lower level of SR-BII and a lower level of overall SCARB1, compared to young women not carrying this allele.
Considering that type 2 diabetes status is characterized by changes in the concentration of plasma lipids, modifications in lipoprotein size, and composition [2,36,37] that may be important modulators of the SCARB1 expression; the aims of the current work were: 1) to assess the phenotypic effect on lipid and lipoprotein concentrations of the genetic variant (rs838896) at SCARB1 in T2DM patients who reside in Juana Koslay, San Luis, Argentina, 2) to examine the association of the genetic variation at the SCARB1 locus with the type 2 diabetes status in these patients, 3) to estimate whether diabetes status modifies the association between the SCARB1 variation and the examined phenotypic traits, and 4) to examine the influence of this genetic variant (rs838896) on mRNA expression in peripheral leucocytes.
2. RESEARCH DESIGN AND METHODS
2.1. Subjects
The present study was carried out in accordance to the guidelines of the Helsinki Declaration. A total of 106 volunteers (66 patients with type 2 diabetes and 40 healthy age-matched controls) participated in this investigation. Criteria published by the American Diabetes Association were used to diagnose T2DM [38]. These patients reside in Juana Koslay, San Luis, Argentina. The protocol for this study was approved by the local Institutional Review Board, and a written informed consent was obtained from each patient to be enrolled. During an initial interview with each patient, they were asked for diseases, medication and smoking histories. Exclusion criteria included liver, kidney and thyroid diseases, as well as the use of anti-lipemic drugs.
2.2. Anthropometric and Clinical Data
For each subject enrolled, height (meters) and weight (Kg) measurements were acquired. Height and weight were measured to the nearest 0.5 cm and 0.1 kg, respectively. The body mass index (BMI) was calculated as weight divided by height squared (Kg/m2).
2.3. Blood Sampling
Venous blood samples for glucose, lipid, and lipoprotein analysis were collected into EDTA-containing (1 g/liter) tubes from all subjects after a 12-h overnight fast at the beginning of the study. Blood was also collected to extract DNA and RNA (see below).
2.4. Biochemical Measurement
Fasting plasma glucose (FPG) was measured by using a glucose oxidase method with a commercial enzymatic kit (Wiener Lab, Rosario, Argentina) and Glycated hemoglobin (HbA1c) concentration was measured with a coupled ionic-exchange chromatography/spectrophotometric assay (BioSystems, Barcelona, Spain) in a Bayer Express Plus Chemistry Analyzer (Bayer Diagnostics, Siemens, Germany). Total cholesterol (TC), triglycerides (TG) and HDL-c concentrations were measured using commercial kits by following manufacturer’s instructions ((Wiener Lab, Rosario, Argentina) in a Bayer Express Plus Chemistry Analyzer (Bayer Diagnostics, Siemens, Germany). Low density lipoprotein-cholesterol (LDL-c) was calculated with the Friedewald formula: LDL-c= total cholesterol (mg/dL) – HDL-c (mg/dL) – triglycerides (mg/dL)/5 [39].
2.5. Genotyping and SNP Analysis
Genomic DNA was isolated from diabetic patients and healthy volunteers using conventional protocol by Qiagen kits (Qiagen, Inc., Valencia, CA). DNA concentration was detected by UV-VIS spectroscopy and diluted to a final concentration of 20 ng/μL. The SCARB1 polymorphism (rs838896) was analyzed by Tetra Primer AMRSPCR [40]. Two pairs of primers were used, one which amplifies a fragment of 431 bp, common to both alleles (outer primers: forward 5’ GTAGATAGTGAGCTTTGACATGGAGGCCAG 3’ and reverse 5’ ACTTCCGTTCCCTCCTGATAGCAGCT 3’) and another pair specific for the SNP (inner primers): forward 5’ TTGGAACTGATTCTGGGAGTGACAGTGG 3’ for the G allele and reverse 5’ GATTCCCCTCTTTAAAAGCCTCCAAAGG 3’ for the C allele. Nucleotide sequence and SNP details were obtained from NCBI website (http://www.ncbi.nlm. nih.gob). Primers were designed ‘‘in silico’’ in a free access web (http://cedar.genetics.soton.ac.uk) and then checked for specificity (http://blast.ncbi.nlm.nih.gov/ Blast.cgi). Each PCR reaction was performed containing 200 ng of genomic DNA, 1 pmol of each outer primer and 5 pmol of each inner primer (1:5 ratio of outer to inner primer concentration) , 200 μM dNTPs, 2.5 mM MgCl2, 1X buffer, and 1 unit of AmpliTaq Gold DNA polymerase (Perkin-Elmer). PCRs were carried out in 35 μL reactions. The template DNA was denatured for 3 minutes at 95˚C before undergoing 35 cycles of denaturation for 1 minute at 95˚C, primer annealing for 1 minute at 68˚C, and extension for 1 minute at 72˚C, and final extension at 72˚C for 3 minutes. The resultant products obtained after PCR were separated by electrophoresis on 2.5% agarose gel containing GelRed. This resulted in 3 DNA fragments of: 431 bp common to both alleles (outer primers amplification), 292 bp for the C allele and 195 bp for the G allele. The image was visualized and photographed under UV transillumination. Allele frequencies for this SNP were calculated by allele counting. Randomly selected 20% of samples were re-genotyped for cross validating initial genotypes. In case of unclear genotyping results, the samples were repeated again in duplicates until clear genotype was available. Unclear genotyping results, even after repetition was excluded from the study. No genotyping error was observed during cross validation.
2.6. Measurement of Gene Expression
Total RNA was isolated from leucocytes using the TRIzol reagent following manufacturer’s instructions (Life Technologies, Carlsbad, CA). Agarose gel electrophoresis and GelRed staining confirmed the purity and integrity of isolated RNA. RNA concentration was assessed by spectrophotometric measurements at 260/280 nm. Then, ten micrograms of total RNA were reversetranscribed with 200 units of Moloney-murine leukemia virus (MMLV) reverse transcriptase (Promega) using random hexamers as primers in 20-μL reaction mixture following the instructions provided by the manufacturer. The reverse transcription reactions were performed using a GeneAmp PCR system 2400 (PerkinElmer, Wellesley, MA) with the following conditions: 65˚C for 10 min, 37˚C for 60 min, and 90˚C for 5 min.
Given the existence of alternative splice forms of SCARB1 involving inclusion/exclusion of exon 12, primers able to detect both isoforms were used. Primers were constructed from the published cDNA (http://www. ncbi.nlm.nih.gob) sequence of SCARB1 (Forward 5’ CTGCGTCCTGCTGCTGGTCC 3’ and Reverse 5’ GGCTCACGGTGTCCTCAGGA 3’) and the house-keeping gene beta-Globin (Forward 5’ CAACTTCATCCACGTTCACC 3’ and Reverse 5’ GAAGAGCCAAGGACAGGTAC 3’), used as internal control. SCARB1 primers were designed to generate a 197 bp fragment for the SR-BI transcript (exon 12 including) and a 68 bp fragment for the SR-BII transcript (exclusion of exon 12) (Figure 1).
PCR was used to quantify human overall SCARB1 products (SR-BI and SR-BII isoforms) and beta-Globin expression from the cDNA samples. PCRs were carried out in 35 μL reactions. Each amplification reaction included 20 ng of cDNA, 20 pmol of each PCR primer and 1 unit of AmpliTaq Gold DNA polymerase (Perkin-Elmer,
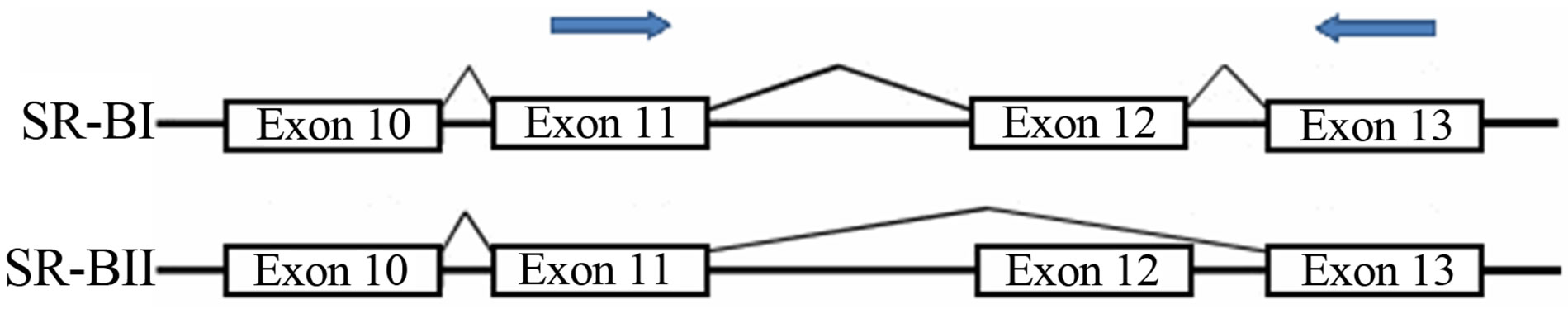
Figure 1. Schematic illustration of the location of primers (arrows) used for RT-PCR.
Waltham, MA). These reactions were performed in a buffer 1X containing 1.5 mmol/L MgCl2, 50 mmol/L KCl, 20 mmol/L Tris-HCl (pH 8.4), and 200 pmol/L of each deoxynucleotide triphosphate. cDNA and control preparations were amplified using the following conditions: 95˚C for 3 minutes followed by 35 cycles of: denaturation for 1 minute at 95˚C, primer annealing for 1 minute (at 62˚C for SCARB1 and 55˚C for beta-Globin), and extension for 1 minute at 72˚C; followed by a final extension at 72˚C for 3 minutes. We controlled for DNA contamination by running three randomly selected, RNA control samples that were not converted to cDNA and no-cDNA/RNA sample in each plate. No observable amplification was detected. The amplification is predicted to generate a 197 bp fragment for the SR-BI transcript and a 68 bp fragment for the SR-BII transcript. PCR products were separated on a 2.5% agarose gel containing GelRed. The image was visualized and photographed under UV transillumination. The intensity of each band was measured using NIH Scion Image 1.6.3 software and reported as the values of band intensity units.
2.7. Statistical Analysis
Chi square test was used to check adjustment of the data to the Hardy-Weinberg equilibrium and to compare the allelic frequencies between controls and diabetic subjects. Comparison of allele frequencies and genotype distributions between case and control samples were done by Pearson’s chi-square test. To analyze the association between SCARB1 genotypes, clinical and biochemical parameters a Student t-test was used when variables were continuous, whereas a Fisher’s exact test was used for the categorical variables. A p < 0.05 was considered to be statically significant.
3. RESULTS
3.1. Subjects Characteristics
One hundred and six individuals were included in this study, 66 of them were diabetic and 40 were healthy controls. Table 1 shows the demographic characteristics of both groups. 51.4% were women and 48.6% were men, age distribution was not different between the groups with a media age of 56.3 years old. The average value of BMI was 25.3 kg/m2 for the control group and 31.2 kg/m2 for the diabetic one. There was a significant difference in weight and BMI between both groups. Diabetic female subjects were more obese than diabetic male subjects.
