American Journal of Plant Sciences
Vol.4 No.8A(2013), Article ID:35902,14 pages DOI:10.4236/ajps.2013.48A003
Biodiversity Assessment of Sugar Beet Species and Its Wild Relatives: Linking Ecological Data with New Genetic Approaches*
![]()
1Faculdade de Ciências, Universidade de Lisboa, BioFIG, Campo Grande, Lisboa, Portugal; 2Instituto de Investigação Científica Tropical, JBT & CIFC/BIOTROP, Travessa Conde da Ribeira, Lisboa, Portugal; 3Universidade do Porto, CIBIO, Centro de Investigação em Biodiversidade & Recursos Genéticos, Vairão, Portugal.
Email: mromeiras@yahoo.co.uk
Copyright © 2013 Filipa Monteiro et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received June 7th, 2013; revised July 7th, 2013; accepted July 31st, 2013
Keywords: Crop Wild Relatives; Beta vulgaris subsp. maritima; Single Nucleotide Polymorphisms (SNPs); Western Mediterranean Region; EcoTILLING
ABSTRACT
The value of crop wild relatives has long been acknowledged and this wild resource has been used to improve crop performance with clear economic benefits. Sugar beet (Beta vulgaris subsp. vulgaris) is the most economically valuable crop species in the order Caryophyllales, B. vulgaris subsp. maritima being the ancestor of the cultivated beets. The wild species of the genus Beta s.l. are commonly found in coastal areas of Europe and Mediterranean Region, where a rich genetic heritage still exists. Broadening the genetic base of sugar beet by introgression with wild relatives is a growing need regarding the maintenance of ecologically important traits. Since wild relatives have adapted to specific habitats, they constitute an important source of novel traits for the beet breeding pool. So, we conducted a broader research project aiming to delimit taxa and identify priority locations to establish genetic reserves of the wild Beta species occurring in Portugal (Western Iberian Peninsula). The aim of this study was: 1) to identify and characterize the main habitats of these wild Beta species; and 2) to present a review of some genetic tools available for future application in sugar beet breeding. In this review, we have focused on EcoTILLING as a molecular tool to assess DNA polymerphisms in wild populations of Beta and identify candidate genes related to drought and salt tolerance, as well as addressed some issues related to next-generation sequencing (NGS) technologies as a new molecular tool to assess adaptive genetic variation on the wild relatives of sugar beet.
1. Introduction
Crop Wild Relatives (CWR) are species closely related to crops that hold greater genetic variation in nature than their cultivated relatives selected by man, thus representing an important reservoir of genetic resources for crop improvement [1-3]. The CWR are primary source of germplasm bearing novel resistant traits to pests and diseases, and abiotic stresses, such as drought and salinity, and have been extensively used in breeding programs of major crops, namely wheat (Triticum aestivum L.), maize (Zea mays L.), rice (Oryza sativa L.), barley (Hordeum vulgare L.), potato (Solanum tuberosum L.), and cassava (Manihot esculenta Crantz) [4]. The conservation of wild relatives of common crops is therefore an imperative condition to preserve a wider gene pool which can be unlocked to improve future food security.
The gene pool provided by CWR specifically adapted to marginal or stress environments constitutes an invaluable resource for improved crop breeding under the new challenges, namely to adapt crops to grow in the presently changing climate. As recognized, the domestication process has often caused genetic bottlenecks, leaving crop plants with limited genetic diversity [5], and thus poorly equipped to respond to the biotic and abiotic pressures that are rapidly evolving and having an increasingly detrimental effect on crop productivity. In contrast, the wild relatives of crops are expected to harbor much higher levels of genetic diversity and specifically, beneficial adaptive genes/alleles underlying processes of local adaptation. The use of CWR genes to improve crop performance is not by far a new concept, but dating back more than 60 years [1]. During this period, significant developments are achieved, each, in its own right, making important contributions to the knowledge base capable to provide a predictive framework that will underpin conventional and novel plant breeding strategies.
1.1. Europe as an Important Centre for Crop Wild Diversity
Kell et al. [6] created the first comprehensive list of CWR for Europe and the Mediterranean using a broad definition of a CWR (i.e., any species belonging to the same genus or closely related genera of a cultivated plant species), and found that approximately 80% of the flora of the region consisted of crops and their wild relatives. Due to this huge diversity hotspot, in Europe there are a number of initiatives to study and catalogue the CWR species, such as the “International Database for Beta” (IDBB) (http://idbb.jki.bund.de), “Beta Working Group” from the European Cooperative Programme for Plant Genetic Resources (ECPGR) (http://www.ecpgr.cgiar.org/networks/sugar_starch_fibre_crops/beta.html), European platform “An Integrated European in situ Management Work Plan: Implementing Genetic Reserves and On Farm Concepts” (AEGRO) (http://aegro.jki.bund.de/aegro/), or the CWR Global Portal (www.cropwildrelatives.org). In 2005, the European Community funded a project, Plant Genetic Resources (PGR) Forum (www.pgrforum.org), to provide access to CWR data to a broad user community [7]. Furthermore, the recently published European Red List of Vascular Plants [8] identified that at least 16% of the CWR species assessed at European level for which sufficient data were available, were threatened or were likely to become threatened in the near future, and that many more were threatened at national level.
The CWR do not spread evenly across the world, but are concentrated in relatively small regions often referred to as “centres of food crop diversity” [9]. Europe is one of the most important centres for CWR diversity, namely including oats (Avena sativa L.), apple (Malus domestica Borkh.), annual meadow grass (Festuca pratensis Huds.), white clover (Trifolium repens L.), and sugar beet (Beta vulgaris L. subsp. vulgaris) [10]. Among these crops, sugar beet is considered within the top ten of the world rank in economic importance, growing on about 5.2 Mha in 38 countries, and supplying around 20% of the 167 Mt sugar produced annually, with sugar cane (Saccharum officinarum L.) supplying most of the remainder [11]. Half of its production is originated from countries in the European Union, complementing sugar cane production from tropical climates [12].
1.2. Objective of This Study
The interest in the wild relatives of important crops for breeding in the face of environmental change means that a species-level understanding of the identity and relationships of these taxa is of more than marginal interest [13]. So, research was within a broader research project aiming to delimit taxa, select appropriate wild accessions, and identify priority locations in which to establish genetic reserves of the wild Beta species occurring in Portugal (SW Europe). The aim of this study was 1) to identify ecotypes of these wild Beta species; and 2) to present a review of some genetic tools available for future application in sugar beet breeding. In this review, we have focused on EcoTILLING as a molecular tool to assess DNA polymorphisms in wild populations and identify candidate genes related to drought and salt tolerance, as well as addressed some issues related to next-generation sequencing (NGS) technologies, as a new molecular tool to assess the genetic variation of sugar beet CWR species.
2. Wild Beet from Portugal Mainland
Beta vulgaris (Family Amaranthaceae) includes the cultivated beets (subsp. vulgaris) (leaf beet, garden beet, fodder and sugar beet) and the wild “sea beet” (subsp. maritima), fully cross-compatible with each other [14]. Sea beet is the most widespread taxon within the genus Beta, and can be found quite easily along the seashores of the Mediterranean Sea and the European Atlantic Ocean [11]. In east Atlantic coasts of Europe, this species displays a wide latitudinal range, which ranges from southern Norway and southern Sweden, to the South of Portugal.
