Agricultural Sciences
Vol.4 No.5(2013), Article ID:32093,6 pages DOI:10.4236/as.2013.45032
Isolation and expression analysis of CBF4 from Vitis amurensis associated with stress
![]()
1College of Horticulture, Nanjing Agricultural University, Nanjing, China
2Department of Horticulture, Heilongjiang Academy of Agricultural Science, Harbin, China; *Corresponding Authors: tjm266@sina.com, zzhang@njau.edu.cn, dongchanggy@126.com
Copyright © 2013 Chang Dong et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 21 March 2013; revised 26 April 2013; accepted 15 May 2013
Keywords: Vitis amurensis; CBF4; Stress; Transcript Accumulation
ABSTRACT
The C-repeat Binding Factor/Dehydration Responsive Element Binding factor (CBF/DREB) responsive pathway is involved in plant response to abiotic stress. In this study, we report the isolation of VaCBF4 (a complete cDNA) and its promoter from Vitis amurensis through rapid amplification of cDNA ends and genome walking techniques, respectively. The CBF4 transcript accumulation of V. amurensis increased under cold, salinity, and abscisic acid (ABA) and salicylic acid (SA) treatments, whereas that of Vitis vinifera showed a different change. The transcript levels of VaCBF4 in the roots, stems, leaves, and petioles under cold, salinity, and ABA and SA treatments were up-regulated in a timedependent manner. The presence of the cis-elements MBC, ABRE, and as-2-box in the VaCBF4 promoter further confirmed that this promoter is a component of the CBF transduction pathway, which is involved in plant response to multistress.
1. INTRODUCTION
Low temperature, high salinity, and drought are environmental factors that limit the geographical distribution and affect the growth, development, and productivity of plants. Plants respond and adapt to these stresses at the molecular, physiological, and biochemical levels. The expression levels of various genes in plants are activated by these stresses [1,2]. The products of these genes regulate the gene expression and signal transduction in response to stress. The cis-elements involved in stress response were analyzed to elucidate the molecular mechanisms of gene expression in response to stress. The C-repeat/Dehydration Responsive Element (CRT/DRE) with the core CCGAC sequence is a cis-acting element in the promoter that regulates gene expression in response to cold and drought in Arabidopsis [3,4]. A similar motif was identified as the Low-Temperature Responsive Element (LTRE) in cold-inducible genes [5].
At present, the most understood genetic system with an important role in cold acclimation is the Arabidopsis CBF (CRT/DRE-binding transcription factors) cold response pathway [6]. Exposing Arabidopsis plants to low temperature or drought results in the rapid induction of a small family of transcriptional factors known as CBF1, CBF2, CBF3(or as DREB1B, DREB1C, and DREB1A, respectively) [7], and CBF4 [8]. CBF/DREB-type proteins belong to the AP2/ERF (APETALA 2/Ethylene Response Factors) DNA-binding protein family. Moreover, the constitutively over-expression of CBF genes induces the expression of downstream genes and improves the freezing and salt tolerance of transgenic plants [9].
Despite the high conservation of CBF residues in plants, some structural and regulatory differences were observed in the CBF cold response pathway among plant species. For example, the CBF regulon of dwarf apple was found to be induced more than that of Arabidopsis [10]. A previous work on blueberry showed that CBF genes from different species have dramatically different levels of induction under low temperature [11]. Although recent breakthroughs have increased the knowledge on the molecular basis of the CBF cold pathway in herbaceous species, little is known in woody plants. To date, data on CBF genes from perennial plants include functional analysis of CBF from cherry [12], eucalyptus [13], blueberry [11], and apple [10].
Grape is one of the most important fruit crops in the world, and its production has been expanding as more varieties and better quality wines become available. However, low temperature limits the geographical distribution and affects the growth, development, and productivity of this crop. Vitis amurensis is a freezing-tolerant wild grape species native to Northern China. This species can survive below 40˚C [14]. V. amurensis is one of the most widely used species for rootstock and winemaking in grape cultivation. However, little is known about the freezing tolerance of V. amurensis at the molecular level.