As shown in Table 2 FPG and HbA1c concentrations were higher in the diabetic group when compared with age-matched control subjects. Total cholesterol, TG and LDL-c were increased in the diabetic group, while HDL-c was significantly lower in these patients, when
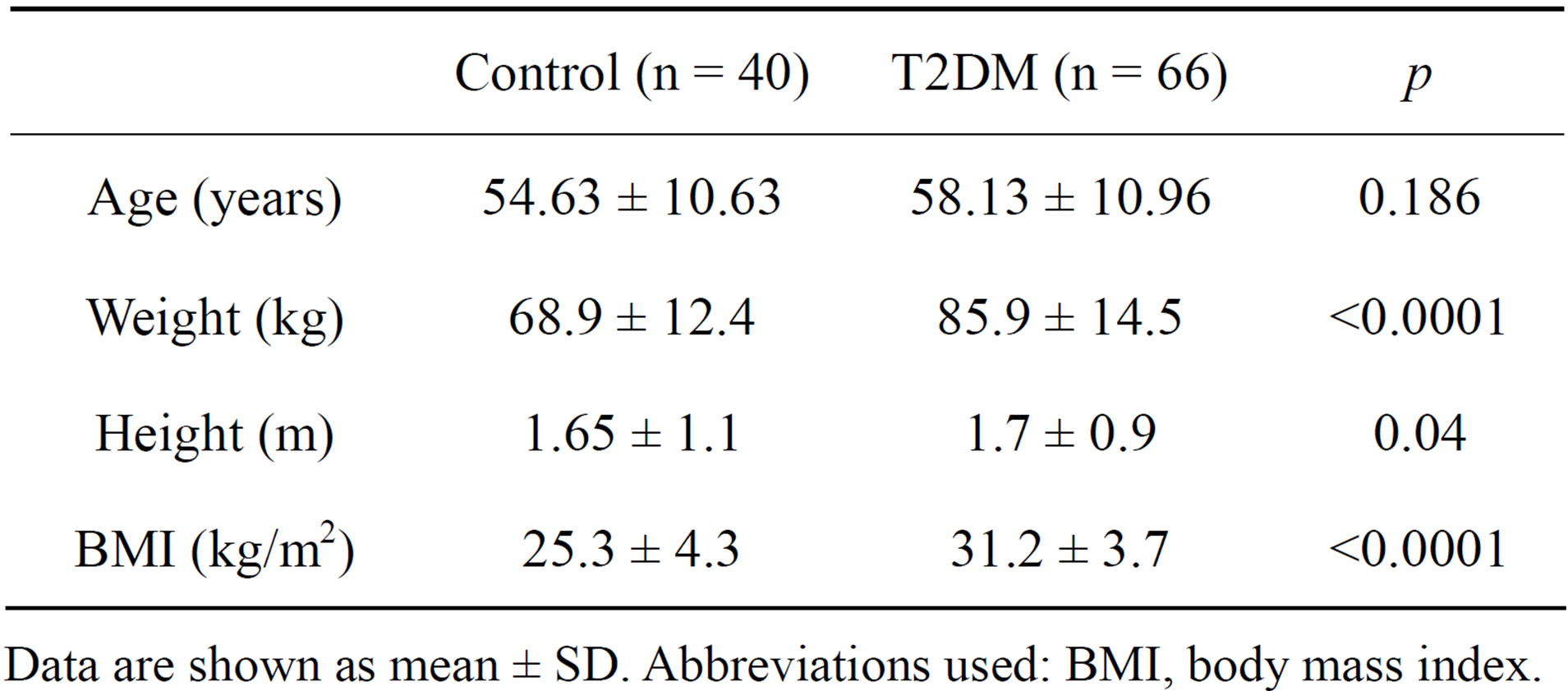
Table 1. Anthropometric characteristics in control and T2DM patients.
Data are shown as mean ± SD. Abbreviations used: BMI, body mass index.
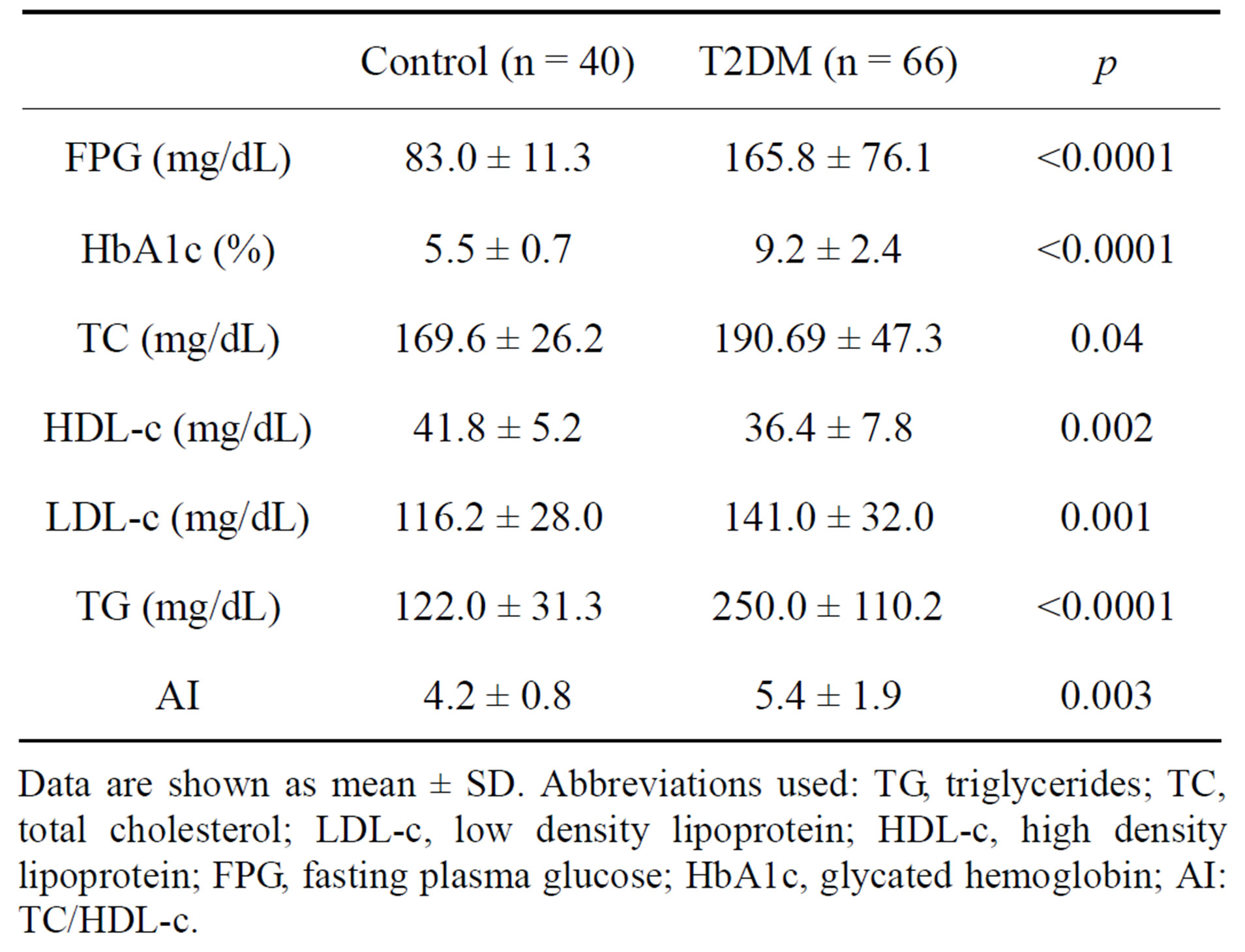
Table 2. Biochemical characteristics in control and T2DM patients.
Data are shown as mean ± SD. Abbreviations used: TG, triglycerides; TC, total cholesterol; LDL-c, low density lipoprotein; HDL-c, high density lipoprotein; FPG, fasting plasma glucose; HbA1c, glycated hemoglobin; AI: TC/HDL-c.
compared to controls. LDL-c was lower in diabetic female than in diabetic male subjects. The atherogenic index (AI) was higher in the diabetic patients, what suggests a risk for cardiovascular diseases in these patients.
3.2. Genotype Frequencies
During the amplification reaction, a 431 bp region of SCARB1 was amplified with Outer forward primer and Outer reverse primer that served as internal control for the quality of the PCR amplification. The allele-specific amplification (Forward inner/Reverse outer and Forward outer/Reverse inner) resulted in the 195 bp PCR product specific for the G allele and the 292 bp PCR products specific for the C allele (Figure 2).
Table 3 shows the absolute number and frequency (%) of controls and T2DM patients having each genotype. The distribution of genotypes was as expected from the Hardy-Weinberg equilibrium because no statistically significant differences (p > 0.05) were detected between the observed and the expected frequencies in any case. We did find statistically significant differences between diabetes status and controls in the genotype and allele frequencies of the SCARB1 variants. The frequency of the C allele was significantly higher in diabetic subjects than that in controls (0.5 vs. 0.30), and this SNP showed strong association with T2DM [OR = 2.429 with 95% CI (1.359 - 4.339), p = 0.004].
