Open Journal of Modelling and Simulation
Vol.05 No.01(2017), Article ID:73291,15 pages
10.4236/ojmsi.2017.51003
Identifying Critical Parameters in SIR Model for Spread of Disease
Sarbaz H. A. Khoshnaw1, Najem A. Mohammad2, Rizgar H. Salih3
1Department of Mathematics, University of Raparin, Ranya, Kurdistan Region, Iraq
2Department of Mathematics, Koya University, Koy Sanjaq, Kurdistan Region, Iraq
3Faculty of Science, University of Raparin, Ranya, Kurdistan Region, Iraq

Copyright © 2017 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: October 30, 2016; Accepted: January 2, 2017; Published: January 5, 2017
ABSTRACT
Calculating analytical approximate solutions for non-linear infectious disease models is a difficult task. Such models often require computational tools to analyse analytical approximate methods which appear in some theoretical and practical applications in systems biology. They represent key critical elements and give some approximate solutions for such systems. The SIR epidemic disease model is given as the non-linear system of ODE’s. Then, we use a proper scaling to reduce the number of parameters. We suggest Elzaki transform method to find analytical approximate solutions for the simplified model. The technique plays an important role in calculating the analytical approximate solutions. The local and global dynamics of the model are also studied. An investigation of the behaviour at infinity was conducted via finding the lines and singular points at infinity. Model dynamic results are computed in numerical simulations using Pplane8 and SimBiology Toolbox for Mathlab. Results provide a good step forward for describing the model dynamics. More interestingly, the simplified model could be accurate, robust, and used by biologists for different purposes such as identifying critical model elements.
Keywords:
Mathematical Modelling, Epidemic SIR Disease Model, Stability Analysis, Analytical Approximate Solutions, Sensitivity Analysis

1. Introduction
A system of kinetic equations can be applied to describe the behaviours of biochemical kinetics. The system is obtained from the reaction mechanisms by the law of mass action. The rate constant of any given elementary reaction, known as the proportionality constant, depends on the reaction condition (temperature, solvent, PH, etc.). The reaction conditions are generally held by biochemists to avoid dealing with higher-order complexities [1] [2] [3] .
An arbitrary number 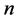 of elementary reactions can be expressed as follows:
of elementary reactions can be expressed as follows:
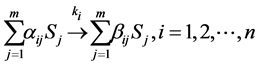 (1)
(1)
where 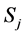 is the
is the  species and
species and 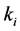 is the rate constant of step
is the rate constant of step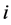 . The non-negative integers
. The non-negative integers 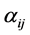 and
and 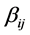 are the stoichiometric coefficients for species j occurring as a reactant and product, respectively, in the
are the stoichiometric coefficients for species j occurring as a reactant and product, respectively, in the 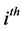 reaction step. The rate of product of species j can be given by:
reaction step. The rate of product of species j can be given by:
 (2)
(2)
where 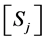 is the concentration of
is the concentration of 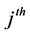 species and
species and 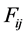 is the rate of change of
is the rate of change of 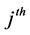 species in reaction
species in reaction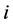 . A reaction rate
. A reaction rate  is given by the law of mass action as follows:
is given by the law of mass action as follows:
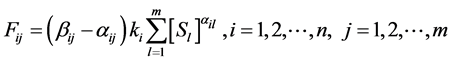 (3)
(3)
The readers can see further details about the chemical kinetics in [4] - [12] .
The idea of sensitivity analysis has been used in dynamical analysis of biochemical kinetics and systems biology models. The concept of sensitivity analysis theory in application to chemical kinetic problems was given by Rabitz in [13] . The local and global methods of sensitivity analysis in chemical kinetics were further studied in [14] . There are also some applications of sensitivity analysis of systems biology models in [15] - [21] . More recently, the methods of local sensitivity analysis have been used to identify the critical model elements for a kinetic model of nuclear receptor signalling, the reader can see further details about the method in [10] [11] . This method is applied to determine which variable or parameter is sensitive to a particular condition which is defined by a variable or parameter. The system of ODEs discussed here is:
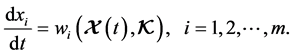 (4)
(4)
The model input 