In this study, the VaCBF4 gene was isolated from V. amurensis. We also compared VaCBF4 gene expression in the leaves of cold-tolerant and cold-sensitive grapes with that in tissues of V. amurensis at different times of low temperature, high salinity, and abscisic acid (ABA) and salicylic acid (SA) treatments, and then confirmed its transcript patterns under stress.
2. MATERIAL AND TREATMENTS
Sixty-day-old in vitro samples of wild V. amurensis and Vitis vinifera’ Manicure Finger’ grown in Murashige and Skoog (MS) medium under a 16 h light/8 h dark regime at 25˚C were subjected to the following treatments for stress-responsive gene expression: cold stress by transferring to 4˚C, salinity stress by supplying 200 mM NaCl, and hormonal treatments by supplying ABA (10 μM) or SA (5 μM) directly to the plants at different times.
2.1. Isolation of VaCBF4
Total RNA from non-stressed or cold-stressed wild samples in vitro was extracted using the CTAB method (Lin et al., 2008) and converted to cDNA using Primer Mix according to the instructions of Rever Tra Ace RT Kit (Toyobo, Japan). The partial coding sequence of the CBF4 genes was amplified using gene-specific primers, C4-F and C4-R (Table 1). The primers were designed based on the genome of Vitis (http://www.vitisgenome.it). The 3’ regions of the corresponding cDNA fragments were obtained by 3’ rapid amplification of cDNA ends (RACE) using the cDNA amplification kit (Clontech, CA, USA) with the gene-specific primers 16326 and C4-rF2 for the first step and 16324 and C4-rF3 for the second step (Table 1). Amplification was carried out under the following conditions: 94˚C for 4 min; 36 cycles of 94˚C for 30 s, 60˚C for 45 s, 64˚C for 60 s, and 72˚C for 60 s; and a final extension at 72˚C for 10 min.
The isolation of the promoter sequence was carried out using a Universal Genome Walker kit (UGWK, Clontech, USA) on 10 ng of V. amurensis genomic DNA digested by four blunt-end-generating restriction enzymes (EcoR V, DraI, SacI, and StuI). After purification, the restriction fragments were ligated with Genome Walker adaptors (Ad1 and Ad2, Table 1). The polymerase chain reaction (PCR) products were amplified on each restriction fragment set using the primers C4-R1 and ada1 for the first step PCR and C4-R2 and ada 2 for the second step PCR (Table 1). The PCR products were cloned and sequenced by the pMD19-T vector.
2.2. Sequence Analysis
Homology searches were performed using BLAST with default parameters at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignment of CBFs was performed using Crustal 1.83, and shading was carried out using DNAMAN software (Version 5.2.2, Lynnon Biosoft). The CBF protein sequences necessary for constructing a phylogenetic tree were retrieved from the GenBank database and depicted using Crustal 1.83. Phylogenetic analysis was based on the neighbor-joining method using Mega 4.1 software with 1000 bootstrap replicates and substitution model of Poisson correction. Promoter sequence analysis was performed using PLACE (http://www.dna.affrc.go.jp) and Plant CARE.
2.3. Real-time Reverse Transcription Quantitative-PCR (RT-qPCR)
Total RNA from different treatments and tissues were digested with RNase-free DNAase I (TaKaRa, Japan) to remove genomic DNA contamination. cDNA fragments were synthesized as aforementioned. Transcript level was determined on an ABI PRISM 7300 (Applied Biosystems, Foster City, CA, USA). For each reaction, 20 ng of cDNA was used with SYBR Premix Ex Taq mix (TaKaRa, Japan), including primers C4rt1 and C4rt2 for the CBF4 gene. All experiments were run in triplicates with different synthesizedc DNA fragments. A positive control was provided through a parallel analysis using the grape malate dehydrogenase gene as an internal reference [15], and was calculated relative to a calibrator using the formula 2−△△Ct.