3.3. Genotype Associations with Lipid Traits
Table 4 shows plasma concentrations of lipids and lipoproteins by the SCARB1 polymorphism in controls and T2DM. Because of the small number of subjects, homozygotes for the minor C allele and heterozygotes were grouped as carriers of the C allele (GC and CC). In controls, no differences in lipids concentrations were
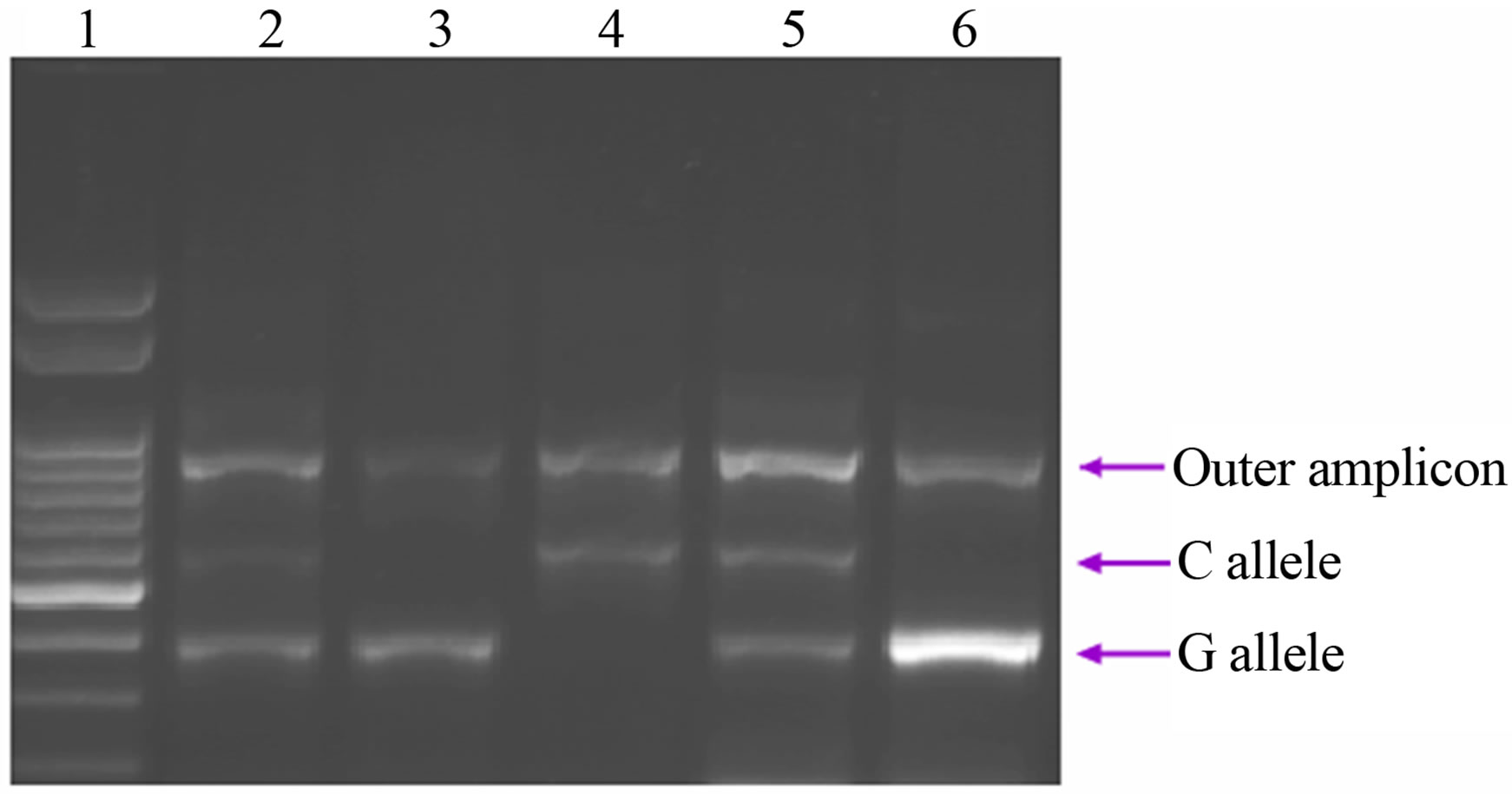
Figure 2. Representative agarose gel of Tetra Primer ARMS-PCR amplification of DNA. Lane 1 shows the molecular weight marker; Lanes 2 and 5 show a typical G/C genotype; Lanes 3 and 6 show a typical G/G genotype; and Lane 4 shows a typical C/C genotype.
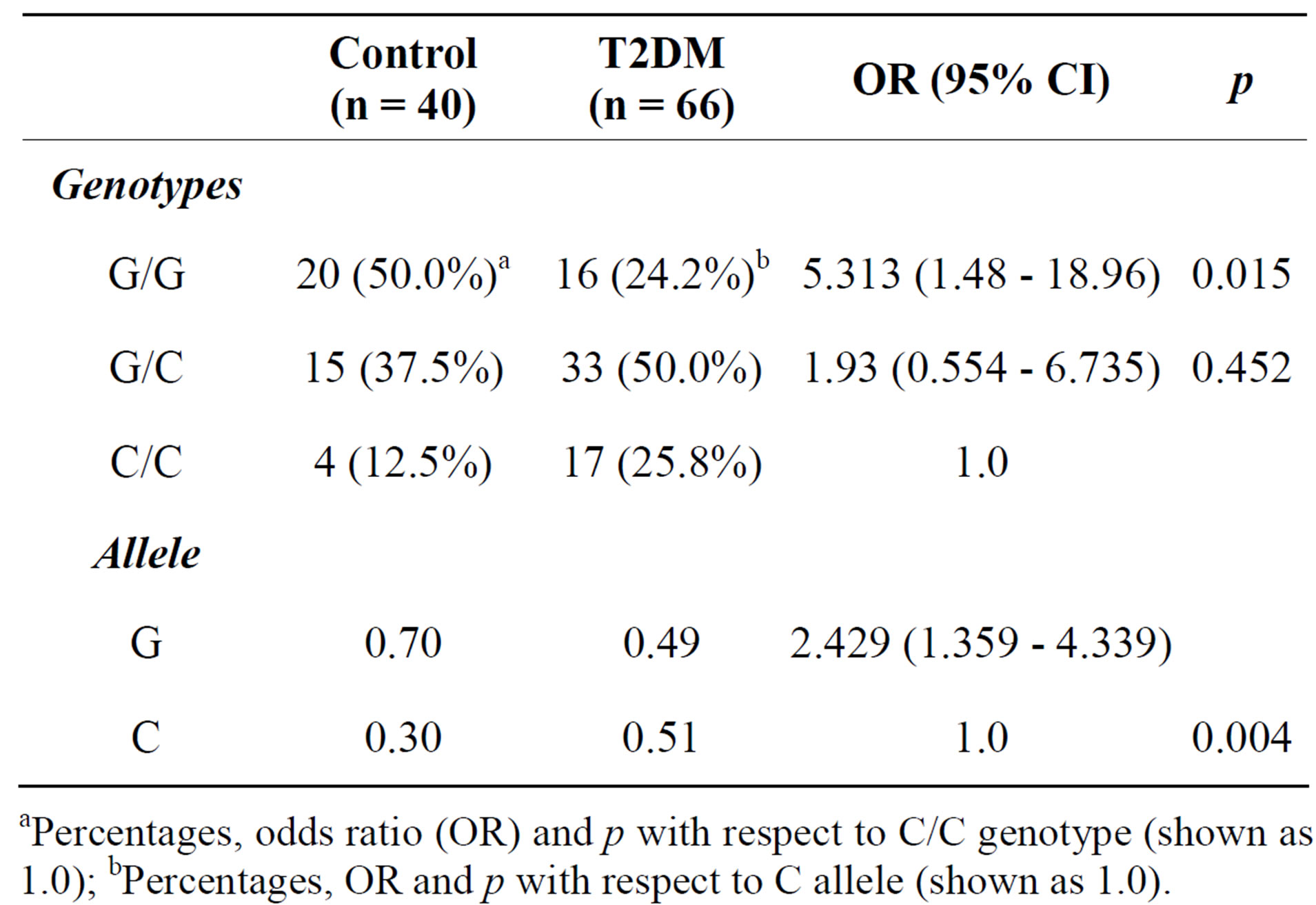
Table 3. Distribution of SCARB1 genotypes and allele frequencies in controls and T2DM patients.
aPercentages, odds ratio (OR) and p with respect to C/C genotype (shown as 1.0); bPercentages, OR and p with respect to C allele (shown as 1.0).
detected in carriers of the C allele of intron 11 polymorphism (rs838896), whereas in diabetics the presence of the C allele was associated to an increase in LDL-c and slight decrease in HDL-c when compared to the GG homocygotes.
3.4. mRNA Expression Levels and Presence of Alternative Splice Variants
Given the existence of alternative splice forms of SCARB1 involving inclusion/exclusion of exon 12, we hypothesized that a SNP located within the associated haplotype block could modulate splicing efficiency. As different isoforms of SCARB1 probably might modulate receptor function, the expression of SCARB1 isoforms was investigated in human leucocytes by RT-PCR using primers designed to detect both SR-BI and SR-BII transcripts. We quantified the amounts of the full-length SR-BI isoform (including exon 12), the alternative SR-BII isoform (skipping exon 12) and overall SCARB1 mRNA (all splice forms) in human leucocytes from 66 T2DM and 40 controls.
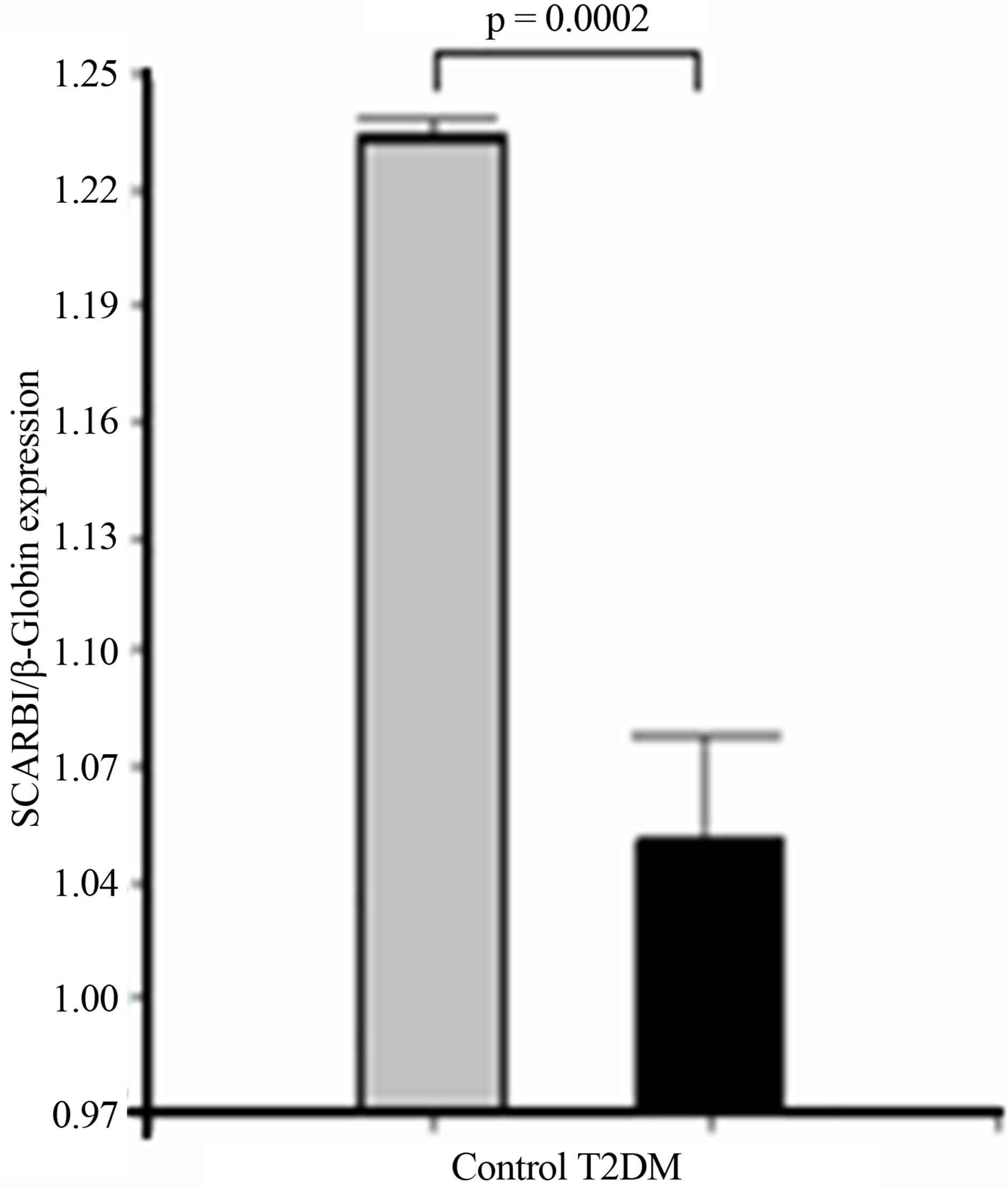
As shown in Figure 3, overall SCARB1 expression was significantly higher in leucocytes from controls when compared to type 2 diabetic patients (p = 0.0002).
