Agricultural Sciences
Vol. 3 No. 7 (2012) , Article ID: 24395 , 5 pages DOI:10.4236/as.2012.37109
Bioinformatics analysis of metallothionein gene from Agaricus bisporus, Ganoderma lucidum, Taiwanofungus camphoratus and Paxillus involutus
![]()
1Xuzhou Key Laboratory of Modern AgroBiotechnology, Xuzhou Vocational College of Bioengineering, Xuzhou, China; *Corresponding Author: xzqck@yahoo.com.cn
2Department of Agriculture and Landscape Engineering, Xuzhou Vocational College of Bioengineering, Xuzhou, China
3Office of Science and Education, Xuzhou Agricultural Commission, Xuzhou, China
Received 15 July 2012; revised 20 August 2012; accepted 9 October 2012
Keywords: Metallothionein Gene; Agaricus bisporus; Ganoderma lucidum; Taiwanofungus camphoratus; Paxillus involutus
ABSTRACT
Metallothionein (MT) gene has an important role in the detoxification of toxic metals especially some heavy metals. Based on the published sequences of MT genes from Agaricus bisporus, Garnoderma lucidum, Taiwanofungus camphoratus and Paxillus involutus as a type of the edible or medical mushroom in NCBI database, their molecular structures, physiological functions and evolutionary relationship were analyzed using the bioinformatics methods. Results showed that AbMT, GlMT, TcMT and PiMT genes contained complete open reading frame (ORF) and their theoretical points of the last three encoding proteins were higher than 7.0. AbMT, GlMT, TcMT and PiMT were relatively rich in random coil and extended strand, but transmembrane helices and signal peptides were not found. 4 MTs were mainly localized in cell nucleus (over 60%) and their cellular functions might have some relation to central intermediary metabolism. Multiple sequence alignment indicated relatively high identity (more than 52%) and short genetic distances (lower than 0.900) among 4 MT nucleotide sequences. Abundant genetic diversity and strong codon bias were found based on the halotype diversity, average number of nucleotide differences, nucleotide diversity, effective number of codons, codon bias index and scaled chi-square. Simultaneously, we deduced that 4 MT genes during molecular evolution were under positive selection. The present study might provide basis for further investigation of MTs molecular mechanisms and genetic laws.
1. INTRODUCTION
Metallothioneins (MTs) are a multigenic family of low-molecular weight (ranging from 6 to 7 kDa), cysteine-rich (ranging from 20% to 33%), metal-binding proteins (7 metals/MT) that are widespread in animals, higher plants, eukaryotic microorganisms and some prokaryotes [1-5]. They are divided into three different classes on the basis of their cysteine content and structure. The Cys-Cys, Cys-X-Cys and Cys-X-X-Cys motifs (in which X denotes any amino acid) are characteristic and invariant for metallothioneins [6]. Although the precise function of MTs remains elusive, many studies reveal that they play a variety of important biological roles in maintaining intracellular metal homeostasis, eliminating metal toxification and protecting against intracellular oxidative damages during the acute phase response [7-10]. In addition, the presence of MTs in the nuclear can protect the DNA from the damage induced by oxidative stress [11].
Edible mushrooms have occupied an increasingly important place in several parts of the world, because of their delicacy, abundance and also as a substitute for the seafoods [12,13]. In recent years, heavy metal pollution has become one of the serious public concerns and environmental problems [14,15]. However, certain mushrooms are known to accumulate heavy metals. Further studies showed that heavy metal concentrations in mushroom are considerably higher than those in agricultural crop plants, vegetables and fruits [16]. This suggests that heavy metal pollution of the mushroom has been drawn more and more attention. To our delight, an important hallmark of MTs is their induction by multiple heavy metal species at the transcriptional level. Therefore the MT genes serve as a valuable model for investigating the mechanism of cellular response to heavy metals. The clarification of heavy metal-dependent gene regulation will in turn contribute to our understanding of the physiological roles of MTs [17].
As a type of edibel or medical mushroom, Agaricus bisporus, Ganoderma lucidum, Taiwanofungus camphoratus, Paxillus involutus are causing serious problems as heavy metal contamination. So in the present study, we completed the bioinformatics analysis of MT genes and their encoding proteins from these four mushrooms. The results will give some theoretical foundations for molecular mechanisms and genetic laws of MT genes in fungi.