3. RESULTS
3.1. Identification of the VaCBF4 Gene from V. amurensis
We constructed a cDNA admixture template from the cold-treated and non-treated in vitro leaves to identify the gene related to low temperature. A CBF4-like partial sequence was found, suggesting that the corresponding gene was probably induced by low temperature. Given that this sequence contained the intact 5’ end of the Open Reading Frame (ORF) of a putative CBF4 gene, we can

Table 1. Primers used in this study.
obtain the rest of the sequence by 3’ RACE assay. A fragment of 513 bp was amplified through nested PCR. Based on the sequence analysis, we obtained a cDNA sequence designated as VaCBF4 (GenBank accession No. HM231149). VaCBF4 contains an ORF of 654 bp, a 5’ untranslated region (UTR) of 22 bp, and a 3’ UTR of 278 bp. Moreover, this cDNA contains a single ORF of 218 amino acids and encodes a putative protein with a predicted molecular mass of 24.23 kDa and a pI of 5.4. The obtained amino acid encodes an AP2/ERF DNA binding protein (Figure 1). We found putative nuclear localization signals (NLS) from the N-terminus to the C-terminus, as well as the domains AP2, DSAWR, and LWSY in the C-terminal region.
3.2. Phylogenetic Analysis of VaCBF4
A BLAST search was performed using the deduced amino acid sequence of VaCBF4. Several homologous sequences of VaCBF4 in higher plants were found in the GenBank database. Alignment analysis and domain comparison indicated that VaCBF4 shared high homology to other proteins in the AP2/ERF domain (Figure 1). Through systematic phylogenetic analysis, AP2/ERF proteins were classified into 15 plant CBF proteins using the neighbor-joining phylogenetic method (Figure 2). The results showed that the sequence from V. amurensis belong to the CBF4 cluster of CBF proteins.
3.3. Sequence Analysis of the VaCBF4 Promoter
The VaCBF4 promoter was obtained using the UGWK method (Figure 2). The result revealed that a typical TATA-box was located at −108 bp upstream of the ATG translational initiation codon. Furthermore, cis-acting elements involved in stress-related responses were found
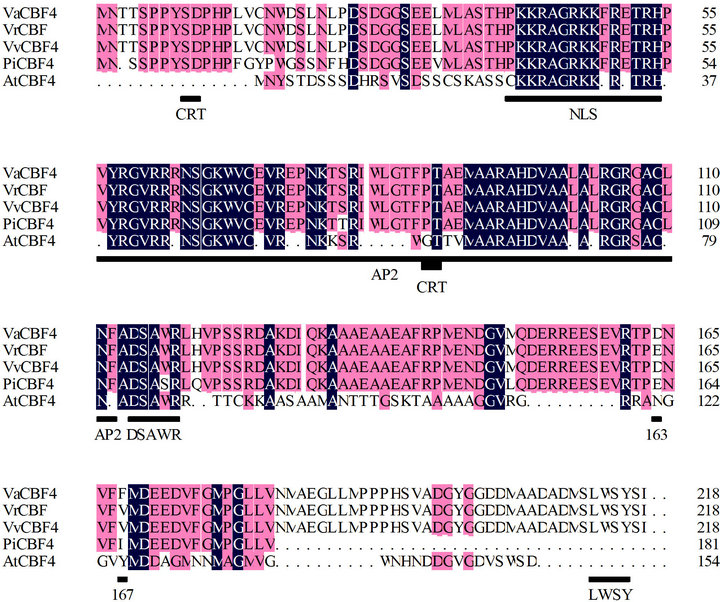
Figure 1. Multiple alignment of VaCBF4 with the CBF4 proteins of Vitaceae, Parthenocissus, and Arabidopsis. The amino acid sequences are from NCBI [AtCBF4 (np_200012), PiCBF4 (ABU55674), VrCBF4 (AY706986), and VvCBF4 (XP_00228 0097)]. The nuclear localization signal, AP2, DSAWR, and LWSY domains are indicated by bars.
in the VaCBF4 promoter. These elements, which include ACE, as-2-box, MeJA-like, MYB, ARE, MYC (MYC recognition site), LTRE, and ABRE, are necessary for stress responsive expression under abiotic stress.