Figure 4 shows a comparative graph of the percentage expression based on the total expression of SR-BI and SR-BII in controls and diabetics. In normal subjects the highest percentage of expression corresponds to SR-BI (77% ± 2%) compared to SR-BII (21% ± 3%) showing an extremely significant difference, while in diabetics although there is a trend to increased expression of SR-BI (55% ± 11%) it showed no significant differences in the expression of SR-BII (45% ± 11%). Diabetic subjects showed overexpression of SR-BII isoform and a decresed expression of SR-BI when compared to controls.
In order to determine the functional consequences of SNP (rs838896), we carried out genetic association analysis of the associated SNP and expression of SCARB1. We then compared relative levels of each splice form (ratio to overall SCARB1 expression) in T2DM and controls, and found no differences by genotype. We then compared relative levels of each splice form in men ver-
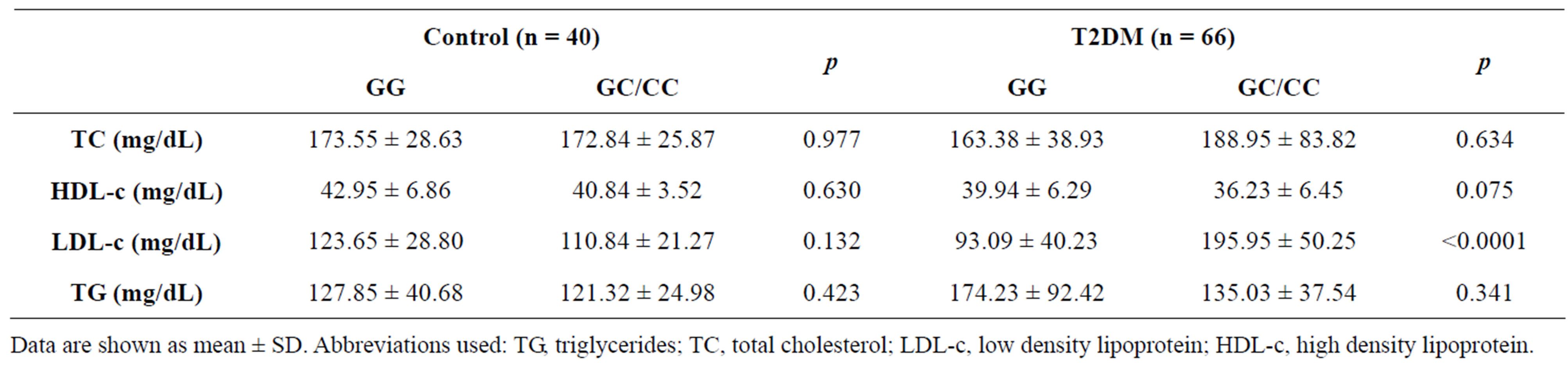
Table 4. Plasma levels of lipids and lipoproteins by the SCARB1 polymorphism in controls and T2DM.
Data are shown as mean ± SD. Abbreviations used: TG, triglycerides; TC, total cholesterol; LDL-c, low density lipoprotein; HDL-c, high density lipoprotein.
 (a)
(a)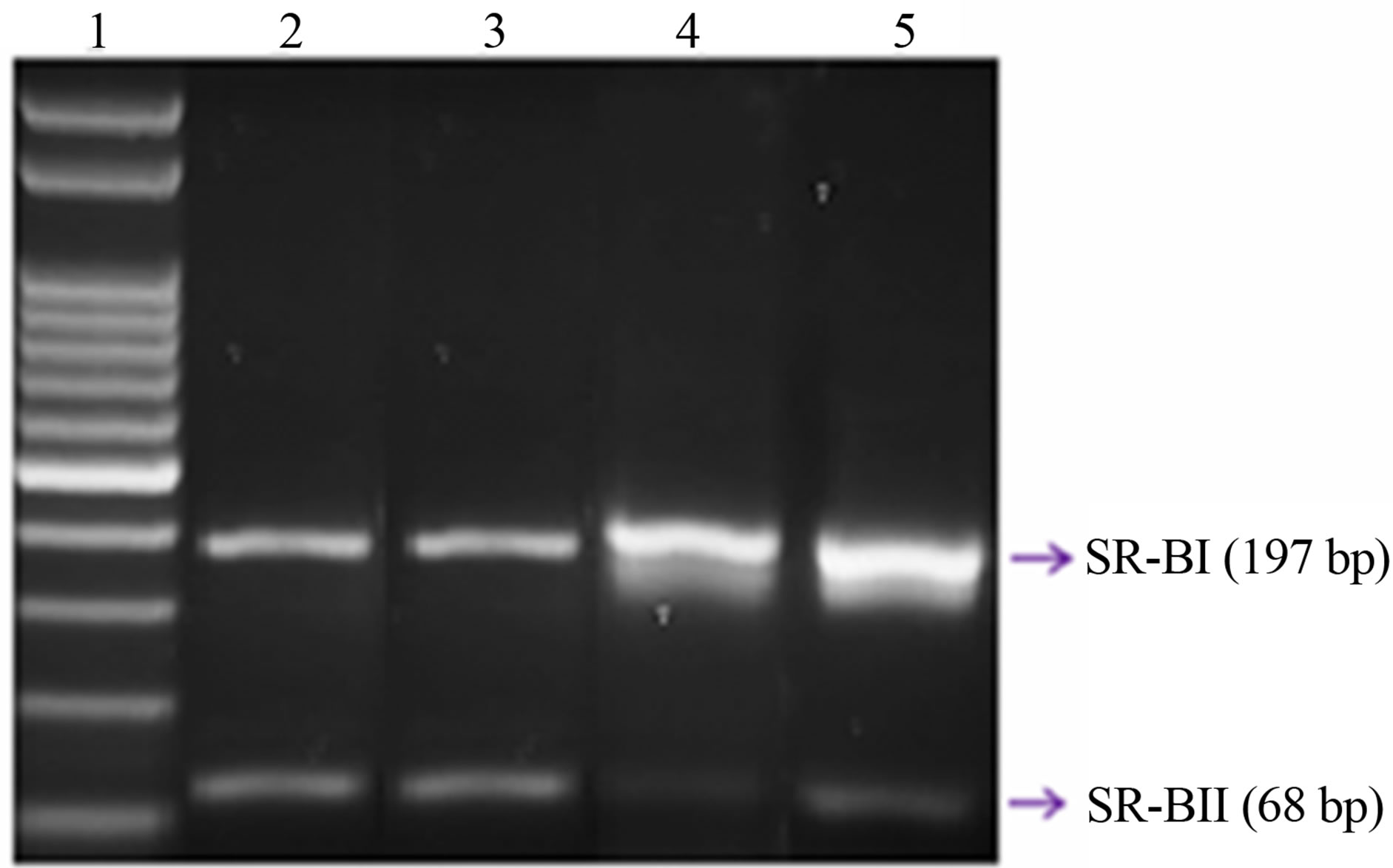 (b)
(b)
Figure 3. Expression of overall SCARB1. (a) The bar graphs represent the densitometric analysis of SCARB1 expression in leucocytes of controls and type 2 diabetic patients; (b) Representative agarose gel: Lane 1 shows the molecular weight marker; Lanes 2 and 3: T2DM; and Lanes 4 and 5: Control. The expression of SCARB1 isoforms was assessed by RT PCR, using specific primers, and it was normalized against the expression of the housekeeping gene beta-Globin, as reported in Material and Methods. Data are expressed as mean ± SEM.
sus women, but no significant associations were found (data not shown).
3.5. Correlation and Lineal Regression Studies
Because SCARB1 mRNA was reduced in the diabetic group, we performed correlations to determine whether this reduction was associated to metabolic factors that

Figure 4. Relative levels of each splice form (ratio to overall SCARB1 expression) in T2DM and controls.
are important for insulin resistance. When the data from all diabetic patients were pooled together, interestingly SCARB1 was negatively correlated to LDL-c levels (r = −0.89, p < 0.0001), FPG (r = −0.944, p = 0.001) and there was a trend to correlate with HDL-c levels (r = −0.29, p = 0.08).
A subsequent linear regression analysis revealed that there was a significant and negative correlation between overall SCARB1 expression and LDL-c levels and the changes of the SR-BI and SR-BII isoforms expression levels in T2DM. Furthermore there was a strong positive correlation among the expression of SR-BII and LDL-c levels and a negative relationship between SR-BI and LDL-c (Figure 5).
4. DISCUSSION
No large population studies examining the association between genetic polymorphisms at the SCARB1 gene and diabetic status have been reported to date. The studies of the SCARB1 gene have been limited to a few candidate SNPs in exon 1, intron 5, and exon 8 [34].
In this study we analyzed the G/C change at intron 11 (rs838896) and confirmed the association of the minor C allele at SCARB1 with type 2 diabetes in an Argentinean small city [OR = 2.429 with 95% CI (1.359 - 4.339), p = 0.004]. Further studies will be needed to evaluate the impact of SCARB1 polymorphism on genetic predisposetion to T2DM in the Argentinean population.