2. MATERIALS AND METHODS
2.1. Sequence Retrieval
MT gene sequences of A. bisporus, G. lucidum, T. camphoratus and P. involutus were retrieved from the National Center for Biotechnology Information (NCBI) which can be accessed using the URL: http://www.ncbi.nlm.nih.gov/, then named AbMT (GenBank accession no. AJ271695.1), GlMT (EF489399.1), TcMT (DQ520933.1) and PiMT (AY525379.1), respectively. Their corresponding amino acid sequences designated as AbMT (CAB85689.1), GlMT (ABP02008.1), TcMT (ABF69031.1) and PiMT (AAS19463.1) were also obtained.
2.2. Bioinformatics Analysis
Molecular structures and physicochemical properties were obtained by ProtParam tool (http://web.expasy.org/protparam/). The signal peptide was predicted using SignalP 4.0 Server (http://www.cbs.dtu.dk/services/SignalP/). The potential protein subcellular localization, protein function, conserved domains(CDD), secondary structure and transmembrane domains were predicted using PSORT II Prediction (http://psort.hgc.jp/form2.html), ProFun 2.2 Server (http://www.cbs. dtu.dk/services/ProtFun/), Conserved Domain Database (CDD) (http://www.ncbi.nlm.nih.gov/ Structure/cdd/cdd.shtml), a self-optimized method for protein secondary structure prediction (SOPM) (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopm.html) and TMHMM Server (version 2.0) (http://www.cbs.dtu.dk/services/TMHMM-2.0/), respecttively. Multiple sequence alignment was carried out using ClustalX (version 1.83) software, respectively. Homology and genetic distances were calculated using DNAStar (version 5.01) and MEGA (version 3.1) software. The genetic diversity analysis was carried out using DnaSP software (version 4.0).
3. RESULTS AND DISCUSSION
3.1. Analysis of Molecular Structures and Physicochemical Properties
Structures and properties of nucleotide and corresponding amino acid sequences of MT genes from A. bisporus, G. lucidum, T. camphoratus and P. involutus were obtained by ProtParam tool (Table 1). GlMT, TcMT and PiMT genes contained complete ORF but lacing 5’- untranslated region (UTR) and 3’ UTR. Then ORF and 5’ UTR have been found in AbMT gene but lacing 3’ UTR. So we found that amino acid residues were more than the other three proteins. Other than the AbMT, theoretical isoeletric points of the other three proteins in Table 1 were higher than 7.0. The result showed that these proteins belonged to basic protein excepting AbMT. According to available reports, MT as acidic and basic protein tends to be in plant body and animal body, respectively. For example, MTs from Camellia oleifer [18], Limonium sinense [19] and Tamarix androssow [20], whereas MTs from Musca domestica [21], Hyriopsis cumingii [22], Argopecten irradians [23] and so forth. The reason is well worth further research. Hence, these physicochemical indices in Table 1 may be pertinent to determine that they are a group gene with significant functional association and close genetic relation.
3.2. Analysis of Secondary Structure, Subcellular Localization and Some Physiological Functions
The secondary structures of AbMT, GlMT, TcMT and PiMT amino acid sequences were predicted using SOPM online server. Alpha helix, extended strand, beta turn and random coil were found only in AbMT and GlMT, while the first one was not predicted in TcMT and PiMT (Table 2). Thus, the current phenomenon was very worth regarding and further research. The secondary structure analysis showed 4 MT proteins were relatively rich in random coil and extended strand. The very high coil structure (over 50%) was attributed to create links in polypeptide chain and disrupting ordered secondary structure [24].