The general form of the local sensitivity is given as a Jacobian matrix as follows

where the matrices 

The initial conditions of the Equation (5) are determined by the input parameter 

The idea of ELzaki transform was introduced by Tarig Elzaki for solving linear ordinary differential equations with constant coefficients. Then, this became more popular and works as a powerful tool in mathematics for solving some complicated equations, for example, solving high order ordinary differential equations with variable coefficients [22] , applying ELzaki transform for solving the integro-differential equations [23] , showing some relationships between Laplace transform and the ELzaki transform [24] , solving linear systems of integro-differential equations with constant coefficients [25] , a solution for nonlinear systems of differential equations using a mixture of Elzaki transform [26] .
The SIR model, first published by Kermack and McKendrick in 1927 [27] , is surly the most well known mathematical model for the spread of an infectious disease. Then, the model was further developed mathematically in [28] [29] . Here, people are divided into three classes: susceptible S, infective I and removed R. Removed individuals are no longer susceptible nor infective for whatever reason. For instance, they have recovered from the disease or they have been vaccinated. They may have been isolated from the rest of the people [30] [31] [32] . After that further explanation of the model was proposed by the authors of these papers [33] [34] [35] [36] [37] .
The main problem in this study is identifying the critical model parameters. Therefore, the aim in this work is to apply some mathematical tools to simplify and analyse the model and then identifying the model elements (variables and parameters). We proposes a number of steps of model analysis, which plays in reducing the number of elements and in calculating analytical approximate solutions of the SIR model. The proposed steps and their advantages are simply given. The first step is that a proper scaling is used in order to minimize the number of elements. This becomes a good step forward for simplifying the original model. Another step is calculating some analytical approximate solutions using Elzaki transformation. Furthermore, using local sensitivity method is another important step in this study. This helps us to identify critical model parameters of the reduced model. Finally, studying the behaviour at infinity of the reduced model provide us an understanding of global dynamics and drawing the global phase portrait of the system.
2. The SIR Epidemic Disease Model
The SIR model may be diagrammed as in Figure 1. The chemical reactions of the model are given

where 


Figure 1. Scheme of the SIR infectious diseases model. Geometric shapes here represent compartments, and arrows indicate flux between the compartments.
with initial conditions

Equation (7) has a conservation law

where 
The dimensionless SIR equations are then given by

where 
Therefore, the conservation law (9) takes the form

In addition, the Equation (10) is then reduced to

with initial conditions 

Many nonlinear systems of differential equations have not exact solutions. Calculating analytical approximate solutions for such systems is a difficult task. The SIR epidemic disease model is given as the non-linear system of ODE’s. Equation (12) can not be solved analytically. Therefore, calculating some analytical approximate solutions for the system provides more information about the behaviours of the model dynamics. Applying the idea of Elzaki transform, we can obtain some analytical approximate solutions of the Equation (12). Take Elzaki transform of Equation (12) to get:

Take the inverse Elzaki transform of Equation (13), the recursive relations are then given by

where

For

For

where,

For

where
In general, a series form of the model solution can be given

3. Proposed Steps of Model Analysis and Simplification
In this work, we propose some steps of model analysis and simplification for the SIR epidemic disease model. The suggested steps are used for minimizing the number of elements (variables and parameters), calculating analytical approximate solutions and identifying critical model parameters. The proposed steps in this study are presented below:
1) Define chemical mechanisms of SIR Model.
2) Define a kinetic model of biochemical reactions as a system of ODEs using the mass action law.
3) Eliminate some variables based on the stoichiometric conservation laws, and use proper scaling for the kinetic equations and determine the minimal number of parameters.
4) Apply Elzaki transform method to calculate analytical approximate solutions of the reduced model.
5) Simulate the reduced model dynamics for different parameter values using Pplane8 for Mathlab. Analyse the reduced models to identify the critical model parameters by the local sensitivity analysis in numerical simulations using the SimBiology Toolbox for Mathlab.
6) Study the global behaviours of the reduced model at infinity.
The above steps can be simply presented in the following flowchart:
The flowchart of proposed steps of model analysis and simplification, the steps are presented in the order of their application.
4. Local Behaviours of the Simplified Model
We use Pplane8 for Mathlab to study the stability analysis of the Equation (12) and to compute numerical simulations in 
Furthermore, we identify the model interacting and numerical simulations in two and three dimensional planes, for different values of