A series of population studies examining the association of polymorphisms in the SR-BI gene with different lipoprotein parameters has suggested that SCARB1 genetic variability plays a significant role in lipoprotein
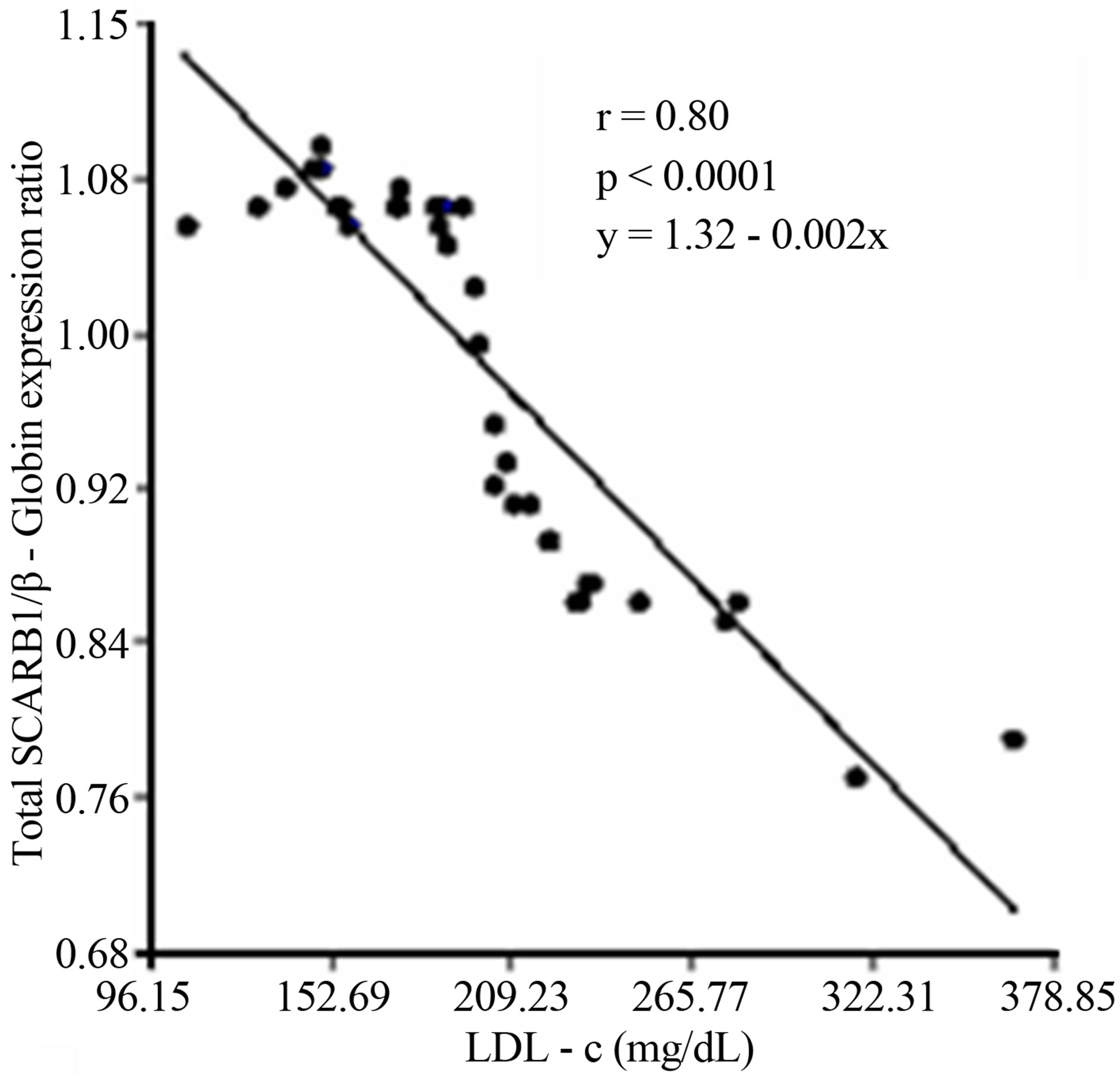 (a)
(a)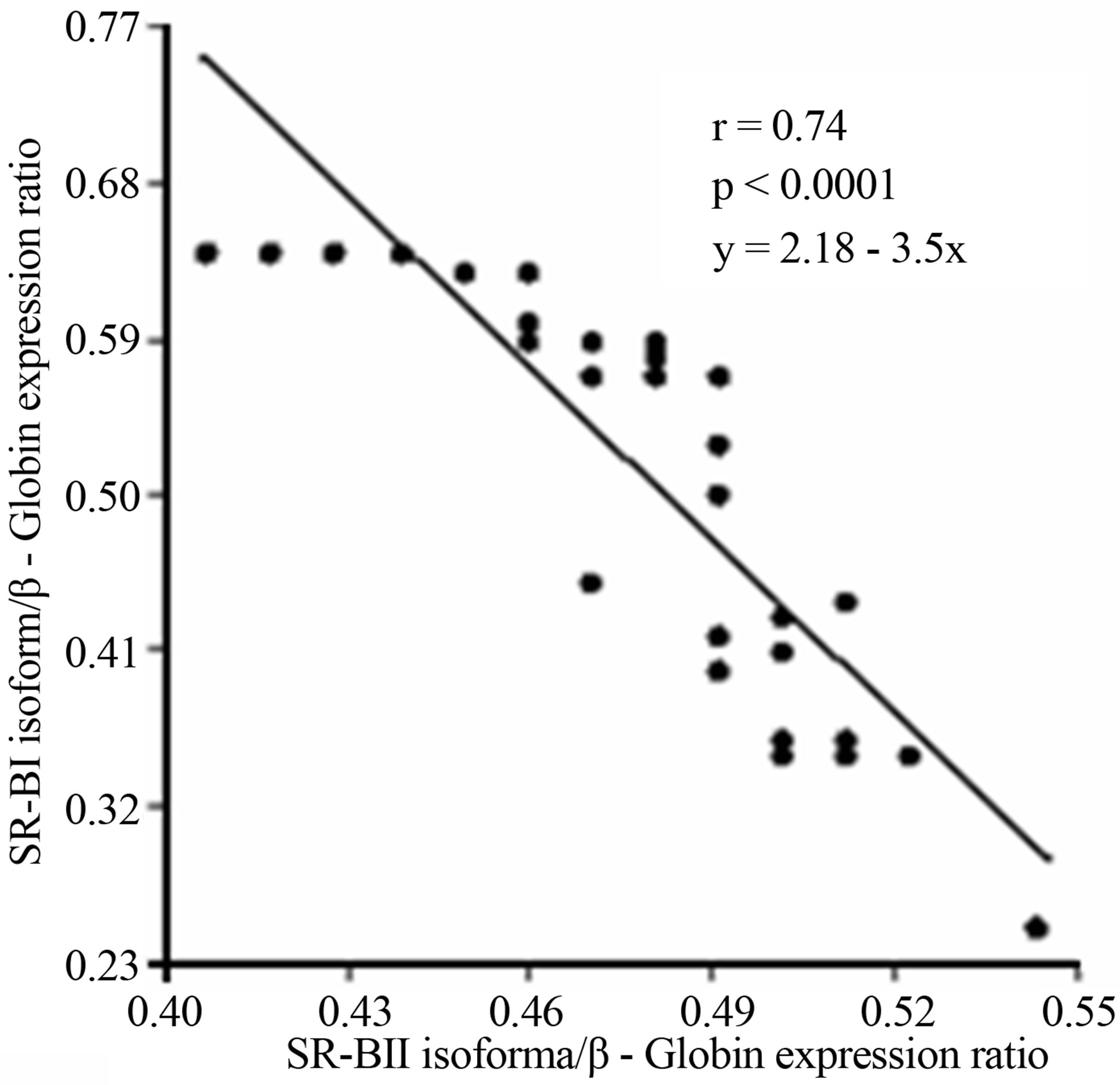 (b)
(b)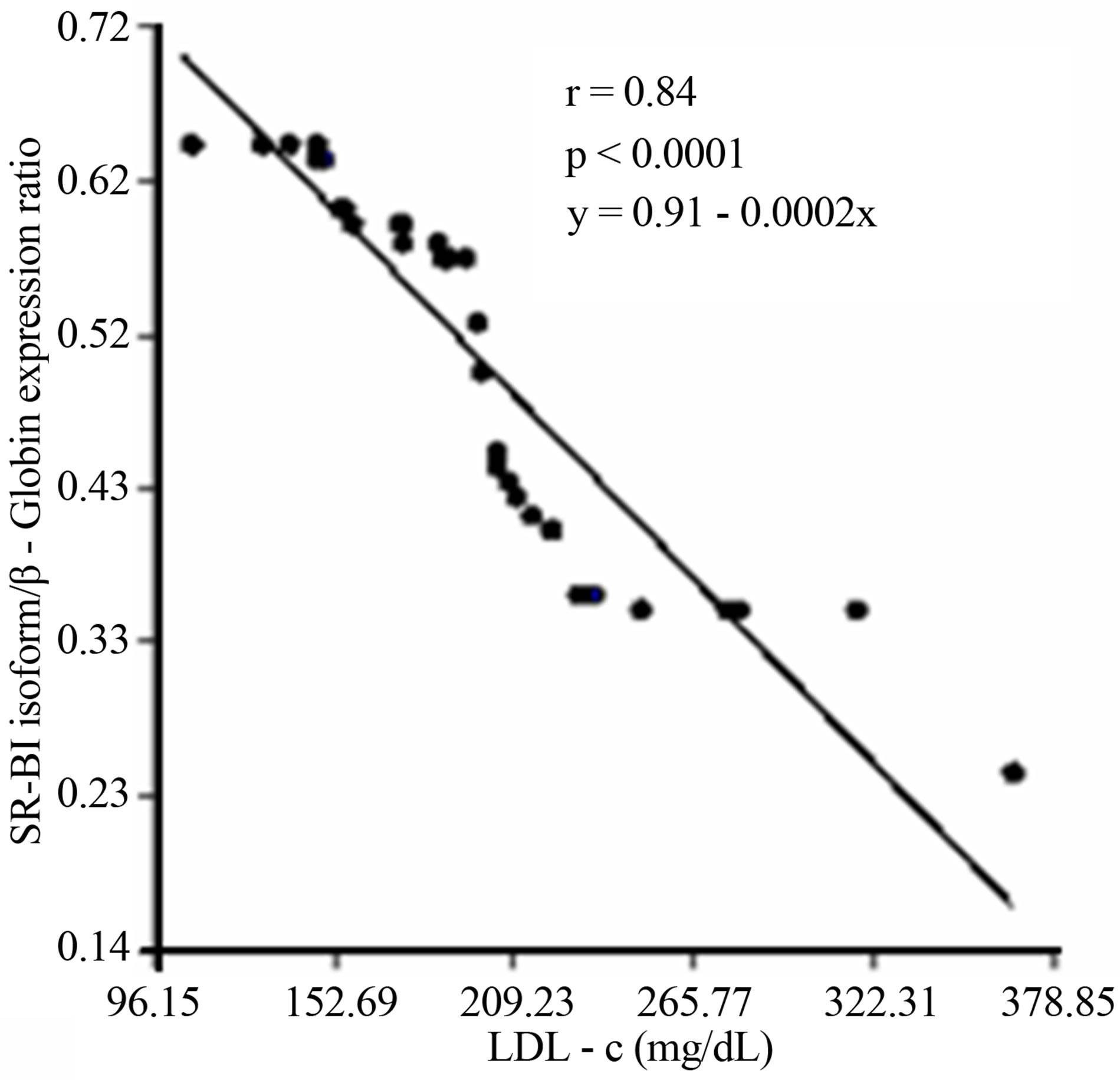 (c)
(c)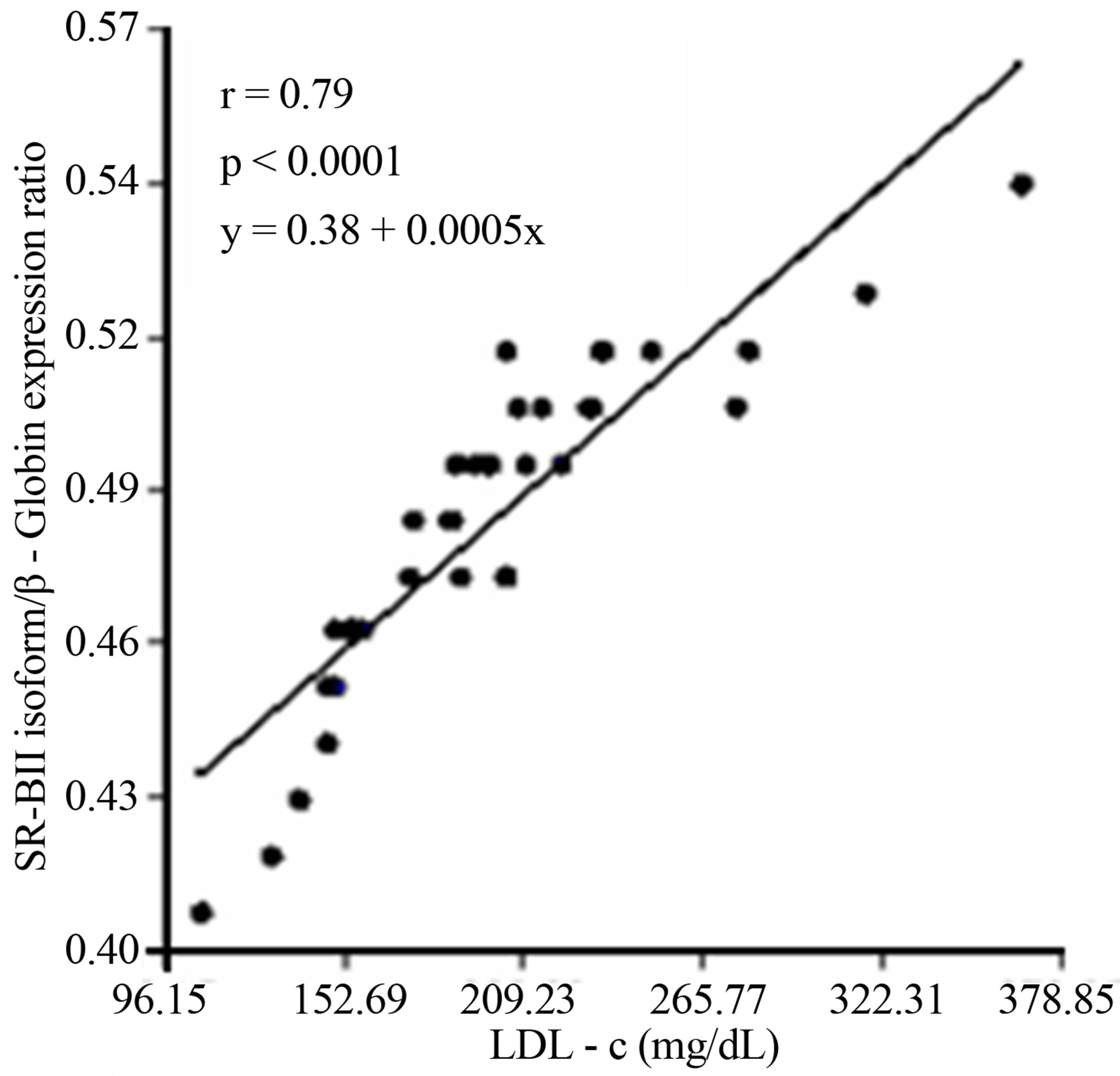 (d)
(d)
Figure 5. (a) Lineal regression analysis between the expression of total SCARB1 and LDL-c levels; (b) Lineal regression analysis between the expression of SR-BI and SR-BII isoforms; (c) Lineal regression analysis between the expression of SR-BII isoform and LDL-c levels; (d) Lineal regression analysis between the expression of SR-BI isoform and LDL-c levels.
metabolism in humans [23,25,36,41]. Furthermore, considering that type 2 diabetes is characterized by low plasma HDL-c concentrations, high triglycerides, a preponderance of small, dense LDL, an increase in oxidized lipoproteins, as well as by other insulin-resistance related parameters that may modulate the expression of the SR-BI gene [20,36]; we have examined the interaction effects between diabetes status and SR-BI gene variants determining plasma lipids and lipoproteins.
The specific contribution of the SCARB1 locus to the homeostasis of lipid metabolism in humans remains mostly undefined. In our study, we have found significant associations between carriers of the C allele at the intron 11 polymorphism (rs838896) and elevated LDL-c concentrations as compared with GG homozygous diabetic subjects. Earlier experimental studies demonstrated that the SR-BI receptor could bind both HDL-c and LDL-c lipoproteins in a nonreciprocal cross competition. Analyses in cultured cells have demonstrated that SR-BI binds other classes of lipoproteins in addition to HDL-c. Hamster SR-BI was originally identified through its ability to bind modified human LDL-c [42] and the human SR-BI, binds VLDL in addition to LDL-c and HDL-c [43]. These studies suggested a role for SR-BI in LDL-c metabolism in humans.
In studies in vivo, alterations in hepatic SR-BI expression have been associated to changes in plasma concentrations of apoB-containing lipoproteins. Sustained, highlevel expression of SR-BI in livers of transgenic mice results in reduced plasma concentrations of LDL-c and apoB [44,45], as well as decreased VLDL and IDL/LDL particle size [45].
The increase in LDL-c concentrations that we observed in diabetic carriers of the C allele is more compatible with a reduced expression of the SR-BI than with an overexpression of this gene variant. These observations, and the fact that the G/C change at intron 11 (rs838896) do not result in a change in the amino acid sequence, suggest that the intron 11 SNP could be in linkage disequilibrium with critical regulatory regions in intron 11 of SCARB1, or alternatively with another functional variant at a neighboring locus [46].
In the present study, we examined the expression of SCARB1 from Type 2 diabetes patients. Hepatocytes and macrophages are appropriate samples for the evaluation of cholesterol transport; however collecting these specimens is not practical in human subjects. Thus, we measured mRNA levels in circulating leucocytes, where mononuclear leucocytes would become macrophages in peripheral tissues. Powell et al. reported that the basic regulatory mechanism in liver and mononuclear leucocytes is similar [47].
In this report, we demonstrated that diabetic patients had significantly reduced expression of overall SCARB1 mRNA in circulating leucocytes cells compared to controls. The hyperglycemia present in T2DM patients downregulated SCARB1 mRNA expression. This suppressive effect has also been reported in HepG2 cells [48] and Caco-2/15 cell line [49] following exposure to high glucose concentrations.
Decreased SCARB1 expression resulted in markedly increased plasma LDL-c concentrations in T2DM subjects. A possible explanation for the increase of LDL-c is that LDL-c lipoproteins may be a poor substrate for SR-BI receptor, because LDL-c is not taken up as efficiently as HDL-c via SR-BI. Studies in vitro have demonstrated that LDL-c can serve as a substrate for selective uptake by SR-BI. However, lipid transfer mediated by SR-BI from LDL-c particles appeared to be less efficient when compared with HDL-c [50,51].