In Portugal mainland the following species can be found: B. vulgaris subsp. maritima, B. macrocarpa, and Patellifolia patellaris, formerly considered as B. patellaris. The genus Patellifolia was formerly included in section Procumbentes of the Beta genus in several taxonomic classifications [e.g. 15,16]. In the present paper, we will use the taxonomic system as described by Hohmann et al. [17], which was based on molecular studies indicating that the section Procumbentes should be separated from the genus Beta. Table 1 gives the distribution of Beta and Patellifolia species, which occur in the West Mediterranean Region, including the Macaronesian Re-
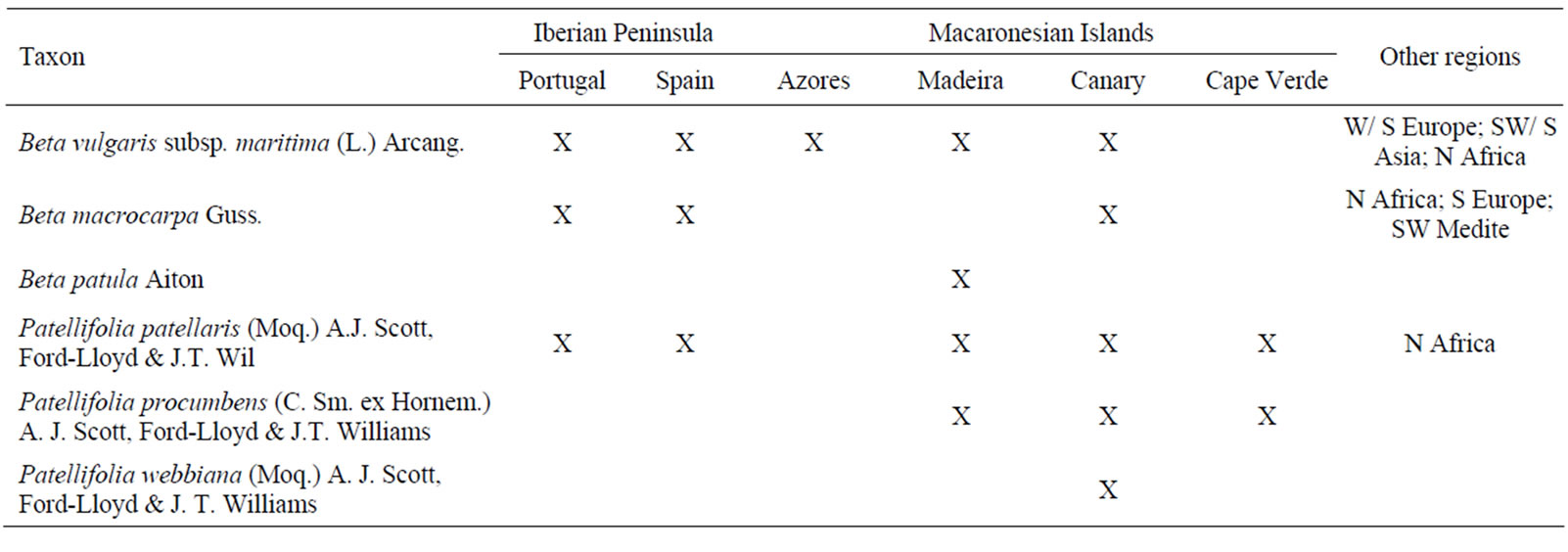
Table 1. Distribution of Beta and Patellifolia species, which occurs in west mediterranean region and macaronesian islands.
gion (i.e. Azores; Madeira and Selvages; Canaries; and Cape Verde Islands). Although a rich genetic heritage of wild Beta s.l. species still exists in the coastal areas of Europe, Mediterranean Region and in the Macaronesian Islands, many of the natural habitats where these plants occur are currently threatened by habitat reduction and loss, or fragmentation and disturbance, and global changes leading to the restriction of the natural distribution range and the genetic diversity decrease [2].
2.1. Geographical Distribution and Ecology
In order to update the information about the distribution of Beta spp. populations, field surveys were carried out in Portugal mainland, since 2008 and during five years, within a broad national Portuguese research project led by one of the authors [see 18]. As a result of this project, it was possible to obtain a thorough characterization of the Beta genus in Portugal mainland, namely in taxonomic, chorology, and ecological aspects. For each species, the most relevant populations were identified in association with the ecological characterization of their habitats and soil uses. In addition, herbarium material obtained from the following herbaria: AVE, COI, ELVE, LISC, LISI, LISU, LISE, MADJ, MADM, P (acronyms according to [19]), was also studied, and a new inventory of the species distribution for Portugal was produced. Based on this new data, the distribution of the Beta species is provided for B. vulgaris subsp. maritima, B. macrocarpa and P. patellaris. This survey is particularly relevant to understand the present distribution and occurrence of the most important populations within the range of climatic factors, such as precipitation (Figure 1(a)) and temperature (Figure 1(b)), found in Portugal, and their implication to the distribution of these plant species as a response to changing climatic conditions. Important populations of B. vulgaris subsp. maritima were identified, and our results reveal that this species is relatively common and widespread, occurring in natural habitats like cliff coasts, sand beaches and salt marshes, and often also in ruderal sites (Figure 1(e)). Most of these populations are well preserved and this species is currently not endangered. However, a different picture was found for the other species. P. patellaris is confined to a small population in inaccessible cliffs, the most south-western point of Portugal mainland (Figure 1(c)) and B. macrocarpa, which was recently included in the European Red List of Vascular Plants [8], is confined to south areas of Portugal, namely to the salt marshes and sandy soils of the Natural Park of Ria Formosa (Figure 1(d)), holding an enormous risk of genetic erosion.
2.2. Target Populations
During the field surveys in south regions of Portugal (in Ria Formosa), it was possible to found sympatric populations of B. vulgaris subsp. maritima and B. macrocarpa, distributed in areas where drought and soil erosion are inter-related environmental problems. Since the South of Portugal is characterized by hot summers, reaching temperatures up to 35˚C, ideal conditions are provided to screen Beta populations under field drought selective pressure, where putative drought tolerant populations most probably survive. A previous study using a flow cytometry analysis was recently published by Castro et al. [20], providing novel insights on the cytogenetic diversity of wild Beta species from Portugal mainland. The results obtained based on the study of more than one hundred individuals of B. vulgaris subsp. maritima and thirty individuals of B. macrocarpa (always growing in sympatry with B. vulgaris subsp. maritima in the South of Portugal), revealed a great cytological diversity, including tetraploid and hexaploid individuals that may constitute potential optimal resources for crop improvement of cultivated beets. The localized presence of these mixed-ploid populations of B. vulgaris subsp. maritima
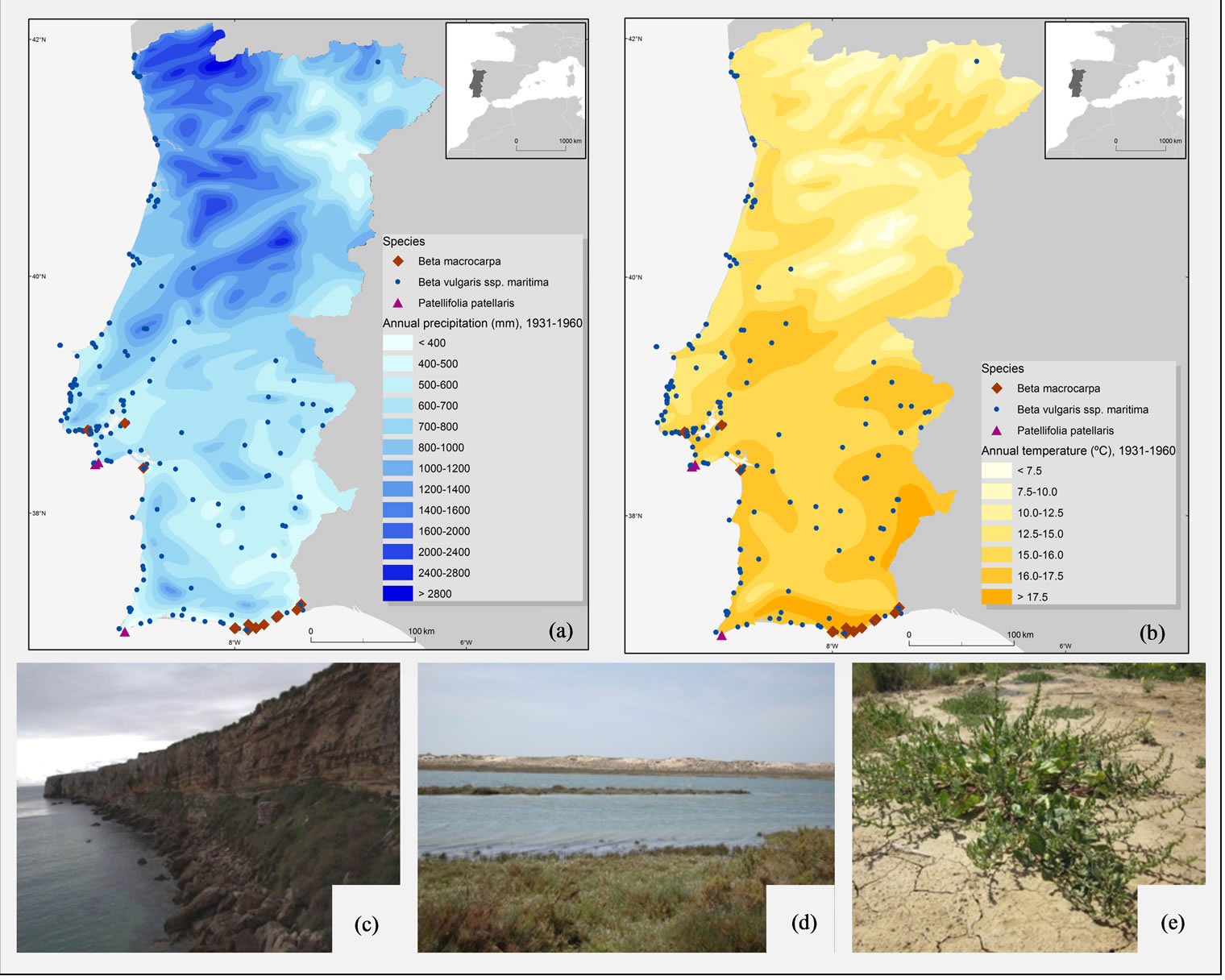
Figure 1. Distribution of Beta vulgaris subsp. maritima, B. macrocarpa and Patellifolia patellaris in Portugal mainland, related to (a) precipitation and (b) temperature; and some characteristic habitats (c) coastal cliffs of Sagres, where P. patellaris occurs, (d) salt-marshes and sandy soils of Ria Formosa, where mixed populations of B. vulgaris subsp. maritima and B. macrocarpa can be found, and (e) B. vulgaris subsp. maritima growing in dry soils of Castro Marim.