In addition, we calculate the local sensitivity of state variables 




Figure 2. Dynamics of the Equation (12) and numerical simulations using Pplane8 for Mathlab in 

Figure 3. Dynamics of the simplified model and numerical simulations using Pplane8 for Mathlab in two dimensional plane, for different values of parameter
they are less sensitive to the given parameter 




5. The Global Behaviours of the Simplified Model
This section is devoted to examine the global phase portrait for Equation (12) by studying the isoclines and the behaviours at infinity. The isoclines are the lines with equal slope. These lines are an important role in sketching the phase portrait. It is easy to know where the trajectories have vertical and horizontal tangent lines by finding the
isoclines for 













Here, we study the direction of trajectories in the quadrants. In the first quadrant,
Figure 4. Dynamics of the simplified model and numerical simulations using Pplane8 for Mathlab in three dimensional plane, for different values of parameter
Figure 5. The local sensitivity of state variables 



since 













The second required one is the line at infinity. It is a projective line that is added to the affine plane. Finding them including singular points and studying the behaviours at infinity of the reduced Equation (12) is very important to an understanding its global dynamics. For this purpose, the two below nonlinear change of variables are used individually.


Salih in [38] have used above transformation in three dimensional case. The points 



Applying the nonlinear change of variables (20) on the reduced Equation (12) and after rescaling of the variables, the new system is obtained

The above system has two singular points 




with a positive 


with two negative eigenvalues 

The system below is obtained by applying the nonlinear change of variables Equation (21) on Equation (12) after rescaling of variables