Our results suggest that the marked downregulation of overall SCARB1 and its impact on LDL-c lipoproteins may lessen the anti-atherogenic properties normally associated to elevated reverse cholesterol transport. Although high LDL-c values were associated to low expression of SCARB1 in T2DM leucocytes, there are some characteristics of SR-BI receptor that might cause difficulty in the interpretation and discussion of this result. First, SR-BI has multiple functions and has a dual role in cholesterol metabolism, participating in the intake and efflux of cholesterol traffic among cells and HDL-c particles [52], as well as contributing to the capture of apoBcontaining lipoproteins, LDL and VLDL [14,53]; second, SR-BI displays specific expression profiles among several tissues that may result from different mechanisms of transcription regulation [54]; and third, there is limited information explaining the factors that modulate the SCARB1 mRNA expression in humans tissues in vivo.
Diabetes also induces some changes in lipid and lipoproteins that may be important modulators. In this regard, it has been described that elevated triglyceride content that frequently occurs in type 2 diabetes may impair the beneficial effects of the SR-BI overexpression [55]. On the other hand, increasing evidence indicates that SR-BI may play additional roles that might be of particular importance in type 2 diabetes. Thus, the SR-BI, as a scavenger receptor, can also bind oxidized LDL that adds support to its antiatherogenic properties [56,57].
Although there are no studies in humans examining the association between the SR-BI expression and atherosclerosis, several results from animal models are consistent in their results showing a decreased atherosclerosis associated to a higher SCARB1 expression [9]. We have shown that the SCARB1 locus is significantly associated to LDL-c concentrations. Our data suggest that, through its association with lipid risk factors and more specifically, with the kinetics of reverse cholesterol transport, the SR-BI may modulate the risk of atherosclerosis in the general population and especially in diabetic subjects.
Given the existence of alternative splice forms of SRBI involving inclusion/exclusion of exon 12, we hypothesized that a SNP (rs838896) located within the associated haplotype block may modulate splicing efficiency. We quantified the amounts of the full-length SRBI isoform (including exon 12), the alternative SR-BII isoform (skipping exon 12) and overall SCARB1 mRNA (all splice forms) in leucocytes from controls and T2DM individuals. In normal subjects the highest percentage of expression corresponds to SR-BI compared to SR-BII, consistent with other studies [35]; while in diabetics the expression of SR-BI showed no differences when compared to SR-BII. Although there are no studies in humans examining the expression of both isoforms in T2DM, we conclude that the sustained hyperglycemia promotes an overexpression of SR-BII isoform.
In order to determine the functional consequences of SCARB1 polymorphism (rs838896), we carried out genetic association analysis of the associated SNP and expression of SCARB1 isoforms. Our data do not support an effect of this SNP on relative amounts of SR-BI or SR-BII in leucocytes. In order to determine whether the rs838896 variant is the cause of the different expression levels of SCARB1 isoforms, follow up functional studies using cell culture and other systems need to be done.
In the other hand, estrogen is known to have a profound impact on serum lipid levels, resulting in a decrease in LDL-c and triglycerides, affording younger women relative protection from coronary artery disease. Estrogen is also a potent regulator of SR-BI receptor, influencing the relative expression of SR-BI and SR-BII isoforms. Zhang et al. [58] proposed that estrogen regulation of alternative splicing of SR-BI in the rat occurs via regulatory splicing factors that interact with regulatory sequences in intron 11 of SCARB1 [46,59]. Similarly, treatment of HepG2 liver cell lines with estradiol has been shown to result in a downregulation of SR-BI and up-regulation of the SR-BII splice form [59,60].
Chiba-Falek et al. [35] have detected sex-specific expression of SCARB1 isoforms in vivo, in human liver tissue, and identified polymorphisms in intron 11 of the gene that modify this effect. They also found these same genetic variants to influence triglyceride and HDL-c levels in an endogenous estrogen-dependent manner in a human population study. Our findings do not agree with those of Chiba-Falek due to the patients included in the current study. It should be noted that this was an elderly population of men and women, with relatively low levels of estradiol.
Furthermore, our data also revealed that there is a strong and negative correlation among the expression of SR-BII isoform and LDL-c levels in T2DM. SR-BII is reported to be a minor splice variant in human liver and has been shown to be less efficient at reverse cholesterol transport [61]. We conclude that SR-BII have reduced capacity to de-lipidate LDL-c particles due to its less efficient activity on LDL-c particle catabolism.