TMHMM Server online predicted that, there was no transmembrane helices in AbMT, GlMT, TcMT and PiMT proteins, implying that they may exert catalytic function
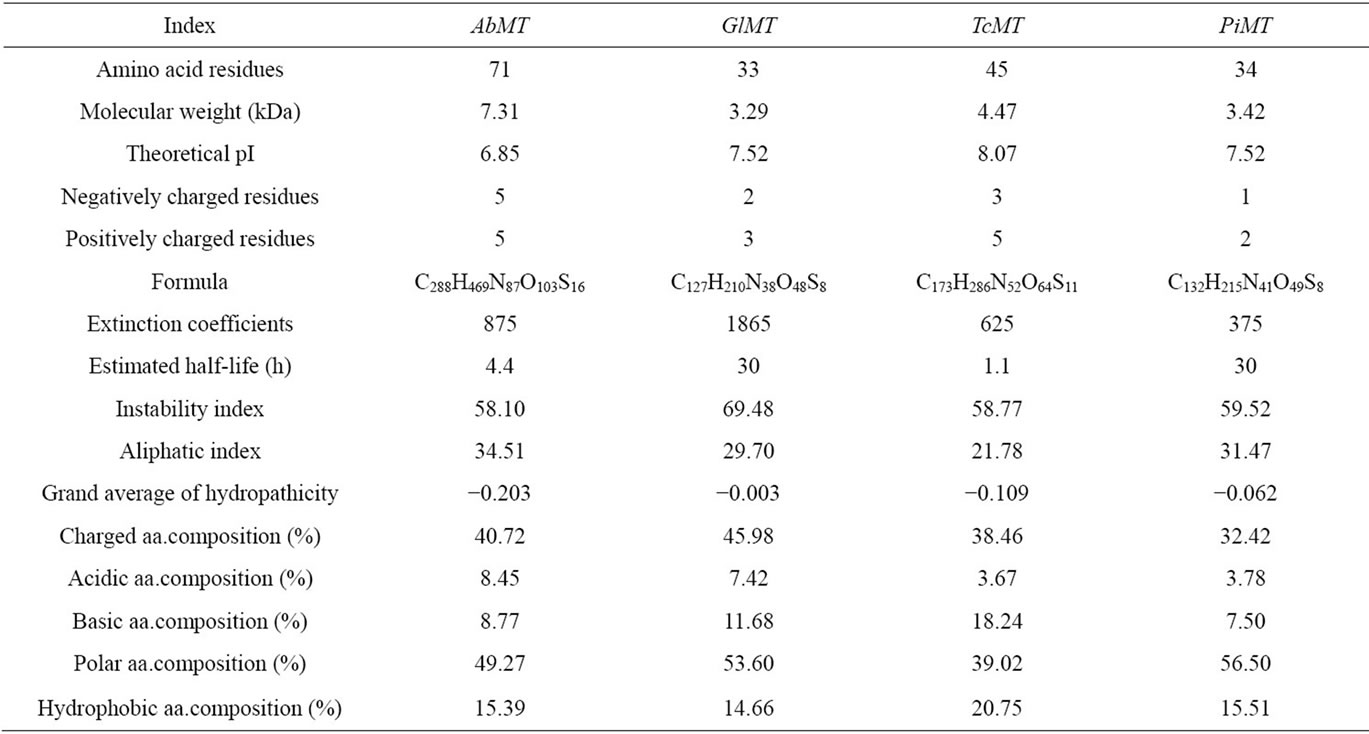
Table 1. Molecular structures and physicochemical properties of AbMT, GlMT, TcMT and PiMT amino acid sequences.
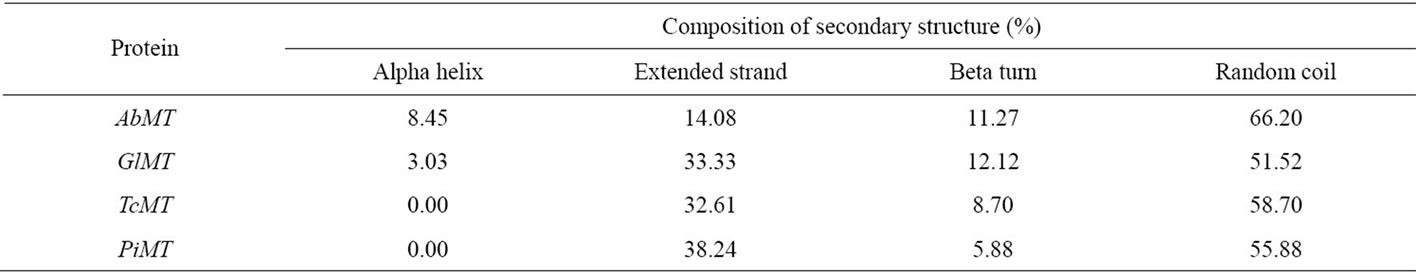
Table 2. Composition of secondary structure of AbMT, GlMT, TcMT and PiMT amino acid sequences.
directly in cytosol without transportation. Simultaneously, the signal peptide was not identified the four amino acid sequences using SignalP 4.0 online server. Server Determining subcellular localization is important as a first step towards studying its physiological function. With the help of PSORT II Prediction, sub-cellular localization analysis of 4 proteins demonstrated that MT was mainly localized in cell nucleus (over 60%), other were cytoplasm, mitochondrion, plasma membrane (other than AbMT) and so forth.