3.4. Differential Accumulation of Vitis CBF4 under Low Temperature Stress
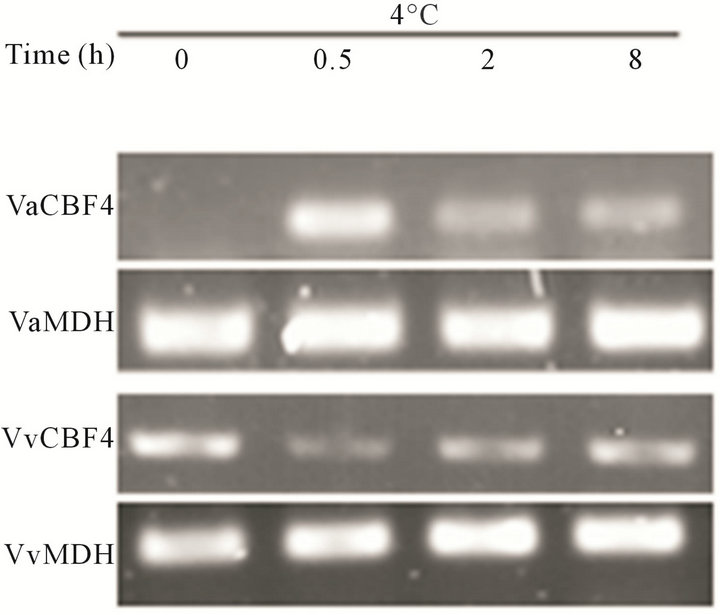
The CBF4 gene expression in the leaves was compared in vitro with that in the relatively cold-hardy wild V. amurensis and the relatively cold-sensitive V. vinifera cultivar “Manicure Finger” (Figure 3(a)). V. amurens reached its maximum midwinter cold hardiness level at <−40˚C, whereas “Manicure Finger” was cold hardy at
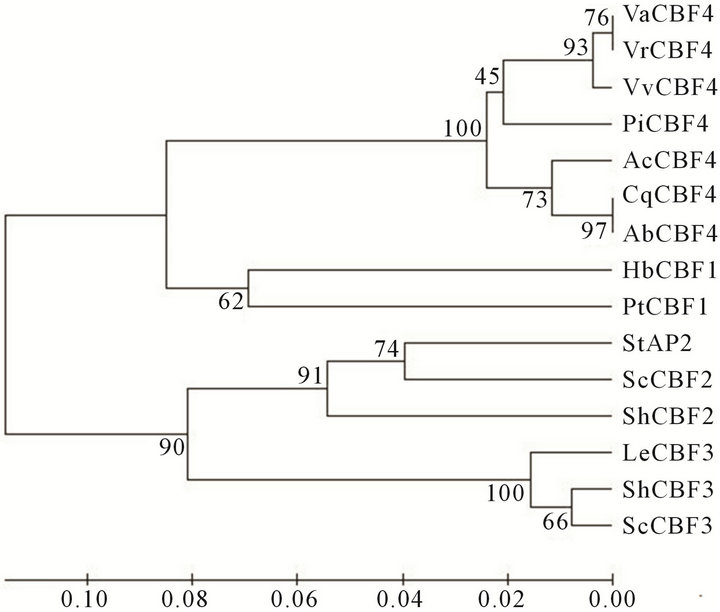
Figure 2. Bootstrap phylogenetic analysis of the selection of plant CBF proteins from NCBI. The amino acid sequences are VvCBF4 (XP_002280097), AbCBF4 (ABU55675), VrCBF (AAW58104), PiCBF4 (ABU55674), AcCBF4 (ABU55676), CqCBF4 (EF150912), HbCBF1 (AAY43213), PtCBF1 (ABO48363), ScCBF2 (ACB45094), ShCBF2 (ACB45080), ShCBF3 (ACB45078), and ScCBF3 (ACB45092).
 (a)
(a) (b)
(b)
Figure 3. Transcript accumulation of Vitis CBF4 under natural and low temperature stresses. (a) An expression difference of CBF4 in the cold-hardy V. amurensis (Va) and the less cold-hardy V. vinifera “Manicure Finger” (Vv); (b) Expression of organism in V. amurensis.
approximately −7˚C. Under cold treatment, the expression of CBF4 in V. amurensis was not substantially different from that in V. vinifera. However, a significant difference in transcription was found between the two species under natural conditions. Unlike the transcript accumulation of VvCBF4, the transcript accumulation of VaCBF4 in the leaves, roots, stems, and petioles was not observed under natural conditions. This finding was further confirmed by quantitative RT-PCR (Figure 3(b)).