System (25) has only two singular points 



which it has one negative eigenvalue 


which it has two positive eigenvalues 

6. Conclusion
Mathematical presentations and numerical simulations of infectious disease models are crucial topics in systems biology. Describing the dynamics of such systems often requires some techniques of model analysis. We studied an epidemic disease model called SIR model with three species and two parameters. We proposed a number of steps of
Figure 6. Global phase portraits of the simplified Equation (12); the green solid curves depict the line at infinities, the green dotted lines depict unstable singular points and the red dotted line depict stable singular point.
model analysis. The suggested steps of model analysis here importantly play in reducing the number of elements and in calculating analytical approximate solutions of the SIR model. The simplified model helps to study the full model of SIR in different ways. Firstly, identifying the critical model parameters of the reduced model becomes much easier compared to the full model. Secondly, the mathematical representation of the reduced model can help to integrate experimental knowledge into a coherent picture. Furthermore, studying the behaviour at infinity of the reduced model helped us to understand its global dynamics and to draw the global phase portrait of the system. Finally, the reduced model could be accurate, robust, and applied by biologists for various purposes. The proposed techniques here of model analysis will be applied to a wide range of complex infectious disease models in systems biology.
Acknowledgements
We thank the Editor and the referee for their comments. We also thank both Raparin and Koya universities for providing us with wonderful facilities. This support is greatly appreciated.
Cite this paper
Khoshnaw, S.H.A., Mohammad, N.A. and Salih, R.H. (2017) Identifying Critical Parameters in SIR Model for Spread of Disease. Open Journal of Modelling and Simulation, 5, 32-46. http://dx.doi.org/10.4236/ojmsi.2017.51003
References
- 1. Briggs, G.E. and Haldane, J.B.S. (1925) A Note on the Kinetics of Enzyme Action. Biochemical Journal, 19, 338-339.
https://doi.org/10.1042/bj0190338 - 2. Semenoff, N. (1939) On the Kinetics of Complex Reactions. The Journal of Chemical Physics, 7, 683-699.
https://doi.org/10.1063/1.1750515 - 3. Segel, L.A. and Slemrod, M. (1989) The Quasi-Steady-State Assumption: A Case Study in Perturbation. SIAM Review, 31, 446-477.
https://doi.org/10.1137/1031091 - 4. Gorban, A.N., Radulescu, O. and Zinovyev, A.Y. (2010) A Symptotology of Chemical Reaction Networks. Chemical Engineering Science, 65, 2310-2324.
https://doi.org/10.1016/j.ces.2009.09.005 - 5. Gorban, A. and Radulescu, O. (2008) Dynamic and Static Limitation in Multiscale Reaction Networks, Revisited. Advances in Chemical Engineering, 34, 103-173.
https://doi.org/10.1016/S0065-2377(08)00003-3 - 6. Gorban, A.N. and Karlin, I.V. (2003) Method of Invariant Manifold for Chemical Kinetics. Chemical Engineering Science, 58, 4751-4768.
https://doi.org/10.1016/j.ces.2002.12.001 - 7. Rao, S., Van der Schaft, A., Van Eunen, K., Bakker, B.M. and Jayawardhana, B. (2014) A Model Reduction Method for Biochemical Reaction Networks. BMC Systems Biology, 8, 1-11.
https://doi.org/10.1186/1752-0509-8-52 - 8. Radulescu, O., Gorban, A.N., Zinovyev, A. and Noel, V. (2012) Reduction of Dynamical Biochemical Reaction Networks in Computational Biology. arXiv preprint arXiv:1205.2851.
- 9. Radulescu, O., Gorban, A.N., Zinovyev, A. and Lilienbaum, A. (2008) Robust Simplifications of Multiscale Biochemical Networks. BMC Systems Biology, 2, 86-97.
https://doi.org/10.1186/1752-0509-2-86 - 10. Khoshnaw, S.H.A. (2015) Model Reductions in Biochemical Reaction Networks. Ph.D. thesis, Department of Mathematics, University of Leicester, UK.
- 11. Khoshnaw, S.H. (2015) Reduction of a Kinetic Model of Active Export of Importins. AIMS Conference on Dynamical Systems, Differential Equations and Applications, Madrid, 7-11 July 2015, 705-722.
https://doi.org/10.3934/proc.2015.0705 - 12. Khoshnaw, S.H. (2013) Iterative Approximate Solutions of Kinetic Equations for Reversible Enzyme Reactions. Natural Science, 5, 740-755.
https://doi.org/10.4236/ns.2013.56091 - 13. Rabitz, H. (1981) Chemical Sensitivity Analysis Theory with Applications to Molecular Dynamics and Kinetics. Computer and Chemistry, 5, 167-180.
https://doi.org/10.1016/0097-8485(81)80104-0 - 14. Rabitz, H., Kramer, M. and Dacol, D. (1983) Sensitivity Analysis in Chemical Kinetics. Annual Review of Physical Chemistry, 34, 419-461.
https://doi.org/10.1146/annurev.pc.34.100183.002223 - 15. Ihekwaba, A.E.C., Broomhead, D.S., Grimley, R.L., Benson, N. and Kell, D.B. (2004) Sensitivity Analysis of Parameters Controlling Oscillatory Signalling in the NF-kappaB Pathway: The Roles of IKK and IkappaBalpha. Systems Biology, 1, 93-103.