and B. macrocarpa, might indicate a higher trend for the occurrence of potential increased levels of heterozygosity in this geographic area that could be exploited as a resource to breeding programs for assessing novel traits with economic interest. So, the introgression of such germplasm from target wild populations adapted to specific environments would be a putative way to increase the genetic variation for characters that could contribute to drought tolerance in sugar beet. It is evident from the above results that the southeastern coastal areas of Portugal mainland should be investigated further as potential sites for in situ conservation of these highest priority beet wild relatives. These populations already occur within protected areas, namely in the “Natural Park of Ria Formosa” and should be prioritized for inclusion in the CWR genetic reserve network on the basis that they have already been afforded some degree of protection, even if only by default. These results provide data that will be helpful to the future conservation, utilization and management of these wild relatives, as well as to future molecular population genetic assessments.
3. Analysis and Review of the Current State of Knowledge
3.1. Advances in Molecular Research towards Sugar Beet Breeding
The importance of Beta spp. to man dates back as long ago as the late Mesolithic period, from evidence that it was already part of the human diet in that age [21]. However, beets grown exclusively for sucrose are of relatively recent origin. Economic production of sucrose was accelerated by edict in Napoleonic France under British blockade of sucrose from tropically grown sugar cane [22]. The first sugar beets with higher levels of sucrose were selected from a white fodder beet variety, the White Silesian beet [23], and this variety is still considered to be the primary source of sugar beet germplasm grown today [16].
As sugar beet was probably selected from one single cultivated population, the genetic base of the crop is supposedly narrow, and the wild Beta species yield a huge repertoire of novel genes related to ecologically important traits [24]. In this context, it must be noted that the degree of relatedness of the wild relatives to the crop (B. vulgaris subsp. vulgaris) is a very important concept to be used, being in the primary gene pool (GP1) the species that belongs to Section Beta (i.e. B. vulgaris subsp. maritima; B. vulgaris subsp. adanensis; B. macrocarpa; B. patula), in the secondary gene pool (GP2) the species that belongs to section Corollinae (i.e. B. corolliflora, B. macrorhiza, B. lomatogona, B. nana) and in the tertiary gene pool (GP3) the wild relatives of the genus Patellifolia (formerly included in section Procumbentes of the Beta genus). In particular B. vulgaris subsp. maritima, the closest wild relative of cultivated beets, and B. macrocarpa that belongs to the gene pool 1, both found in Portugal mainland, constitute highly suitable targets for screening genetic resources useful to incorporate into breeding strategies.
By the 1980s, public and private plant breeders began to consider more seriously the wild sea beet as a genetic resource in which to find resistance to the increasing pressure from insects, nematodes, and diseases, and as a source of genes for greater productivity [16]. Breeding programs devoted to sugar beet have long been in place with successful results on the introgression of many traits of interest identified in other Beta species, particularly those related with disease resistance [25-29]. Much of the research pertaining to sugar beet breeding and genetics has been summarized in Biancardi et al. [29] and Draycott [30]. In particular, significant advances on dissecting genetic important traits were achieved through the construction of high density genetic linkage maps in B. vulgaris that integrate different marker types, for gene mapping and cloning, and development of markers for breeding assistance [e.g. 31-35]. As an example, resistance to the beet cyst nematode (Heterodera schachtii Schmidt), a major pest in the cultivation of sugar beet, has been one of the most coveted agronomic traits retrieved from the wild relatives, that lacking in cultivated Beta species, could be found in the wild species P. patellaris and related species P. procumbens and P. webbiana (GP3). Identification and cloning of the resistance gene Hs1pro-1 from P. procumbens [36] was thus one of the first breakthroughs in Beta research, extending later to the cloning of a 2nd tightly linked resistance gene, Hs1-1 [35]. In the last years, fine-tunning of molecular resources is being exponentially obtained towards the accurately identification of the genes underlying such traits, and while genetic markers are used to chromosomally assign and finemap the trait, a physical map is needed to access the genomic region between the markers that flank the trait. In such framework, candidate gene discovery might be supported by integration with genome sequences of other species via map anchors of known sequence [37]. In the same study, Dohm et al. [37] generated an extended high-resolution and high-density genetic map and constructed a genome-wide physical map of sugar beet, providing comprehensive genomic information that covers all nine chromosomes, and thus means enabling future high-resolution trait mapping, gene identification, and cross-referencing to regions sequenced in other plant species.
Although the main research goals in sugar beet, deriving from breeder’s interests and concerns, have been focused on sugar content, beet yield, bolting control, seed quality, and pest and disease resistance, a growing attention is being paid to other traits as a consequence of the future climatic changes. Even more so when the Mediterranean is expected to be one of the most affected regions in the near future, with a substantial decrease in precipitation and a pronounced warming. The impact of drought has already been recognized as a major cause of yield losses in sugar beet [38,39], although the selection for increased drought tolerance has not been a breeding priority until recently [40]. Dry regions, such as those found in the South of Portugal, are outstanding areas to assess the degree of genotypic variation for drought resistance, across many types of environments, including the huge areas of salt marshes, and putative adaptations to soil salinity. Beyond the required deep knowledge of the genetic variability of the wild populations, the ecological characterization of their habitats per se, may indicate that a species possess appropriate physiological, ecological or behavioral adaptations to successfully colonize that particular environment. As a result, the wild beet species growing naturally in those environments will undoubtedly contribute with novel genes to sugar beet improvement for the “new” traits of interest.
3.2. Genetic Relationships within the Beta vulgaris Complex
While there is no doubt of the value of CWRs for sustainable agriculture and future food security, there is still a poor knowledge on the extent of the wild genetic diversity and crop-wild relationships in a way that could bridge conservation–use needs. The B. vulgaris complex is of particular interest since fully cross-compatible crop, wild and weed forms can be found in sympatric situations in some parts of Europe [41,42], resulting in unwanted recurrent hybridizations in both directions. In factconspecific weeds commonly found infesting the sugar beet production fields and leading to severe agronomic problems, are the proved effect of such phenomenon, having arisen from hybridization between cultivated lines and wild individuals in seed production areas [41].
The effect of such uncontrolled gene flow and introgression is a subject of great concern for the management and conservation of Beta genetic resources, as well as for sugar beet sustainable production. In one hand, it can lead to loss of local adaptation of native populations, which can be of primary importance during periodic disturbances or extreme environmental conditions, and, on the other hand, to evolution of more aggressive weeds with increased competitiveness. Moreover, the likelihood of transgene escape from crop to wild relatives [43,44] should be taken into account with extreme care [42].
Up to now, investigations on the evolutionary dynamics of genetic diversity within the B. vulgaris complex were thus mainly focused on assessing genetic relationships among the different forms of beet [41,42,45,46], with particular emphasis on crop-to-wild gene flow [47, 48]. Using a panel of polymorphic markers comprising nuclear microsatellite loci, mitochondrial minisatellite loci and one chloroplastic PCR-RFLP marker, Fénart et al. [42] found similar levels of nuclear genetic diversity in weedy beets and sea beets, as opposed to the narrowness of cultivar accessions, but a very strong genetic divergence between wild sea beet and other relatives, which was unexpected given the recent evolutionary history and the full cross-compatibility of all taxa. Also, the different forms of beet clustered into genetic distinctive groups concordant with the classification of cultivated, wild and weedy forms based on their geographical location, phenotype or domesticated status despite the very recent domestication process of sugar beet [42]. High genetic diversity with no isolation-by-distance genetic structure has been commonly found among weed beet populations [41,42,47], although a strong spatial genetic structure was detected in high-density, highly outcrossing populations [48]. This implies that once established, weed beet populations can sustain a high level of genetic variation and thus provide a likely basis for potential rapid evolutionary change. Moreover, characteristics of particular source populations within initial native ranges are likely to shape the evolution of invasive populations [49]. This is particularly true as inland wild beets already adapted to the margins of arable fields and other anthropogenic habitats, through short life-span and first-year flowering, can be found [48]. On the other hand, crop-towild gene flow has been implicated in the increased likelihood of extinction of wild relatives by displacing native wild allelic diversity [47]. Altogether these studies highlight the need for careful survey and management of both sugar beet fields and seed production areas where wild and cultivated relatives co-occur, as both a measure to conserve wild populations from genetic swamping and to ensure crop high-yielding production.