Accumulations of each transcript in the roots, stems, leaves, and petioles were analyzed by quantitative RTPCR to further determine the acquisition mechanisms of cold acclimation in relation to VaCBF4 gene expression. As shown in Figure 4, VaCBF4 exhibited different expression patterns in the organs under low temperature treatment. Accumulations of VaCBF4 transcript in all organs were inactivated by low temperature prior to treatment at 0.5 h compared with the corresponding controls. During the subsequent chilling period, the transcript level of VaCBF4 in the stems and petioles slightly increased at the early stage of treatment and then reached the peak on day 2 or day 3. By contrast, the transcript level of VaCBF4 in the roots sharply reached the peak at 4 h and then gradually decreased thereafter. However, the mRNA expression in the leaves was slightly detected at 0.5 h and then remained almost constant after over 3 d of treatment at 4˚C.
3.5. Expression Pattern of VaCBF4 in Salinity Stress
We analyzed the expression of the VaCBF4 gene under high salinity stress to obtain more information about its expression under stress. The results showed that VaCBF4 expression was induced by salinity throughout the treatment period (Figure 5). However, distinct organs dis-
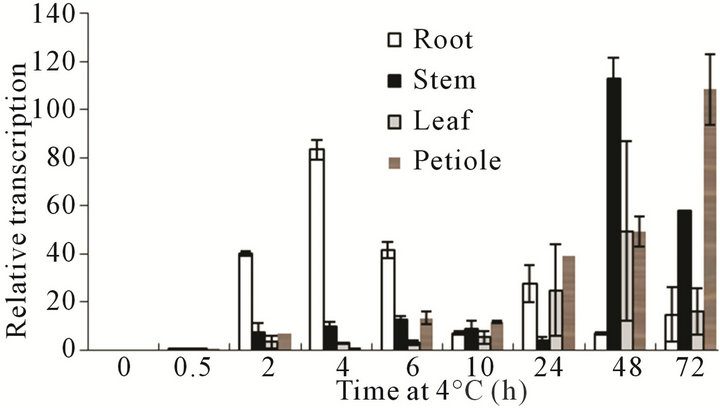
Figure 4. Changes in the mRNA accumulation of VaCBF4 in the roots, stems, leaves, and petioles under 4˚C treatment.
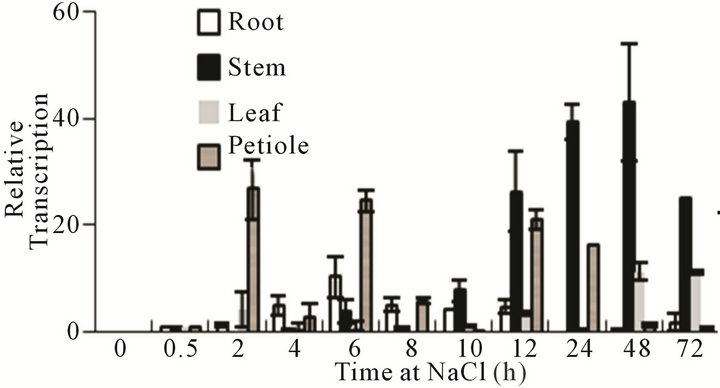
Figure 5. Changes in the mRNA accumulation of VaCBF4 in the roots, stems, leaves, and petioles during under 200 mM NaCl treatment.
played marked differences in gene expression levels during the treatment period. The expression of VaCBF4 in the stems and leaves reached the peak after approximately 2 d of treatment. The peak transcript accumulation in the roots was observed at 6 h, whereas that in the petioles was wavy.