https://doi.org/10.1049/sb:20045009 - 16. Hu, D. and Yuan, J.M. (2006) Time-Dependent Sensitivity Analysis of Biological networks: Coupled MAPK and PI3K Signal Transduction Pathways. The Journal of Physical Chemistry A, 110, 5361-5370.
https://doi.org/10.1021/jp0561975 - 17. Wu, W.H., Wang, F.S. and Chang, M.S. (2008) Dynamic Sensitivity Analysis of Biological Systems. BMC Bioinformatics, 9, S17.
https://doi.org/10.1186/1471-2105-9-s12-s17 - 18. Perumal, T.M. and Gunawan, R. (2011) Understanding Dynamics Using Sensitivity Analysis: Caveat and Solution. BMC Systems Biology, 5, 41.
https://doi.org/10.1186/1752-0509-5-41 - 19. Charzynska, A., Nalecz, A., Rybinski, M. and Gambin, A. (2012) Sensitivity Analysis of Mathematical Models of Signalling Pathways. Journal of Biotechnology, Computational Biology and Bionanotechnology, 93, 291-308.
- 20. Sumner, T., Shephard, E. and Bogle, I. (2012) A Methodology for Global-Sensitivity Analysis of Time-Dependent Outputs in Systems Biology Modelling. Journal of the Royal Society Interface, 9, 2156-2166.
https://doi.org/10.1098/rsif.2011.0891 - 21. Azam, M., Bhatti, A., Arshad, A. and Babar, M. (2013) Sensitivity Analysis of Wnt Signaling Pathway. 10th International Bhurban Conference on Applied Sciences and Technology (IBCAST), Islamabad, 15-19 January 2013, 122-127.
https://doi.org/10.1109/ibcast.2013.6512143 - 22. Elzaki, T.M. (2011) On the Elzaki Transform and Ordinary Differential Equation with Variable Coefficients. Advances in Theoretical and Applied Mathematics, 6, 13-18.
- 23. Elzaki, T.M. and Ezaki, S.M. (2011) Solution of Integro-Differential Equations by Using ELzaki Transform. Global Journal of Mathematical Sciences: Theory and Practical, 3, 295-303.
- 24. Elzaki, T.M. (2011) On the Connections between Laplace and Elzaki Transforms. Advances in Theoretical and Applied Mathematics, 6, 1-11.
- 25. Elzaki, T.M. and Ezaki, S.M. (2011) On the Solution of Integro-Differential Equation Systems by Using ELzaki Transform. Global Journal of Mathematical Sciences: Theory and Practical, 3, 13-23.
- 26. Elzaki, T.M. (2012) A Solution for Nonlinear Systems of Differential Equations Using a Mixture of Elzaki Transform and Differential Transform Method. International Mathematical Forum, 7, 625-630.
- 27. Kermack, W.O. and McKendrick, A.G. (1927) A Contribution to the Mathematical Theory of Epidemics. Proceedings of the Royal Society of London A, 115, 700-721.
https://doi.org/10.1098/rspa.1927.0118 - 28. Kermack, W.O. and McKendrick, A.G. (1933) Contributions to the Mathematical Theory of Epidemics. III. Further Studies of the Problem of Endemicity. Proceedings of the Royal Society of London A, 141, 94-122.
https://doi.org/10.1098/rspa.1933.0106 - 29. Capasso, V. and Serio, G. (1978) A Generalization of the Kermack-McKendrick Deterministic Epidemic Model. Mathematical Biosciences, 42, 43-61.
https://doi.org/10.1016/0025-5564(78)90006-8 - 30. Hethcote, H.W. (2000) The Mathematics of Infectious Diseases. SIAM Review, 42, 599-653.
https://doi.org/10.1137/S0036144500371907 - 31. Diekmann, O. and Heesterbeek, J. (2000) Mathematical Epidemiology of Infectious Diseases: Model Building, Analysis and Interpretation. John Wiley & Sons, Hoboken.
- 32. Stone, L., Shulgin, B. and Agur, Z. (2000) Theoretical Examination of the Pulse Vaccination Policy in the SIR Epidemic Model. Mathematical and Computer Modelling, 31, 207-215.
https://doi.org/10.1016/S0895-7177(00)00040-6 - 33. Korobeinikov, A. and Maini, P.K. (2004) A Lyapunov Function and Global Properties for SIR and SEIR Epidemiological Models with Nonlinear Incidence. Mathematical Biosciences and Engineering, 1, 57-60.
https://doi.org/10.3934/mbe.2004.1.57 - 34. Korobeinikov, A. (2007) Global Properties of Infectious Disease Models with Nonlinear Incidence. Bulletin of Mathematical Biology, 69, 1871-1886.
https://doi.org/10.1007/s11538-007-9196-y - 35. Keeling, M.J. and Rohani, P. (2008) Modelling Infectious Diseases in Humans and Animals. Princeton University Press, Princeton.
- 36. Vynnycky, E. and White, R. (2010) An Introduction to Infectious Disease Modelling. Oxford University Press, Oxford.
- 37. Harko, T., Lobo, F.S. and Mak, M. (2014) Exact Analytical Solutions of the Susceptible-Infected-Recovered (SIR) Epidemic Model and of the SIR Model with Equal Death and Birth Rates. Applied Mathematics and Computation, 236, 184-194.
https://doi.org/10.1016/j.amc.2014.03.030 - 38. Salih, R.H. (2015) HOPF Bifurcation and Center Bifurcation in Three Dimensional Lotka-Volterra Systems. PhD Thesis, Plymouth University, Plymouth.
- 39. Neto, A.L. (1345) Algebraic Solutions of Polynomial Differential Equations and Foliations in Dimension Two. In: Gomez-Mont, X., Seade, J.A. and Verjovski, A., Eds., Holomorphic Dynamics, Springer, Berlin, 192-232.




