5. CONCLUSION
To our knowledge, the results from this study provide novel evidence that hyperglycemia may affect reverse cholesterol transport by controlling SCARB1 expression in diabetic patients. The linear regression analysis revealed that there is a strong and negative correlation between the changes of SCARB1 expression and LDL-c levels. We conclude that the sustained hyperglycemia promotes overexpression of SR-BII isoform, which is less efficient in reverse cholesterol transport and leads to elevate LDL-c concentrations in T2DM patients.
REFERENCES
- Howard, B.V., Cowan, L.D., Go, O., Welty, T.K., Robbins, D.C. and Lee, E.T. (1998) Adverse effects of diabetes on multiple cardiovascular disease risk factors in women. The Strong Heart Study. Diabetes Care, 21, 1258-1265. http://dx.doi.org/10.2337/diacare.21.8.1258
- Taskinen, M.R. (2002) Diabetic dyslipidemia. Atherosclerosis, 3, 47-51.
- Franceschini, G. (2001) Epidemiologic evidence for highdensity lipoprotein cholesterol as a risk factor for coronary artery disease. American Journal of Cardiology, 88, 9-13. http://dx.doi.org/10.1016/S0002-9149(01)02146-4
- Erkelens, D.W. (2001) Insulin resistance syndrome and type 2 diabetes mellitus. American Journal of Cardiology, 88, 38-42. http://dx.doi.org/10.1016/S0002-9149(01)01883-5
- Ford, E.S., Giles, W.H. and Dietz, W.H. (2002) Prevalence of the metabolic syndrome among US adults: Findings from the third National Health and Nutrition Examination Survey. JAMA, 287, 356-359. http://dx.doi.org/10.1001/jama.287.3.356
- Stein, O. and Stein, Y. (1999) Atheroprotective mechanisms of HDL. Atherosclerosis, 144, 285-301. http://dx.doi.org/10.1016/S0021-9150(99)00065-9
- Silver, D.L., Jiang, X.C., Arai, T., Bruce, C. and Tall, A.R. (2000) Receptors and lipid transfer proteins in HDL metabolism. Annals of the New York Academy of Sciences, 902, 103-111. http://dx.doi.org/10.1111/j.1749-6632.2000.tb06305.x
- Acton, S., Rigotti, A., Landschulz, K. T., Xu, S., Hobbs, H. H. and Krieger, M. (1996) Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science, 271, 518-520. http://dx.doi.org/10.1126/science.271.5248.518
- Krieger, M. (2001) Scavenger receptor class B type I is a multiligand HDL receptor that influences diverse physiologic systems. Journal of Clinical Investigation, 108, 793-797.
- Rigotti, A., Miettinen, H. E. and Krieger, M. (2003) The role of the high-density lipoprotein receptor SR-BI in the lipid metabolism of endocrine and other tissues. Endocrine Reviews, 24, 357-387. http://dx.doi.org/10.1210/er.2001-0037
- Mardones, P., Quinones, V., Amigo, L., Moreno, M., Miquel, J. F., Schwarz, M., Miettinen, H. E., Trigatti, B., Krieger, M., VanPatten, S., Cohen, D. E. and Rigotti, A. (2001) Hepatic cholesterol and bile acid metabolism and intestinal cholesterol absorption in scavenger receptor class B type I-deficient mice. Journal of Lipid Research, 42, 170-180.
- Trigatti, B.L., Rigotti, A. and Braun, A. (2000) Cellular and physiological roles of SR-BI, a lipoprotein receptor which mediates selective lipid uptake. Biochimica et Biophysica Acta, 1529, 276-286. http://dx.doi.org/10.1016/S1388-1981(00)00154-2
- Brodeur, M.R., Luangrath, V., Bourret, G., Falstrault, L. and Brissette, L. (2005) Physiological importance of SRBI in the in Vivo metabolism of human HDL and LDL in male and female mice. Journal of Lipid Research, 46, 687-696. http://dx.doi.org/10.1194/jlr.M400165-JLR200
- Van Eck, M., Hoekstra, M., Out, R., Bos, I.S., Kruijt, J.K., Hildebrand, R.B. and Van Berkel, T.J. (2008) Scavenger receptor BI facilitates the metabolism of VLDL lipoproteins in vivo. Journal of Lipid Research, 49, 136-146. http://dx.doi.org/10.1194/jlr.M700355-JLR200
- Hu, L., Hoogt van der, C.C., Espirito Santo, S.M., Out, R., Kypreos, K.E., van Vlijmen, B.J., Van Berkel, T.J., Romijn, J.A., Havekes, L.M., van Dijk, K.W., et al. (2008) The hepatic uptake of VLDL in lrp-ldlr-/-vldlr-/- mice is regulated by LPL activity and involves proteoglycans and SR-BI. Journal of Lipid Research, 49, 1553-1561. http://dx.doi.org/10.1194/jlr.M800130-JLR200
- Eckhardt, E.R., Cai, L., Sun, B., Webb, N.R. and Westhuyzen van der, D.R. (2004) High density lipoprotein uptake by scavenger receptor SR-BII. Journal of Biological Chemistry, 279, 14372-14381. http://dx.doi.org/10.1074/jbc.M313793200
- Webb, N.R., Connell, P.M., Graf, G.A., Smart, E.J., de Villiers, W.J., de Beer, F.C. and Westhuyzen van der, D.R. (1998) SR-BII, an isoform of the scavenger receptor BI containing an alternate cytoplasmic tail, mediates lipid transfer between high density lipoprotein and cells. Journal of Biological Chemistry, 273, 15241-15248. http://dx.doi.org/10.1074/jbc.273.24.15241
- Kozarsky, K.F., Donahee, M.H., Glick, J.M., Krieger, M. and Rader, D.J. (2000) Gene transfer and hepatic overexpression of the HDL receptor SR-BI reduces atherosclerosis in the cholesterol-fed LDL receptor-deficient mouse. Arteriosclerosis, Thrombosis, and Vascular Biology, 20, 721-727. http://dx.doi.org/10.1161/01.ATV.20.3.721
- Van Eck, M., Twisk, J., Hoekstra, M., Van Rij, B.T., Van der Lans, C.A., Bos, I.S., Kruijt, J.K., Kuipers, F. and Van Berkel, T.J. (2003) Differential effects of scavenger receptor BI deficiency on lipid metabolism in cells of the arterial wall and in the liver. Journal of Biological Chemistry, 278, 23699-23705. http://dx.doi.org/10.1074/jbc.M211233200
- Meigs, J.B., Shrader, P., Sullivan, L.M., McAteer, J.B., Fox, C.S., Dupuis, J., Manning, A.K., Florez, J.C., Wilson, P.W., D’Agostino Sr., R.B. and Cupples, L.A. (2008) Genotype score in addition to common risk factors for prediction of type 2 diabetes. New England Journal of Medicine, 359, 2208-2219. http://dx.doi.org/10.1056/NEJMoa0804742
- Florez, J.C., Hirschhorn, J. and Altshuler, D. (2003) The inherited basis of diabetes mellitus: Implications for the genetic analysis of complex traits. Annual Review of Genomics and Human Genetics, 4, 257-291. http://dx.doi.org/10.1146/annurev.genom.4.070802.110436
- Norris, J.M., Langefeld, C.D., Scherzinger, A.L., Rich, S.S., Bookman, E., Beck, S.R., Saad, M.F., Haffner, S.M., Bergman, R.N., Bowden, D.W. and Wagenknecht, L.E. (2005) Quantitative trait loci for abdominal fat and BMI in Hispanic-Americans and African-Americans: The IRAS Family study. International Journal of Obesity and Related Metabolic Disorders, 29, 67-77. http://dx.doi.org/10.1038/sj.ijo.0802793
- Lewis, C.E., North, K.E., Arnett, D., Borecki, I.B., Coon, H., Ellison, R.C., Hunt, S.C., Oberman, A., Rich, S.S., Province, M.A. and Miller, M.B. (2005) Sex-specific findings from a genome-wide linkage analysis of human fatness in non-Hispanic whites and African Americans: The HyperGEN study. International Journal of Obesity (London), 29, 639-649. http://dx.doi.org/10.1038/sj.ijo.0802916
- Wilson, S.G., Adam, G., Langdown, M., Reneland, R., Braun, A., Andrew, T., Surdulescu, G.L., Norberg, M., Dudbridge, F., Reed, P.W., Sambrook, P.N., Kleyn, P.W. and Spector, T.D. (2006) Linkage and potential association of obesity-related phenotypes with two genes on chromosome 12q24 in a female dizygous twin cohort. European Journal of Human Genetics, 14, 340-348. http://dx.doi.org/10.1038/sj.ejhg.5201551
- Acton, S., Osgood, D., Donoghue, M., Corella, D., Pocovi, M., Cenarro, A., Mozas, P., Keilty, J., Squazzo, S., Woolf, E.A. and Ordovas, J.M. (1999) Association of polymorphisms at the SR-BI gene locus with plasma lipid levels and body mass index in a white population. Arteriosclerosis, Thrombosis, and Vascular Biology, 19, 1734- 1743. http://dx.doi.org/10.1161/01.ATV.19.7.1734
- Yoon, Y., Song, J., Hong, S.H. and Kim, J.Q. (2003) Analysis of multiple single nucleotide polymorphisms of candidate genes related to coronary heart disease susceptibility by using support vector machines. Clinical Chemistry and Laboratory Medicine, 41, 529-534. http://dx.doi.org/10.1515/CCLM.2003.080
- Tai, E.S., Adiconis, X., Ordovas, J.M., Carmena-Ramon, R., Real, J., Corella, D., Ascaso, J. and Carmena, R. (2003) Polymorphisms at the SRBI locus are associated with lipoprotein levels in subjects with heterozygous familial hypercholesterolemia. Clinical Genetics, 63, 53-58. http://dx.doi.org/10.1034/j.1399-0004.2003.630108.x
- McCarthy, J.J., Lewitzky, S., Reeves, C., Permutt, A., Glaser, B., Groop, L.C., Lehner, T. and Meyer, J.M. (2003) Polymorphisms of the HDL receptor gene associated with HDL cholesterol levels in diabetic kindred from three populations. Human Heredity, 55, 163-170. http://dx.doi.org/10.1159/000073986
- Hong, S.H., Kim, Y.R., Yoon, Y.M., Min, W.K., Chun, S.I. and Kim, J.Q. (2002) Association between Hae III polymorphism of scavenger receptor class B type I gene and plasma HDL-cholesterol concentration. Annals of Clinical Biochemistry, 39, 478-481. http://dx.doi.org/10.1258/000456302320314485
- Richard, E., von Muhlen, D., Barrett-Connor, E., Alcaraz, J., Davis, R. and McCarthy, J.J. (2005) Modification of the effects of estrogen therapy on HDL cholesterol levels by polymorphisms of the HDL-C receptor, SR-BI: The Rancho Bernardo Study. Atherosclerosis, 180, 255-262. http://dx.doi.org/10.1016/j.atherosclerosis.2004.12.013
- Hsu, L.A., Ko, Y.L., Wu, S., Teng, M.S., Peng, T.Y., Chen, C.F. and Lee, Y.S. (2003) Association between a novel 11-base pair deletion mutation in the promoter region of the scavenger receptor class B type I gene and plasma HDL cholesterol levels in Taiwanese Chinese. Arteriosclerosis, Thrombosis, and Vascular Biology, 23, 1869- 1874. http://dx.doi.org/10.1161/01.ATV.0000082525.84814.A9
- Morabia, A., Ross, B.M., Costanza, M.C., Cayanis, E., Flaherty, M.S., Alvin, G.B., Das, K., James, R., Yang, A.S., Evagrafov, O. and Gilliam, T.C. (2004) Populationbased study of SR-BI genetic variation and lipid profile. Atherosclerosis, 175, 159-168. http://dx.doi.org/10.1016/j.atherosclerosis.2004.03.014
- Roberts, C.G., Shen, H., Mitchell, B.D., Damcott, C.M., Shuldiner, A.R. and Rodriguez, A. (2007) Variants in scavenger receptor class B type I gene are associated with HDL cholesterol levels in younger women. Human Heredity, 64, 107-113. http://dx.doi.org/10.1159/000101962
- Osgood, D., Corella, D., Demissie, S., Cupples, L.A., Wilson, P.W., Meigs, J.B., Schaefer, E.J., Coltell, O. and Ordovas, J.M. (2003) Genetic variation at the scavenger receptor class B type I gene locus determines plasma lipoprotein concentrations and particle size and interacts with type 2 diabetes: The Framingham Study. Journal of Clinical Endocrinology & Metabolism, 88, 2869-2879. http://dx.doi.org/10.1210/jc.2002-021664
- Chiba-Falek, O., Nichols, M., Suchindran, S., Guyton, J., Ginsburg, G.S., Barrett-Connor, E. and McCarthy, J.J. (2010) Impact of gene variants on sex-specific regulation of human Scavenger receptor class B type 1(SR-BI) expression in liver and association with lipid levels in a population-based study. BMC Medical Genetics, 19, 9. http://dx.doi.org/10.1186/1471-2350-11-9
- Goff Jr., D.C., D’Agostino Jr., R.B., Haffner, S.M., Saad, M.F. and Wagenknecht, L.E. (2000) Lipoprotein concentrations and carotid atherosclerosis by diabetes status: Results from the insulin resistance atherosclerosis study. Diabetes Care, 23, 1006-1011. http://dx.doi.org/10.2337/diacare.23.7.1006
- Tan, K.C., Ai, V.H., Chow, W.S., Chau, M.T., Leong, L. and Lam, K.S. (1999) Influence of low density lipoprotein (LDL) subfraction profile and LDL oxidation on endothelium dependent and independent vasodilation in patients with type 2 diabetes. Journal of Clinical Endocrinology & Metabolism, 84, 3212-3216. http://dx.doi.org/10.1210/jc.84.9.3212
- The Expert Committee of the Diagnosis and Classification of Diabetes Mellitus (1997) Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care, 20, 1183-1197.