3.6. ABA and SA Are Regulatory Factors for VaCBF4
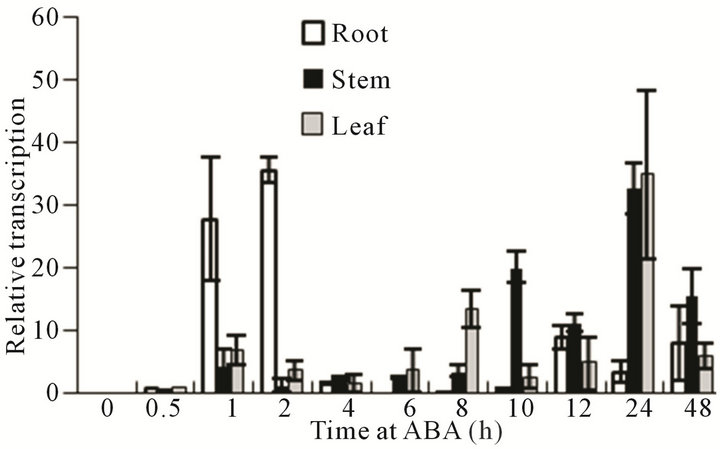
ABA and SA have important functions in physiological and genetic regulation during stress. We hypothesized that the expression pattern of VaCBF4 may be affected by ABA and SA. Therefore, we measured the transcript level of the organs treated with ABA and SA, and compared their expression patterns. The expression of VaCBF4 was induced by exogenous ABA and SA treatments in Figure 6. However, the organs displayed significant differences in gene expression levels. Under ABA and SA treatments, the peak pattern in the roots was found at the early stage, whereas that in the stems was observed at the later stage. However, the expression pattern of VaCBF4 in the leaves under ABA and SA treatments was wavy. The peak VaCBF4 expression in the leaves under ABA treatment appeared at 1, 8, and 24
 (a)
(a)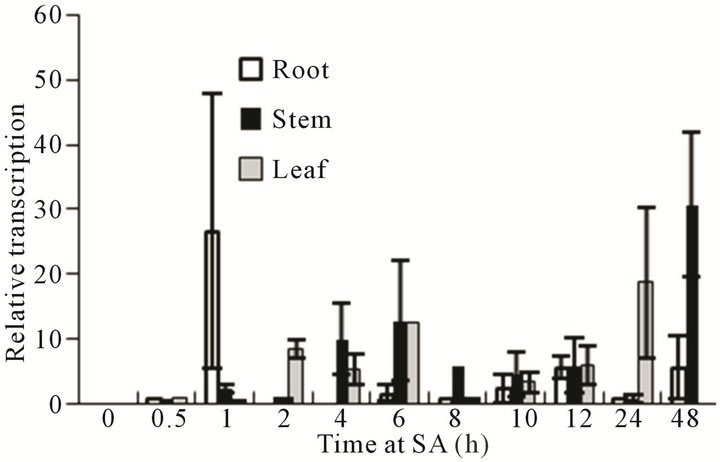 (b)
(b)
Figure 6. Changes in the mRNA accumulation of VaCBF4 in the roots, stems, and leaves under ABA and SA treatments.
h, whereas that in the leaves under SA treatment appeared at 2, 6, and 24 h.
4. DISCUSSION
We isolated and characterized a complete cDNA sequence of CBF4 from V. amurensis. The N-terminal region of VaCBF4 was found to share the conserved CBF4 domains with that of the other known CBF4 (Figure 1). Vitis CBF4 was proven to be a nuclear protein [15]. Moreover, Va CBF4 was able to map to the CBF4 group in the CBF/DREB subfamily through phylogenetic analysis. These data indicated that VaCBF4 is a CBF domain protein that may function as a transcription factor in V. amurensis.
Cold induces the expression of most reported CBF genes, including VaCBF4. VaCBF4 transcripts were detectable in different tissues (roots, stems, leaves, and petioles) under low temperature treatment. These transcripts remain present for a long period, with lower expression in the leaves than in the other tissues. A similarly cold-induced gene expression was reported for the CBF4 of Vitis riparia and V. vinifera [15]. In agreement with the high identity between their amino acid domains, the expression patterns were almost the same between V. amurensis and V. vinifera under low temperature treatment. Interestingly, under natural conditions, the CBF4 expression of V. vinifera was visible in the leaves [15], whereas that of V. amurensis was not detectable in the leaves, stems, roots, and petioles. This finding was further confirmed by quantitative RT-PCR. The important role of CBF4 proteins in the freezing tolerance of V. amurensis may be attributed to the difference in regulon size between V. vinifera and V. amurensis, given the high similarity between VaCBF4 and VvCBF4 (only substitution at position 167 of one negatively charged amino acid, F, for another, V). Haake et al. [8] showed that Arabidopsis CBF4 expression is induced by drought and ABA treatments, but not by cold treatment, under experimental conditions. The freezing-hardy materials may have more CBF4 regulons than the freezing-sensitive materials. However, this assumption needs further investigation. In addition, we showed that the accumulation of CBF4 transcripts from V. amurensis was dramatically induced by salinity and exogenous ABA and SA treatments. Hence, we propose that the CBF4 gene also functions in the signal transduction of high salinity and hormone adaption in V. amurensis. However, the relative transcript levels of VaCBF4 in different tissues exhibited different changes after exposure to different stress treatments.