On a priority course for screening genetic resources and locating adaptive traits, efforts should now be concentrated on evaluating the genetic variation of the coastal wild populations occurring in specific habitats, which have a distinct evolutionary trajectory. Knowledge of the genetic structure of wild populations can provide insight on the fundamental evolutionary influences of selection, mutation, gene flow and genetic drift, reflecting into patterns of population differentiation. There is an important gap on this subject as very few studies have dealt with patterns of genetic variation in wild sea beet. Fievet et al. [50] analyzed wild sea beet populations along the French coasts of the Anglo-Norman gulf and found a significant geographical genetic structuring with evidence of a distance-dependant pattern of dispersal, comprising pollen vs. seed-mediated gene flow, within which some populations are most likely a source of genes, whereas others behave as a sink.
For the knowledge that has been built up on this important species complex, here reviewed, assignment analyses based on multilocus microsatellite genotypes proved to be a powerful tool for tracing contemporary cross-breeding events between wild-cultivated relatives, assessing genetic variation and recently to distinguish sugar beet varieties [51]. Looking for adaptive traits will require however, genomic approaches or candidate gene targeted approaches that could detect population genetic divergence most likely associated to processes of local adaptation. A wide collection of molecular resources and tools were made available for the B. vulgaris complex over the last decade (see The B. vulgaris Resource, http://bvseq.molgen.mpg.de), including about 30 000 sugar beet expressed sequence tags (ESTs) in the NCBI dbEST database, that now can now be used to characterize the wild genetic diversity and locate the desired traits.
4. Linking Ecological Data with Genetic Analyses to a Comprehensive Knowledge of Beet Diversity
Before CWR can be used in any plant-breeding program, their genetic resources have to be evaluated by assessing genetic variability in traits of interest for breeders [52]. The field of agricultural genomics is in the focus of a technological revolution caused by the emergence of DNA sequencing technologies; yet, reverse genetics approaches are still a valuable tool for the detection of differences in target sequences to identify allelic variations in natural populations and in the context of functional genomics.
Besides indicating relationships between individuals or populations, DNA polymorphisms can be directly associated to phenotypic differences or be genetically linked to its causative factor [53]. As a result, distinguishing adaptive variation from neutral mutations are becoming key issues to understand the molecular basis of heritable phenotypic changes and to carry out functional genomics. Single Nucleotide Polymorphisms (SNPs) have emerged as the most widely used genotyping markers due to their abundance in the genome, together with small insertions/ deletions (indels), and the relative ease in determining their frequency [54]. To efficiently identify SNPs, a number of different techniques have been developed namely PCRand sequencing-based methods [reviewed in 55-57], which all have their limitations. Thus, there is an increasing demand for high-throughput genotyping technologies able to increase the ability to determine nucleotide differences putatively associated with traits of agroeconomic interest. A number of studies have been able to link SNPs/indels with phenotypic traits of agronomic interest, such as an 8-bp deletion of a betainealdehyde dehydrogenase gene (OsBADH2) in rice [58] and a SNP with G replaced by A in the same gene of soybean [59] that revealed to be responsible for fragrant phenoltypes.
Under a CWR research perspective, biodiversity assessment using functional genes related to ecologically important traits of agronomic interest as markers, could allow determining their usefulness as a supplementing gene pool for crop improvement [60]. Such targeted approach focused on candidate genes of ecological/adaptive significance has the potential to provide crucial information, which cannot be obtained from a genetic diversity screen based on random anonymous markers. Yet, this is only possible if CWR species and specific populations have been identified as a basis to work on.
4.1. Genotypic Variation for Salinity and Drought Tolerance
Within the goal of broadening sugar beet gene pool for drought and salt tolerance, wild relatives from gene pool 1 and 2 (Beta species) and also from gene pool 3 (Patellifolia species) represent the best sampling source to evaluate adaptive genetic variation, encompassing germplasm from different populations and different regions. As referred above in Section 2.2, in Portugal, representatives of the Beta and Patellifolia genera have successfully colonized adverse environments developing potential resistance/tolerance to stressful abiotic conditions, namely drought and salt stresses, which render them as target populations to be included for in situ characterization as potential donors of genes involved on related adaptive responses. In such environmental conditions, as those described in the present study, expression of a variety of genes is induced in adapted plants not only to assure their survival, but also to assist in maintaining their regular growth and productivity [61]. Under a candidate gene approach, assessment of the natural genetic variation among different CWR species is performed on a set of candidate genes involved in drought and salt tolerance leading to in situ germplasm characterization. Candidate genes involved in drought and salt tolerance were identified and can be selected from evidences on literature [e.g. 62,63]. As drought and salt tolerance share common mechanisms, stress-induced genes with known functions, as well as specific drought- [64] and salt-responsive genes can be targeted [65]. Previous studies on wild relatives of wheat and barley revealed rich genetic diversities for drought and salt tolerances resulting from adaptation to a broad range of environments [66]. A database of allelic variants of the wild relatives of sugar beet will deliver a tool on functional molecular markers linked to drought and salt tolerance that can be applied in marker-assisted selection for sugar beet breeding programs.
4.2. EcoTILLING: As a SNP Discovery Tool to Assess Natural Polymorphisms
Among the candidate gene-based approaches, EcoTILLING has proven to be a fast, reliable and low-cost technique allowing a high-throughput analysis of natural genetic variation [67]. This technique is based on the Targeting Induced Local Lesions in Genomes (TILLING) method, relying on the formation of DNA heteroduplexes between an unknown homozygous DNA and a known sequence by the usage of the endonuclease CEL I [68]. It rapidly screens many samples towards the identification of SNPs and/or small indels in a gene of interest, whose number and position are revealed [67,69]. Due to the reliability of the endonuclease system, only one representative of each haplotype needs to be sequenced to identify the point mutation in the gene of interest. Moreover, EcoTILLING was recently used to detect SNPs and small indels across 375 rice germplasm accessions, targeting several salt tolerance genes [70]. Representative SNPs or indels were assessed in 3’-untranslated region, thus pointing out for the need to determine not only the allelic variants location but also their potential contribution to the regulation of gene expression. Also, in sugar beet, a study reporting EcoTILLING as a reverse genetics approach evaluated the diversity in a panel of 268 B. vulgaris accessions targeted for three flowering time genes, in which associations between nucleotide polymorphisms in one of the genes (BvFL1) with bolting before winter as well as winter hardiness was assessed [71]. In general, EcoTILLING have shown great promise for accelerating the process of the identification of natural resistance alleles, which can be used to breed improved cultivars. Successful detection of SNPs or small indels in target genes in natural populations was achieved in Populus trichocarpa Torr. & A. Gray regarding genes involved on wood quality and pathogen defense [72], and in a collection of Cucumis spp., resulting in the identification of new allelic variants in eIF4E, a factor which controls virus susceptibility [73]. Quarrying for variation in resistance genes help to speed up the process of identifying alleles that could provide immunity to various diseases, for example, allelic variation was examined and identified using EcoTILLING in mlo and Mla powdery mildew resistance genes of barley [74], thus providing several allelic variants that could be exploited to breed cultivars with improved resistance.
Based on the genomic resources now available [EST (http://genomics.msu.edu/sugarbeet/blast.html) for sugar beet on databases and TIGR Gene Indices databases (http://compbio.dfci.harvard.edu/tgi/plant.html, release Beet 4.0)], looking for genes of interest became a task with higher viability, as well as annotating SNP localization in coding regions, non coding regions, start and stop codons, and splice sites by gene models based on genome sequencing data (http://bvseq.molgen.mpg.de/). Data from population analyses of sequence variation can then lead the way into getting insights on the adaptive nature of DNA polymorphisms and engaging follow-up studies to ascertain causal relations between phenotypes and allelic variants. As non-synonymous substitutions can potentially alter protein function and result in a phenotypic change, as opposite to synonymous substitutions, candidate genes from which these allelic variants could be identified among different wild relative species/populations are of major importance to further assess related changes in gene expression.