- Friedewald, W.T., Levy, R.I. and Fredrickson, D.S. (1972) Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clinical Chemistry, 18, 499-502.
- Ye, S., Dhillon, S., Ke, X., Collins, A.R. and Day, I.N. (2001) An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Research, 29, E88. http://dx.doi.org/10.1093/nar/29.17.e88
- Perez-Martinez, P., Ordovas, J.M., Lopez-Miranda, J., Gomez, P., Marin, C., Moreno, J., Fuentes, F., Fernandez de la Puebla, R.A. and Perez-Jimenez, F. (2003) Polymorphism exon 1 variant at the locus of the scavenger receptor class B type I gene: Influence on plasma LDL cholesterol in healthy subjects during the consumption of diets with different fat contents. American Journal of Clinical Nutrition, 77, 809-813.
- Acton, S.L., Scherer, P.E., Lodish, H.F. and Kreiger, M. (1994) Expression cloning of SR-BI, a CD36-related Class B Scavenger Receptor. Journal of Biological Chemistry, 269, 21003-21009.
- Calvo, D., Gomez-Coronado, D., Lasuncion, M.A. and Vega, M.A. (1997) CLA-1 is an 85-kD plasma membrane glycoprotein that acts as a high-affinity receptor for both native (HDL, LDL, and VLDL) and modified (OxLDL and AcLDL) lipoproteins. Arteriosclerosis, Thrombosis, and Vascular Biology, 17, 2341-2349. http://dx.doi.org/10.1161/01.ATV.17.11.2341
- Wang, N., Arai, T., Ji, Y., Rinninger, F. and Tall, A.R. (1998) Liverspecific overexpression of scavenger recaptor BI decreases levels of very low density lipoprotein apoB, low density lipoprotein apoB, and high density lipoprotein in transgenic mice. Journal of Biological Chemistry, 273, 32920-32926. http://dx.doi.org/10.1074/jbc.273.49.32920
- Ueda, Y., Royer, L., Gong, E., Zhang, J., Cooper, P.N., Francone, O. and Rubin, E.M. (1999) Lower plasma levels and accelerated clearance of high density lipoprotein (HDL) and non-HDL cholesterol in scavenger receptor class B Type I transgenic mice. Journal of Biological Chemistry, 274, 7165-7171. http://dx.doi.org/10.1074/jbc.274.11.7165
- Zhang, X., Merkler, K.A. and McLean, M.P. (2008) Characterization of regulatory intronic and exonic sequences involved in alternative splicing of scavenger receptor class B gene. Biochemical and Biophysical Research Communications, 372, 173-178. http://dx.doi.org/10.1016/j.bbrc.2008.05.007
- Powell, E.E. and Kroon, P.A. (1994) Low density lipoprotein receptor and 3-hydroxy-3-methylglutaryl coenzyme A reductase gene expression in human mononuclear leukocytes is regulated coordinately and parallels gene expression in human liver. Journal of Clinical Investigation, 93, 2168-2174. http://dx.doi.org/10.1172/JCI117213
- Murao, K., Yu, X., Imachi, H., Cao, W.M., Chen, K., Matsumoto, K., Nishiuchi, T., Wong, N.C. and Ishida, T. (2008) Hyperglycemia suppresses hepatic scavenger receptor class B type I expression. American Journal of Physiology, Endocrinology and Metabolism, 294, E78- E87. http://dx.doi.org/10.1152/ajpendo.00023.2007
- Ravid, Z., Bendayan, M., Delvin, E., Sane, A.T., Elchebly, M., Lafond, J., Lambert, M., Mailhot, G. and Levy, E. (2008) Modulation of intestinal cholesterol absorption by high glucose levels: Impact on cholesterol transporters, regulatory enzymes, and transcription factors. American Journal of Physiology, Gastrointestinal and Liver Physiology, 295, G873-G885. http://dx.doi.org/10.1152/ajpgi.90376.2008
- Swarnakar, S., Temel, R.E., Connelly, M.A., Azhar, S. and Williams, D.L. (1999) Scavenger receptor class B, type I, mediates selective uptake of low density lipoprotein cholesteryl ester. Journal of Biological Chemistry, 274, 29733-29739. http://dx.doi.org/10.1074/jbc.274.42.29733
- Stangl, H., Hyatt, M. and Hobbs, H.H. (1999) Transport of lipids from high and low density lipoproteins via scavenger receptor-BI. Journal of Biological Chemistry, 274, 32692-32698. http://dx.doi.org/10.1074/jbc.274.46.32692
- Cuchel, M., Lund-Katz, S., de la Llera-Moya, M., Millar, J.S., Chang, D., Fuki, I., Rothblat, G.H., Phillips, M.C. and Rader, D.J. (2010) Pathways by which reconstituted high-density lipoprotein mobilizes free cholesterol from whole body and from macrophages. Arteriosclerosis, Thrombosis, and Vascular Biology, 30, 526-532. http://dx.doi.org/10.1161/ATVBAHA.109.196105
- Rhainds, D., Brodeur, M., Lapointe, J., Charpentier, D., Falstrault, L. and Brissette, L. (2003) The role of human and mouse hepatic scavenger receptor class B type I (SR-BI) in the selective uptake of low-density lipoprotein-cholesteryl esters. Biochemistry, 42, 7527-7538. http://dx.doi.org/10.1021/bi026949a
- Landschulz, K.T., Pathak, R.K., Rigotti, A., Krieger, M. and Hobbs, H.H. (1996) Regulation of scavenger receptor, class B, type I, a high density lipoprotein receptor, in liver and steroidogenic tissues of the rat. Journal of Clinical Investigation, 98, 984-995. http://dx.doi.org/10.1172/JCI118883
- Greene, D.J., Skeggs, J.W. and Morton, R.E. (2001) Elevated triglyceride content diminishes the capacity of high density lipoprotein to deliver cholesteryl esters via the scavenger receptor class B type I (SR-BI). Journal of Clinical Investigation, 276, 4804-4811. http://dx.doi.org/10.1074/jbc.M008725200
- Gillotte-Taylor, K., Boullier, A., Witztum, J.L., Steinberg, D. and Quehenberger, O. (2001) Scavenger receptor class B type I as a receptor for oxidized low density lipoprotein. Journal of Lipid Research, 42, 1474-1482.
- Iwashima, Y., Eto, M., Hata, A., Kaku, K., Horiuchi, S., Ushikubi, F. and Sano, H. (2000) Advanced glycation end products-induced gene expression of scavenger receptors in cultured human monocyte-derived macrophages. Biochemical and Biophysical Research Communications, 277, 368-380. http://dx.doi.org/10.1006/bbrc.2000.3685
- Lopez, D. and McLean, M.P. (2006) Estrogen regulation of the scavenger receptor class B gene: Anti-atherogenic or steroidogenic, is there a priority? Molecular and Cellular Endocrinology, 247, 22-33. http://dx.doi.org/10.1016/j.mce.2005.10.005
- Zhang, X., Moor, A.N., Merkler, K.A., Liu, Q. and McLean, M.P. (2007) Regulation of alternative splicing of liver scavenger receptor class B gene by estrogen and the involved regulatory splicing factors. Endocrinology, 148, 5295-5304. http://dx.doi.org/10.1210/en.2007-0376
- Graf, G.A., Roswell, K.L. and Smart, E.J. (2001) 17betaestradiol promotes the upregulation of SR-BII in HepG2 cells and in rat livers. Journal of Lipid Research, 42, 1444-1449.
- Webb, N.R., Connell, P.M., Graf, G.A., Smart, E.J., de Villiers, W.J., de Beer, F.C. and Westhuyzen van der, D.R. (1998) SR-BII, an isoform of the scavenger receptor BI containing an alternate cytoplasmic tail, mediates lipid transfer between high density lipoprotein and cells. Journal of Biological Chemistry, 273, 15241-15248. http://dx.doi.org/10.1074/jbc.273.24.15241