ProtFun 2.2 Server online analysis showed that cellular function of 4 proteins might have some relation to central intermediary metabolism and this laid some evidence to subcellular localization prediction, because central intermediary metabolite occurred usually within the cell [24]. Non N-glycosylated site was recognized, but some putative O-glycosylated sites were predicted in AbMT (8 sites), GlMT (2 sites), TcMT (2 sites) and PiMT (3sites), playing the important role in recognizing and binding some heavy metal ions. And then CDD recognized the absence of conserved domains in these proteins. Furthermore, protein may contain a total of 8 motifs, whose corresponding amino acid regions carry out specific biochemical functions and respective genetic evolutional information [24].
3.3. Analysis of Multiple Alignment and Evolutionary Relationship
We aligned MT gene sequences among A. bisporus, G. lucidum, T. camphoratus and P. involutus using DNAStar (version 5.01) and MEGA (version 3.1) software. The alignments displayed a relatively high degree of homology (more than 52% identity in all the matches), and a relatively short genetic distances (lower than 0.900), especially C-terminal fragment of the cDNA sequences (Table 3). To investigate the evolutionary relationship of these MT genes, a phylogenetic tree was generated by MEGA (version 3.1) software using the Neighbor-Joining method with 1500 bootstrap replicates (Figure 1). The result showed that the genetic relationship coincided
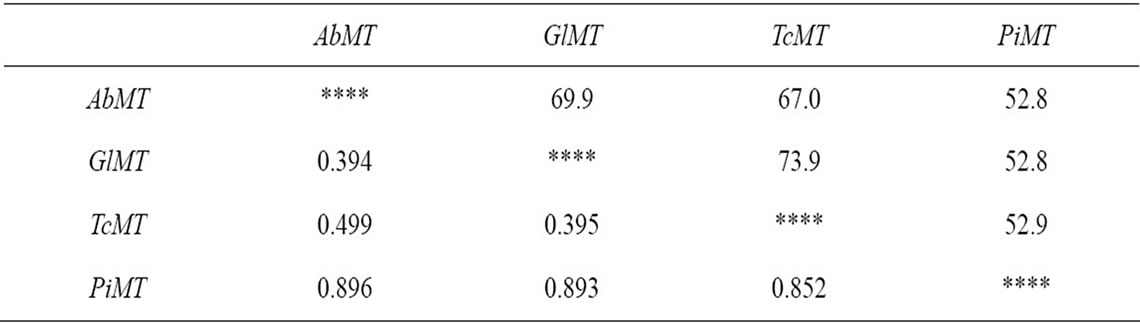
Table 3. Percent identity and genetic distances of AbMT, GlMT, TcMT and PiMT sequences using DNAStar (version 5.01) and MEGA (version 3.1) software.
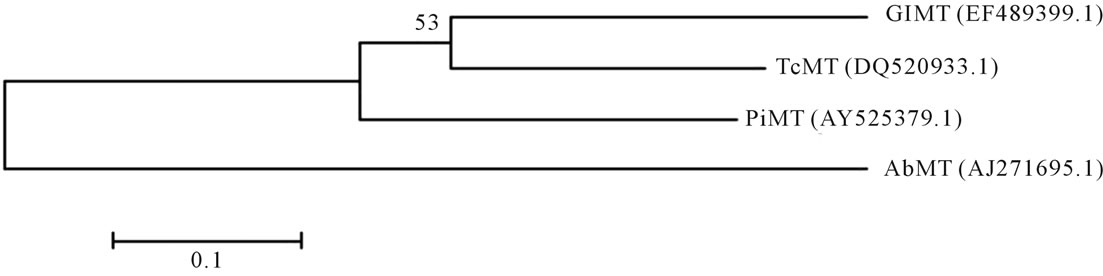
Figure 1. The phylogenetic tree of AbMT, GlMT, TcMT and PiMT was generated from the cDNA sequences using MEGA (version 3.1) software. The reliability of the tree was measured by bootstrap analysis with 1500 replicates.
with the morphological classification and Xie et al. (2007) report [25]. For example, G. lucidum and T. camphoratus belonging to the same order in the morphology were come together in a group. Thus it can be seen that MT gene is one of the most efficient class of molecular marker that has been used widely to detect the deleterious effects of heavy metals [26].