4.3. Real-Time PCR: Uncovering Differential Allelic Expression
SNP allelic variants can play a role in gene regulation, affecting the expression level [reviewed e.g. 75,76]. Regulatory allelic variants may affect the level of gene expression by causing imbalanced expression of alleles, yet this is a phenomenon poorly studied in plants [77-81]. Identification of co-expressed allelic variants with phenotypic effects can provide novel insights into the biological processes underpinning an adaptive trait [82]. Quantitative genotyping, detection of SNPs, allelic discrimination as well as genetic variations can be detected by real-time PCR (qPCR). Specific amplification of allelic variants can rely on the allele-specific expression by amplification refractory mutation system (ARMS) [83] using SYBR®Green. Discrimination power of ARMS technique was further improved using allele-specific forward primers with a deliberate added mismatched nucleotide located near the 3’ terminal region using the same reverse primer, a system named Mismatch Amplification Mutation Assay (MAMA) [84,85]. This method allows discrimination between different allelic variants and the imbalanced allelic expression (AI) can be unveiled by a difference in the ratios measured in cDNA and gDNA, being no reference gene needed in this case as each allele is an internal control of the other [86]. This strategy allows determining an allelic expression profile specific to each genetic variant on the selected SNPs under analysis. Analyzing CWR species differential gene expression in genes putatively related to drought and salt tolerance, as compared with cultivated susceptible species is of great importance, since genotype to phenotype inferences could potentially be addressed, in a way that causal relations between CWR allelic expression profiles and phenotypic variation could be ascertained. Ultimately, it could indicate which CWR populations should be included in future breeding programs concerning drought and salt tolerance introgression on cultivated sugar beet. Taken together, here we present molecular options to link ecological data with genetic analyses as to provide ground for the characterization of genetic variation related with drought and salt stresses in target populations of B. vulgaris subsp. maritima in Portugal mainland. Outline of the workflow proposed is summarized in Figure 2.
5. Directions for Future Research
In the last decades, molecular research has made it possible to shift the paradigm from searching for phenotypes to searching for genes of interest. In fact, given the rapid burst of the next-generation sequencing (NGS) technologies with reduced cost as well as newly available applications being developed, CWR genetics will benefit from the generation of large numbers of genetically informative markers, considering in the first place its major downstream application, marker-assisted selection (MAS). The reported results in this paper were based on extensive field surveys, and provided an indication of natural adaptations to salinity and drought and would be important in determining the direction of research and in designing strategies for future breeding programs. Particularly, the Beta populations that occur in south-eastern regions of Portugal seem to be a highly promising target for further evaluation regarding drought tolerance.
Indeed, these plants have successfully colonized adverse environments and their study can contribute significantly to the knowledge on stress tolerance, namely to drought and salt, and be used in future breeding programs.
Large scale discovery of genome-wide distribute SNPs
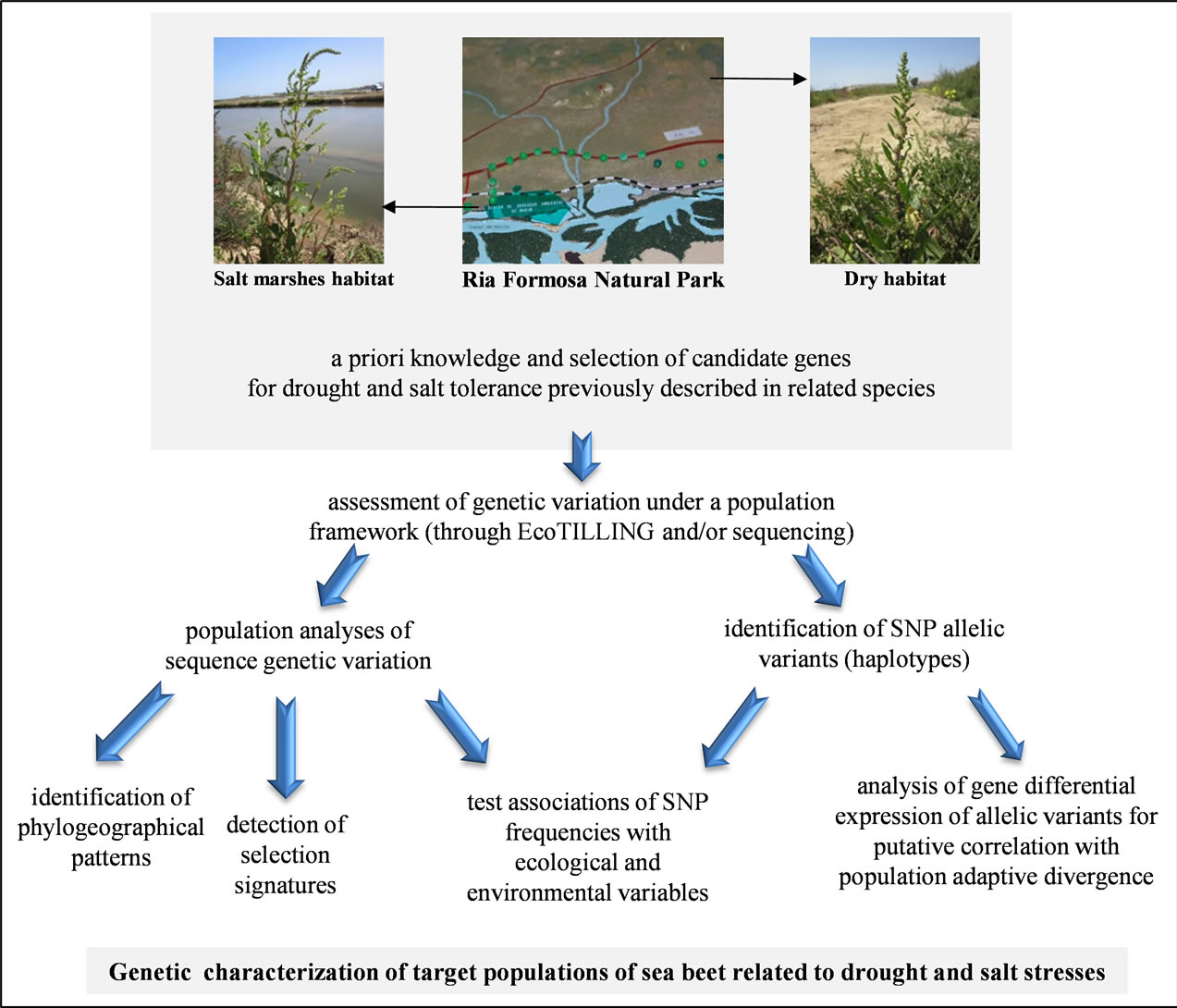
Figure 2. Outline of the workflow proposed for the characterization of the genetic variation related with drought and salt stresses in target populations of Beta vulgaris subsp. maritima in Portugal mainland. The Natural Park of Ria Formosa proposed for screening is indicated by arrows: salt marshes and dry habitats.
can be effectively conducted with NGS technologies on species with or without reference genome, thus enabling studies of non-model organisms and wild populations [87]. Genome reduction methods, such as Reduced Representation Libraries (RRL), Restriction site Associated DNA (RAD) sequencing and complexity reduction of polymorphic sequences (CRoPS) (reviewed in [88]), aim at sequencing only a small fraction of the genome. RAD [89,90] coupled to Illumina sequencing (RADseq) [91] is extremely promising and feasible for detecting SNPs in unknown genomes (e.g. [92]). Applications of RAD sequencing in crop genetics have been performed for the perennial ryegrass (Lolium perenne L.) by constructing a linkage map and quantitative traits loci (QTLs) associated with resistance to stem rust caused by the pathogen Puccinia graminis subsp. graminicola [93]; in lupin (Lupinus angustifolius L.), 30 markers are linked to an anthracnose resistance gene caused by the pathogen Colletotrichum lupini [94], among several other studies (more examples reported in Deschamps et al. [95]). In the context of assessing diversity in nature related with traits probably developed under strong selective pressure, population genomics are used to identify signatures of selection and subsequently loci of adaptive significance and potentially underlying evolutionary change. Given the wide DNA sequencing technologies available, it is advisable to account for both limitations and advantages on selecting the best NGS technology. Deepening the understanding of the molecular connection linking genotype to phenotype will enable sugar beet genomics-assisted breeding. For breeding applications, relevant polymorphisms can be discovered only when novel germplasm is introduced into the breeding pool, thus reinforcing the need to augment the gene pool of crops by recurring to their wild relatives. Ultimately, the selection of CWR species based on genomic tools, rather than selecting random variants with desirable characteristics, will be a promising and still little explored route towards the development of improved crops.