An analysis of regulatory elements in the upstream of the VaCBF4 gene by PLACE and Plant CARE databases covered the putative DNA-binding sites (Figure 7). The promoter sequences of this gene were found to contain several cis-regulatory elements, such as LTRE, ABRE,

Figure 7. Distribution of putative regulatory elements in the VaCBF4 promoter from V. amurensis.
MYC (MYC recognition site), and MBS (MYB bind site). These elements are related to stress response, specifically to cold/dehydration. These cis-regulatory elements are conserved in several plant species. The presence of these conserved motifs confirmed that the VaCBF4 gene was regulated by cold, high salinity, and exogenous ABA and SA treatments. The multiple stressresponse of the VaCBF4 gene also suggests that V. amurensis should be used for further research on molecular mechanism and stress breeding.
5. ACKNOWLEDGEMENTS
This work was supported by the China National Commonwealth (Agriculture) Science and Technology Project (No.200903044.) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
REFERENCES
- Willimsky, G., et al. (1992) Characterization of cspB, a Bacillus subtilis inducible cold shock gene affecting cell viability at low temperatures. Journal of Bacteriology, 174, 6326-6335.
- Liu, J., et al. (2002) Cold signalling associated with vernalization in Arabidopsis thaliana does not involve CBF1 or abscisic acid. Physiologia Plantarum, 114, 125-134. doi:10.1034/j.1399-3054.2002.1140117.x
- Yamaguchi-Shinozaki, K. and Shinozaki, K. (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell Online, 6, 251-264. doi:10.2307/3869643
- Baker, S.S., Wilhelm, K.S. and Thomashow, M.F. (1994) The 5’-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, droughtand ABA-regulated gene expression. Plant Molecular Biology, 24, 701- 713. doi:10.1007/BF00029852
- Jiang, C., Iu, B. and Singh, J. (1996) Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Molecular Biology, 30, 679-684. doi:10.1007/BF00049344
- Chinnusamy, V., Zhu, J.K. and Sunkar, R. (2010) Gene regulation during cold stress acclimation in plants. Methods in Molecular Biology, 639, 39-55. doi:10.1007/978-1-60761-702-0_3
- Medina, J.N., et al. (1999) The Arabidopsis CBF gene family is composed of three ggenes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiology, 119, 463-470. doi:10.1104/pp.119.2.463
- Haake, V., et al. (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiology, 130, 639-648. doi:10.1104/pp.006478
- Ito, Y., et al. (2006) Functional analysis of rice DREB1/ CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant & Cell Physiology, 47, 141-153. doi:10.1093/pcp/pci230
- Yang, W., et al. (2011) Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta, 233, 219-229. doi:10.1007/s00425-010-1279-6
- Polashock, J.J. (2010) Functional identification of a C-repeat binding factor transcriptional activator from blueberry associated with cold acclimation and freezing tolerance. Journal of the American Society for Hortcultural Science, 135.
- Kitashiba, H., et al. (2004) Expression of a sweet cherry DREB1/CBF ortholog in Arabidopsis confers salt and freezing tolerance. Journal of Plant Physiology, 161, 1171-1176. doi:10.1016/j.jplph.2004.04.008
- El Kayal, W. (2006) Expression profile of CBF-like transcriptional factor genes from Eucalyptus in response to cold. Journal of Experimental Botany, 57, 2455-2469. doi:10.1093/jxb/erl019
- Kong, Q.S. (2004) Grape in China. Chinese Agricultural Science and Technique, Beijing.
- Xiao, H., et al. (2008) CBF4 is a unique member of the CBF transcription factor family of Vitis vinifera and Vitis riparia. Plant, Cell & Environment, 31, 1-10.