8 monomorphic sites and 94 polymorphic sites were detected from 4 MT gene sequences by the genetic diversity analysis of DnaSP (version 4.0) software. Singleton variable sites and parsimony informative sites was 74 (amounting to 72.55%) and 20 (amounting to 19.61%), respectively. At the same time, 4 haplotypes were also sorted. Haplotype diversity, average number of nucleotide differences and nucleotide diversity was 1.000, 61.00. and 0.59804, respectively. According to calculation using the total number of mutations, there was no significance (P > 0.10) (Fu and Li’s D* test statistic: −0.36481, Fu and Li’s F* test statistic: −0.55017). Codon usage analysis showed that effective number of codons, codon bias index and scaled chi-square was 35.406, 0.794 and 1.730, respectively. Then a strong codon bias was found among 4 MT gene sequences. Multiple nucleotide sequence alignment indicated that number of synonymous sites, nonsynonymous sites, and synonymous sites and non-coding positions was 23.29, 75.71 and 26.29, respectively. Furthermore, we also found that nonsynonymous sites were over three times as much as synonymous sites. Thus, we deduced that 4 MT gene during molecular evolution were under positive selection according to Guo (1993) and Qiang et al. (2010) report [27,28].
In conclusion, as maintaining intracellular metal homeostasis, eliminating metal toxification and protecting against intracellular,MTs may be inducted by mutiple heavy metals at the transcriptional level and need to pay more attention. In the present study, molecular characteristics and physiological functions of MT genes and their corresponding proteins from A. bisporus, G. lucidum, T. camphoratus and P. involutus were analyzed using some bioinformatics methods to provide basis for further investigation.
4. ACKNOWLEDGEMENTS
The authors would like to thank all the laboratory members for their technical advice and helpful discussion. This work was supported by a grant from Agricultural Three New Engineering of Jiangsu Province (No. SX(2011)380). At the same time, we expressed our gratitude to the anonymous reviewers for helpful suggestions and comments to improve the manuscript.
![]()
![]()
REFERENCES
- Hamer, D.H. (1986) Metallothionein. Annual Review of Biochemistry, 55, 913-951. doi:10.1146/annurev.biochem.55.1.913
- Cherian, M.G., Jayasurya, A. and Bay, B.H. (2003) Metallothioneins in human tumors and potential roles in carcinogenesis. Mutation Research, 533, 201-209. doi:10.1016/j.mrfmmm.2003.07.013
- Ren, H.M., Xu, M.X., He, P.F., Muto, N., Itoh, N., Tanaka, K., Xing, J. and Chu, M.M. (2006) Cloning of crucian carp (Carassius cuvieri) metallothionein-II gene and characterization of its gene promoter region. Biochemical and Biophysical Research Communications, 342, 1297- 1304. doi:10.1016/j.bbrc.2006.02.082
- Huang, G.Y. and Wang, Y.S. (2009) Expression analysis of type 2 metallothionein gene in mangrove species (Bruguiera gymnorrhiza) under heavy metal stress. Chemosphere, 77, 1026-1029. doi:10.1016/j.chemosphere.2009.07.073
- Huang, G.Y. and Wang, Y.S. (2010) Expression and characterization analysis of type 2 metallothionein from grey mangrove species (Avicennia marina) in response to heavy metal stress. Aquatic Toxicology, 99, 86-92. doi:10.1016/j.aquatox.2010.04.004
- Mejáre, M. and Bülow, L. (2001) Metal-binding proteins and peptides in bioremediation and phytoremediation of heavy metals. Trends in Biotechnology, 19, 67-73. doi:10.1016/S0167-7799(00)01534-1
- Schroeder, J.J. and Cousins, R.J. (1990) Interleukin 6 regulates metallothionein gene expression and zinc metabolism in hepatocyte monolayer cultures. Proceedings of the National Academy of Sciences, 87, 3137-3141. doi:10.1073/pnas.87.8.3137
- Haq, F., Mahoney, M. and Koropatnick, J. (2003) Signaling events for metallothionein induction. Mutation Research, 533, 211-226. doi:10.1016/j.mrfmmm.2003.07.014
- Akashi, K., Nishimura, N., Ishida, Y. and Yokota, A. (2004) Potent hydroxyl radical-scavenging activity of drough-induced type-2 metallothionein in wild watermelon. Biochemical and Biophysical Research Communications, 323, 72-78. doi:10.1016/j.bbrc.2004.08.056
- Zhou, G.K., Xu, Y.F., Li, J., Yang, L.Y. and Liu, J.Y. (2006) Molecular Analyses of the Metallothionein Gene Family in Rice (Oryza sativa L.). Journal of Biochemistry and Molecular Biology, 39, 595-606. doi:10.5483/BMBRep.2006.39.5.595
- Cai, L., Koropatnick, J. and Cherian, M.G. (1995) Metallothionein protects DNA from copper-induced but not iron-induced cleavage in vitro. Chemistry & Biology, 96, 143-155. doi:10.1016/0009-2797(94)03585-V
- Hafiz, F., Begum, M., Parveen, S., Nessa, Z. and Azad, A.K.M. (2003) Study of edible mushroom grown on Eucalyptus Camaldulensis trunk and under the soil of Albizzia Procera. Pakistan Journal of Nutrition, 2, 279-282. doi:10.3923/pjn.2003.279.282
- Ita, B.N., Ebong, G.A., Essien, J.P. and Eduok, S.I. (2008) Bioaccumulation potential of heavy metals in edible fungal sporocarps from the Niger Delta Region of Nigeria. Pakistan Journal of Nutrition, 7, 93-97. doi:10.3923/pjn.2008.93.97
- Das, N. (2005) Heavy metal biosorption by mushrooms. Natural Product Radiance, 4, 454-459.