REFERENCES
- R. Hajjar and T. Hodgkin, “The Use of Wild Relatives in Crop Improvement: A Survey of Developments over the Last 20 Years,” Euphytica, Vol. 156, No. 1-2, 2007, pp. 1-13. doi:10.1007/s10681-007-9363-0
- N. Maxted and S. Kell, “Establishment of a Global Network for the in Situ Conservation of Crop Wild Relatives: Status and Needs,” FAO consultancy Report, Commission on Genetic Resources for Food and Agriculture, Rome, 2009, pp. 1-265. http://www.fao.org/docrep/013/i1500e/i1500e18a.pdf
- B. V. Ford-Lloyd, M. Schmidt, S. J. Armstrong, O. Barazani, J. Engels, R. Hadas, K. Hammer, S. P. Kell, D. Kang, K. Khoshbakht, Y. Li, C. Long, B.-R. Lu, K. Ma, V. T. Nguyen, L. Qiu, S. Ge, W. Wei and Z. Zhang, “Crop Wild Relatives—Undervalued, Underutilized and under Threat?” BioScience, Vol. 61, No. 7, 2011, pp. 559- 565. doi:10.1525/bio.2011.61.7.10
- N. Maxted, B. V. Ford-Lloyd, S. L. Jury, S. P. Kell and M. A. Scholten, “Towards a Definition of a Crop Wild Relative,” Biodiversity and Conservation, Vol. 15, No. 8, 2006, pp. 2673-2685. doi:10.1007/s10531-005-5409-6
- S. D. Tanksley and S. R. McCouch, “Seed Banks and Molecular Maps: Unlocking Genetic Potential from the Wild,” Science, Vol. 277, No. 5329, 1997, pp. 1063-1066. doi:10.1126/science.277.5329.1063
- S. P. Kell, H. Knüpffer, S. L. Jury, N. Maxted and B. V. Ford-Lloyd, “Catalogue of Crop Wild Relatives for Europe and the Mediterranean,” University of Birmingham, Birmingham, 2005. http://www.pgrforum.org/cwris/cwris.asp
- S. P. Kell, S. L. Jury, H. Knupffer, B. V. Ford-Lloyd and N. Maxted, “PGR Forum: Serving the Crop Wild Relative User Community,” Bocconea, Vol. 21, 2008, pp. 413- 421.
- M. Bilz, S. P. Kell, N. Maxted and R. V. Lansdown, “European Red List of Vascular Plants,” Publications Office of the European Union, Luxembourg, 2011.
- S. Stolton, T. Boucher, N. Dudley, J. Hoekstra, N. Maxted and S. Kell, “Ecoregions with Crop Wild Relatives are Less Well Protected,” Biodiversity Journal of Life on Earth, Vol. 9, No. 1-2, 2008, pp. 52-55. doi:10.1080/14888386.2008.9712883
- N. Maxted, B. V. Ford-Lloyd and S. P. Kell, “Crop Wild Relatives: Establishing the Context,” In: N. Maxted, B. V. Ford-Lloyd, S. P. Kell, J. Iriondo, E. Dulloo and J. Turok Eds., Crop Wild Relative Conservation and Use, CABI Publishing, Wallingford, 2008, pp. 3-30.
- E. Biancardi, L. W. Panella and R. T. Lewellen, “Beta maritima: The Origin of Beets,” Springer, New York, 2012, p. 250. doi:10.1007/978-1-4614-0842-0
- J. Weiland and G. Koch, “Sugarbeet Leaf Spot Disease (Cercospora beticola Sacc.),” Molecular Plant Pathology, Vol. 5, No. 3, 2004, pp. 157-166. doi:10.1111/J.1364-3703.2004.00218.X
- S. Knapp, M. S. Vorontsova and J. Prohens, “Wild Relatives of the Eggplant (Solanum melongena L.: Solanaceae): New Understanding of Species Names in a Complex Group,” PLoS ONE, Vol. 8, 2013, e57039. doi:10.1371/journal.pone.0057039
- L. W. Panella and R. T. Lewellen, “Broadening the Genetic Base of Sugar Beet: Introgression from Wild Relatives,” Euphytica, Vol. 154, No. 3, 2006, pp. 383-400. doi:10.1007/s10681-006-9209-1
- B. V. Ford-Lloyd, “Sources of Genetic Variation, Genus Beta” In: E. Biancardi, L. G. Campbell, G. N. Skaracis and M. De Biaggi, Eds., Genetics and Breeding of Sugar Beet, Science Publishers, Inc., Enfield, 2005, pp. 25-33.
- J. M. McGrath, L. Panella and L. Frese, “Chapter 1: Beta,” In: C. Kole, Ed., Wild Crop Relatives: Genomic and Breeding Resources, Industrial Crops, Springer-Verlag, Berlin Heidelberg, 2011, pp. 1-28. doi:10.1007/978-3-642-21102-7_1
- S. Hohmann, J. W. Kadereit and G. Kadereit, “Understanding Mediterranean-Californian Disjunctions: Molecular Evidence from Chenopodiaceae-Betoideae,” Taxon, Vol. 55, No. 1, 2006, pp. 67-78.
- M. C. Duarte, “The Wild Relatives of Beta: Genetic Diversity Assessment and Biochemical Studies,” Instituto de Investigação Científica Tropical, I.P. (IICT) (IICT/ MNE), Project Report of the Project PTDC/AGR-AAM/ 73144/2006 Funded by Portuguese Foundation for Science and Technology, 2012.
- P. K. Holmgren, N. H. Holmgren and L. C. Barnett, “Index Herbariorum, Part I: The Herbaria of the World,” The New York Botanic Garden Press, New York, 1990.
- S. Castro, M. M. Romeiras, M. Castro, M. C. Duarte and J. Loureiro, “Hidden Diversity in Wild Beta Taxa from Portugal: Insights from Genome Size and Ploidy Level Estimations Using Flow Cytometry,” Plant Science, Vol. 207, 2013, pp. 72-78. doi:10.1016/j.plantsci.2013.02.012.
- L. Kubiak-Martens, “The Plant Food Component of the Diet at the Late Mesolithic (Ertebølle) Settlement at Tybrind Vig, Denmark,” Vegetation History and Archaeobotany, Vol. 8, No. 1-2, 1999, pp. 117-127. doi:10.1007/BF02042850
- C. Winner, “History of the Crop,” In: D. A. Cooke and R. Scott, Eds., The Sugar Beet Crop: Science into Practice, Chapman and Hall, London, 1993, pp. 1-35.
- H. E. Fischer, “Origin of the ‘Weisse Schlesische Rube’ (White Silesian Beet) and Resynthesis of Sugar Beet,” Euphytica, Vol. 41, No. 1-2, 1989, pp. 75-80. doi:10.1007/BF00022414
- L. Frese, B. Desprez and D. Ziegler, “Potential of Genetic Resources and Breeding Strategies for Base-Broadening in Beta (English),” In: H. D. Cooper, C. Spillane and T. Hodgkin, Eds., Broadening the Genetic Base of Crop Production, IPGRI/FAO, 2001, pp. 295-309. doi:10.1079/9780851994116.0295
- G. H. Coons, “Interspecific Hybrids between Beta vulgaris L. and the Wild Species of Beta,” Journal of the American Society of Sugar Beet Technologists, Vol. 18, 1975, pp. 281-306.
- H. Savitsky, “Hybridization between Beta vulgaris and Beta procumbens and Transmission of Nematode (Heterodera schachtii) Resistance to Sugar Beet,” Canadian Journal of Genetics and Cytology, Vol. 17, No. 2, 1975, pp. 197-209. doi:10.1139/g75-027
- H. Savitsky, “Nematode (Heterodera schachtii) Resistance and Meiosis in Diploid Plants from Interspecific Beta vulgaris X Beta procumbens hybrids,” Canadian Journal of Genetics and Cytology, Vol. 20, No. 2, 1978, pp. 177- 186. doi:10.1139/g78-019
- W. Heijbroek, A. J. Roelands and J. H. de Jong, “Transfer of Resistance to Beet Cyst Nematode from Beta patellaris to Sugar Beet,” Euphytica, Vol. 32, No. 2, 1983, pp. 103- 108. doi:10.1007/BF00021437
- E. Biancardi, L. G. Campbell, G. N. Skaracis and M. De Biaggi, “Genetics and Breeding of Sugar Beet,” Science Publishers, Inc., Enfield, 2005.
- P. Draycott, “Sugar Beet,” Blackwell, Oxford, 2006.
- K. Schumacher, J. Schondelmaier, E. Barzen, G. Steinrücken, D. Borchardt, W. E. Weber, C. Jung and F. Salamini, “Combining Different Linkage Maps in Sugar Beet (Beta vulgaris L.) to Make One Map,” Plant Breeding, Vol. 116, 1997, pp. 23-38. doi:10.1111/j.1439-0523.1997.tb00971.x
- S. J. Rae, C. Aldam, I. Dominguez, M. Hoebrechts, S. R. Barnes and K. J. Edwards, “Development and Incorporation of Microsatellite Markers into the Linkage Map of Sugar Beet (Beta vulgaris spp.),” Theoretical and Applied Genetics, Vol. 100, No. 8, 2000, pp. 1240-1248. doi:10.1007/s001220051430
- V. Laurent, P. Devaux, T. Thiel, F. Viard, S. Mielordt, P. Touzet and M. C. Quillet, “Comparative Effectiveness of Sugar Beet Microsatellite Markers Isolated from Genomic Libraries and GenBank ESTs to Map the Sugar Beet Genome,” Theoretical and Applied Genetics, Vol. 115, No. 6, 2007, pp. 793-805. doi:10.1007/s00122-007-0609-y
- K. Schneider, D. Kulosa, T. R. Soerensen, S. Mohring, M. Heine, G. Durstewitz, A. Polley, E. Weber, Jamsari, J. Lein, U. Hohmann, E. Tahiro, B. Weisshaar, B. Schulz, G. Koch, C. Jung and M. Ganal, “Analysis of DNA Polymorphisms in Sugar Beet (Beta vulgaris L.) and Development of an SNP-Based Map of Expressed Genes,” Theoretical and Applied Genetics, Vol. 115, No. 5, 2007, pp. 601-615. doi:10.1007/s00122-007-0591-4
- F. J. Kopisch-Obuch, G. G. G. Capistrano, A. Müller, H.- J. Harloff, S. L. M. Frerichmann and C. Jung, “Molecular Breeding Research with Sugar Beet,” Tagung der Vereinigung der Pflanzenzüchter und Saatgutkaufleute Pfl anzenzüchter Saatgutkaufl eute Österreichs, 2008, pp. 75- 76.
- D. Cai, M. Kleine, S. Kifle, H. Harloff, N. N. Sandal, K. A. Marcker, R. M. Klein-Lankhorst, E. M. J. Salentijn, W. Lange, W. J. Stiekema, U. Wyss, F. M. W. Grundler and C. Jung, “Positional Cloning of a Gene for Nematode Resistance in Sugar Beet,” Science, Vol. 275, No. 5301, 1997, pp. 832-834. doi:10.1126/science.275.5301.832
- J. C. Dohm, C. Lange, D. Holtgräwe, T. Rosleff Sörensen, D. Borchardt, D. Schulz, H. Lehrach, B. Weisshaar and H. Himmelbauer, “Palaeohexaploid Ancestry for Caryophyllales Inferred from Extensive Gene-Based Physical and Genetic Mapping of the Sugar Beet Genome (Beta vulgaris),” The Plant Journal, Vol. 70, No. 3, 2012, pp. 528-540. doi:10.1111/j.1365-313X.2011.04898.x
- K. W. Jaggard, A. M. Dewar and J. D. Pidgeon, “The Relative Effects of Drought Stress and Virus Yellows on the Yield of Sugar Beet in the UK, 1980-1995,” The Journal of Agricultural Science, Vol. 130, No. 3, 1998, pp. 337-343.
- J. D. Pidgeon, A. R. Werker, K. W. Jaggard, G. M. Richter, D. H. Lister and P. D. Jones, “Climatic Impact on the Productivity of Sugar Beet in Europe, 1961-1995,” Agricultural and Forest Meteorology, Vol. 109, No. 1, 2001, pp. 27-37. doi:10.1016/S0168-1923(01)00254-4
- E. S. Ober and M. C. Luterbacher, “Genotypic Variation for Drought Tolerance in Beta vulgaris,” Annals of Botany, Vol. 89, No. 7, 2002, pp. 917-924. doi:10.1093/aob/mcf093
- F. Viard, J.-F. Arnaud, M. Delescluse and J. Cuguen, “Tracing Back Seed and Pollen Flow within the CropWild Beta vulgaris Complex: Genetic Distinctiveness vs. Hot Spots of Hybridization over a Regional Scale,” Molecular Ecology, Vol. 13, No. 6, 2004, pp. 1357-1364. doi:10.1111/j.1365-294X.2004.02150.x
- S. Fénart, J. F. Arnaud, I. De Cauwer and J. Cuguen, “Nuclear and Cytoplasmic Genetic Diversity in Weed Beet and Sugar Beet Accessions Compared to Wild Relatives: New Insights into the Genetic Relationships within the Beta vulgaris Complex Species,” Theoretical and Applied Genetics, Vol. 116, No. 8, 2008, pp. 1063-1077. doi:10.1007/s00122-008-0735-1
- D. I. Jarvis and T. Hodgkin, “Wild Relatives and Crop Cultivars: Detecting Natural Introgression and Farmer Selection of New Genetic Combinations in Agroecosystems,” Molecular Ecology, Vol. 8, Suppl. 12, 1999, pp. S159-S173. doi:10.1046/j.1365-294X.1999.00799.x
- D. Bartsch and I. Schuphan, “Lessons We Can Learn from Ecological Biosafety Research,” Journal of Biotechnology, Vol. 98, No. 1, 2002, pp. 71-77. doi:10.1016/S0168-1656(02)00087-1
- B. Desplanque, P. Boudry, K. Broomberg, P. SaumitouLaprade, J. Cuguen and H. Van Dijk, “Genetic Diversity and Gene Flow Between Wild, Cultivated and Weedy Forms of Beta vulgaris L. (Chenopodiaceae), Assessed by RFLP and Microsatellite Markers,” Theoretical and Applied Genetics, Vol. 98, No. 8, 1999, pp. 1194-1201. doi:10.1007/s001220051184
- F. Viard, J. Bernard and B. Desplanque, “Crop-Weed Interactions in the Beta vulgaris Complex at a Local Scale: Allelic Diversity and Gene Flow within Sugar Beet Fields,” Theoretical and Applied Genetics, Vol. 104, No. 4, 2002, pp. 688-697. doi:10.1007/s001220100737
- J.-F. Arnaud, S. Fénart, C. Godé, S. Deledicque, P. Touzet and J. Cuguen, “Fine-Scale Geographical Structure of Genetic Diversity in Inland Wild Beet Populations,” Molecular Ecology, Vol. 18, No. 15, 2009, pp. 3201-3215. doi:10.1111/j.1365-294X.2009.04279.x
- J.-F. Arnaud, S. Fénart, M. Cordellier and J. Cuguen “Populations of Weedy Crop-Wild Hybrid Beets Show Contrasting Variation in Mating System and Population Genetic Structure,” Evolutionary Applications, Vol. 3, No. 3, 2010, pp. 305-318. doi:10.1111/j.1752-4571.2010.00121.x
- C. E. Lee and G. W. Gelembiuk, “Evolutionary Origins of invasive Populations,” Evolutionary Applications, Vol. 1, No. 3, 2008, pp. 427-448. doi:10.1111/j.1752-4571.2008.00039.x
- V. Fievet, P. Touzet, J. F. Arnaud and J. Cuguen, “Spatial Analysis of Nuclear and Cytoplasmic DNA Diversity in Wild Sea Beet (Beta vulgaris ssp. maritima) Populations: Do Marine Currents Shape the Genetic Structure?” Molecular Ecology, Vol. 16, No. 9, 2007, pp. 1847-1864. doi:10.1111/j.1365-294X.2006.03208.x
- M. J. M. Smulders, G. D. Esselink, I. Everaert, J. De Riek and B. Vosman, “Characterisation of Sugar Beet (Beta vulgaris L. ssp. vulgaris) Varieties Using Microsatellite Markers,” BMC Genetics, Vol. 11, 2010, p. 41. doi:10.1186/1471-2156-11-41
- B. I. G. Haussmann, H. K. Parzies, T. Presterl, Z. Susic, and T. Miedaner, “Plant Genetic Resources in Crop Improvement,” Plant Genetic Resources, Vol. 2, No. 1, 2004, pp. 3-21. doi:10.1079/PGR200430
- A. Rafalski, “Applications of Single Nucleotide Polymorphisms in Crop Genetics,” Current Opinion in Plant Biology, Vol. 5, No. 2, 2002, pp. 94-100. doi:10.1016/S1369-5266(02)00240-6
- M. W. Ganal, T. Altmann and M. S. Röder, “SNP Identification in Crop Plants,” Current Opinion in Plant Biology, Vol. 12, No. 2, 2009, pp. 211-217. doi:10.1016/j.pbi.2008.12.009
- T. Jehan and S. Lakhanpaul, “Single Nucleotide Polymorphism (SNP)—Methods and Applications in Plant Genetics: A Review,” Indian Journal of Biotechnology, Vol. 5, No. 4, 2006, pp. 435-449.
- C. M. Fusari, V. V. Lia, H. E. Hopp, R. A. Heinz and N. B. Paniego, “Identification of Single Nucleotide Polymorphisms and Analysis of Linkage Disequilibrium in Sunflower Elite Inbred Lines Using the Candidate Gene Approach,” BMC Plant Biology, Vol. 8, 2006, p. 7. doi:10.1186/1471-2229-8-7
- A. Hayward, A. Mason, J. Dalton-Morgan, M. Zander, D. Edwards and J. Batley, “SNP Discovery and Applications in Brassica napus,” Journal of Plant Biotechnology, Vol. 39, No. 1, 2011, pp. 1-12. doi:10.5010/JPB.2012.39.1.049
- L. M. T. Bradbury, T. L. Fitzgerald, R. J. Henry, Q. Jin and D. L. E. Waters, “The Gene for Fragrance in Rice,” Plant Biotechnology Journal, Vol. 3, No. 3, 2005, pp. 363-370. doi:10.1111/j.1467-7652.2005.00131.x
- R. Juwattanasomran, P. Somta, S. Chankaew, T. Shimizu, S. Wongpornchai, A. Kaga and P. Srinives, “A SNP in GmBADH2 Gene Associates with Fragrance in Vegetable Soybean Variety ‘Kaori’ and SNAP Marker Development for the Fragrance,” Theoretical and Applied Genetics, Vol. 122, No. 3, 2011, pp. 533-541. doi:10.1007/s00122-010-1467-6.