- Das, N., Vimala, R. and Karthika, R. (2008) Biosorption of heavy metal—An overview. Indian Journal of Biotechnology, 7, 159-169.
- Zhu, F.K., Qu, L., Fan, W.X., Qiao, M.Y., Hao, H.L. and Wang, X.J. (2011) Assessment of heavy metals in some wild edible mushrooms collected from Yunnan Province, China. Environmental Monitoring and Assessment, 179, 191-199. doi:10.1007/s10661-010-1728-5
- Fuminori, O. (2001) Molecular mechanism of the metallothionein gene expression mediated by metal-responsive transcription factor 1. Journal of Health Science, 47, 513- 519. doi:10.1248/jhs.47.513
- Jiang, Y., Tan, X.F., Zhang, D.Q. and Chen, H.P. (2009) Cloning and sequence analysis of a metallothionein gene from Camellia oleifera. Acta Agriculturae Universitati, 31, 699-705.
- Liu, W.Q., Ni, D.J., Song, L.S., Wu, L.T., Xu, W. and Kong, X.Y. (2006) Cloning and characterization of a metallothionein gene in bay scallop, Argopecten irradians. Oceanologia et Limnologia Sinica, 37, 444-449.
- Zhang, Y., Yang, C.P. and Wang, Y.C. (2007) Cloning and sequence analysis of metallothionein gene from Tamarix androssowii. Bulletin of Botanical Research, 27, 293-296.
- Zhang, D., Ren, G.D., Tang, T., Dong, X.Y. and Liu, F.S. (2010) Cloning, prokaryotic expression and activity detection of the metallothionein gene in Musca domestica. Acta Entomologica Sinica, 53, 379-384.
- Yuan, Y.M., Wang, G.L. and Li, J.L. (2009) Cloning and sequence analysis of metallothionein gene in Hyriopsis cumingii. Chinese Journal of Zoology, 44, 98-104.
- Liu, Y., Chen, S.H., Yin, H.B., Li, L.X., Zhao, J.Q., Zhang, Y., Han, H.L. and Guo, S.L. (2011) Cloning and analysis of metallothionein gene from Limonium sinense Kuntze. Journal of Yantai University (Natural Science and Engineering Edition), 24, 121-125.
- Lei, W., Zhang, J.D. and Qiao, A.M. (2011) Bioinformatics data mining on UDP-glucose: Flavonoid 7-O-glucosyltransferase (UBGAT) genes and their encoding proteins in two plants of genus Scutellaria. African Journal of Biotechnology, 10, 4339-4346.
- Xie, D.Y., Zhu, W., Wu, Z.J., Lin, Q.Y. and Xie, L.H. (2007) Cloning and sequence analysis of metallothionein gene from Garnoderma lucidum. Chinese Agricultural Science Bulletin, 23, 87-90.
- Lin, C.H., John, J.A.C., Ou, L.W., Chen, J.C., Lin, C.H. and Chang, C.Y. (2004) Cloning and characterization of metallothionein gene in ayu Plecoglossus altivelis. Aquatic Toxicology, 66, 111-124.
- Guo, Z.P. (1993) Introduction to population genetics. Agricultural Press, Beijing, 298-332.
- Qiang, C.K., Feng, W.J., Zhao, H., Su, X.L. and Zhou, B.Y. (2010) In silico cloning, sequence analysis and electronic pattern of polyubiquitin gene in the silkworm, Bombyx mori (Lepdoptera: Bombycidae). Genomics and Applied Biology, 29, 783-790.