- P. H. van Tienderen, A. A. de Haan, C. G. van der Linden and B. Vosmantiederen, “Biodiversity Assessment Using Markers for Ecologically Important Traits,” Trends in Ecology & Evolution, Vol. 17, No. 12, 2002, pp. 577-582. doi:10.1016/S0169-5347(02)02624-1
- A. S. Duque, A. M. de Almeida, A. B. da Silva, J. M. da Silva, A. P. Farinha, D. Santos, P. Fevereiro and S. S. Araújo, “Abiotic Stress Responses in Plants: Unraveling the Complexity of Genes and Networks to Survive,” In: K. Vahdati and C. Leslie, Eds., Abiotic Stress-Plant Responses and Applications in Agriculture, InTech, Rijeka, 2013. doi:10.5772/52779
- L. Xiong, K. S. Schumaker and J.-K. Zhu, “Cell Signaling during Cold, Drought, and Salt Stress,” The Plant Cell, Vol. 14, Suppl. 1, 2002, pp. S165-S183. www.plantcell.org/cgi/doi/10.1105/tpc.000596
- J. Z. Zhang, R. A. Creelman and J.-K. Zhu, “From Laboratory to Field. Using Information from Arabidopsis to Engineer Salt, Cold, and Drought Tolerance in Crops,” Plant Physiology, Vol. 135, No. 2, 2004, pp. 615-621. www.plantphysiol.org/cgi/doi/10.1104/pp.104.040295
- M. Hajheidari, M. Abdollahian-Noghabi, H. Askari, M. Heidari, S. Y. Sadeghian, E. S. Ober and G. H. Salekdeh, “Proteome Analysis of Sugar Beet Leaves under Drought Stress,” Proteomics, Vol. 5, No. 4, 2005, pp. 950-960. doi:10.1002/pmic.200401101
- G. Adler, E. Blumwald and D. Bar-Zvi, “The Sugar Beet Gene Encoding the Sodium/Proton Exchanger 1 (BvNHX1) is Regulated by a MYB Transcription Factor” Planta, Vol. 232, No. 1, 2010, pp. 187-195. doi:10.1007/s00425-010-1160-7
- E. Nevo and G. Chen, “Drought and Salt Tolerances in Wild Relatives for Wheat and Barley Improvement,” Plant, Cell & Environment, Special Issue on Drought and Salinity Stress, Vol. 33, No. 4, 2010, pp. 670-685. doi:10.1111/j.1365-3040.2009.02107.x
- L. Comai, K. Young, B. J. Till, S. H. Reynolds, E. A. Greene, C. A. Codomo, L. C. Enns, J. E. Johnson, C. Burtner, A. R. Odden and S. Henikoff, “Efficient Discovery of DNA Polymorphisms in Natural Populations by Ecotilling,” The Plant Journal, Vol. 37, No. 5, 2004, pp. 778-786. doi:10.1111/j.0960-7412.2003.01999.x
- S. Henikoff, B. J. Till and L. Comai, “TILLING. Traditional Mutagenesis Meets Functional Genomics,” Plant Physiology, Vol. 135, No. 2, 2004, pp. 630-636. doi:10.1104/pp.104.041061
- G. Cordeiro, F. G. Eliott and R. J. Henry, “An Optimized Ecotilling Protocol for Polyploids or Pooled Samples Using a Capillary Electrophoresis System,” Analytical Biochemistry, Vol. 355, No. 1, 2006, pp. 145-147. doi:10.1016/j.ab.2006.03.026
- S. Negrão, C. Almadanim, I. Pires, K. L. McNally and M. M. Oliveira, “Use of EcoTILLING to Identify Natural Allelic Variants of Rice Candidate Genes Involved in Salinity Tolerance,” Plant Genetic Resources, Vol. 9, No. 2, 2011, pp. 300-304. doi:10.1017/S1479262111000566
- S. L. Frerichmann, M. Kirchhoff, A. E. Müller, A. J. Scheidig, C. Jung and F. J. Kopisch-Obuch, “EcoTILLING in Beta vulgaris Reveals Polymorphisms in the FLC-Like Gene BvFL1 that Are Associated with Annuality and Winter Hardiness,” BMC Plant Biology, Vol. 13, 2013, p. 52. doi:10.1186/1471-2229-13-52
- E. J. Gilchrist, G. W. Haughn, C. C. Ying, S. P. Otto, J. Zhuang, D. Cheung, B. Hamberger, F. Aboutorabi, T. Kalynyak, L. Johnson, J. Bohlmann, B. E. Ellis, C. J. Douglas and Q. C. Cronk, “Use of Ecotilling as an Efficient SNP Discovery Tool to Survey Genetic Variation in Wild Populations of Populus trichocarpa,” Molecular Ecology, Vol. 15, No. 5, 2011, pp. 1367-1378. doi:10.1111/j.1365-294X.2006.02885.x
- C. Nieto, F. Piron, M. Dalmais, C. F. Marco, E. Moriones, M. L. Gómez-Guillamón, V. Truniger, P. Gómez, J. Garcia-Mas, M. A. Aranda and A. Bendahmane, “EcoTILLING for the Identification of Allelic Variants of Melon eIF4E, a Factor that Controls Virus Susceptibility,” BMC Plant Biology, Vol. 21, 2007, p. 34. doi:10.1186/1471-2229-7-34.
- N. Mejlhede, Z. Kyjovska, G. Backes, K. Burhenne, S. K. Rasmussen and A. Jahoor, “EcoTILLING for the Identification of Allelic Variation in the Powdery Mildew Resistance Genes mlo and Mla of Barley,” Plant Breeding, Vol. 125, No. 5, 2006, pp. 461-467. doi:10.1111/j.1439-0523.2006.01226.x
- J. C. Knight, “Allele-Specific Gene Expression Uncovered,” Trends in Genetics, Vol. 20, No. 3, 2004, pp. 113- 116. doi:10.1016/j.tig.2004.01.001
- L. F. Thomas and P. Sætrom, “Single Nucleotide Polymorphisms Can Create Alternative Polyadenylation Signals and Affect Gene Expression through Loss of Micro RNA-Regulation,” PLoS Computational Biology, Vol. 8, No. 8, 2012, Article ID: e1002621. doi:10.1371/journal.pcbi.1002621
- M. Guo, M. A. Rupe, C. Zinselmeier, J. Habben, B. A. Bowen and O. S. Smith, “Allelic Variation of Gene Expression in Maize Hybrids,” The Plant Cell, Vol. 16, No. 7, 2004, pp. 1707-1716. doi:10.1105/tpc.022087
- N. M. Springer and R. M. Stupar, “Allele-Specific Expression Patterns Reveal Biases and Embryo-Specific Parent-of-Origin Effects in Hybrid Maize,” The Plant Cell, Vol. 19, No. 8, 2007, pp. 2391-2402. www.plantcell.org/cgi/doi/10.1105/tpc.107.052258
- Y. Zhuang and K. L. Adams, “Extensive Allelic Variation in Gene Expression in Populus F1 Hybrids,” Genetics, Vol. 177, No. 4, 2007, pp. 1987-1996. doi:10.1534/genetics.107.080325
- M. von Korff, S. Radovic, W. Choumane, K. Stamati, S. M. Udupa, S. Grando, S. Ceccarelli, I. Mackay, W. Powell, M. Baum and M. Morgante, “Asymmetric AlleleSpecific Expression in Relation to Developmental Variation and Drought Stress in Barley Hybrids,” The Plant Journal, Vol. 59, No. 1, 2009, pp. 14-26. doi:10.1111/j.1365-313X.2009.03848.x
- A. Akkinepalli, N. Ereful, Y. Liu, K. Malabanan, R. Howells, K. Stamati, W. Powell, H. Leung, A. Greenland, I. Mackay and D. Lee, “Snapshots of Gene Expression in Rice: Limitations for Allelic Expression Imbalance Determination,” Genome, Vol. 55, No. 5, 2012, pp. 400-406. doi:10.1139/g2012-023
- J. de Meaux, U. Goebel, A. Pop and T. Mitchell-Olds, “Allele-Specific Assay Reveals Functional Variation in the Chalcone Synthase Promoter of Arabidopsis thaliana that is Compatible with Neutral Evolution,” The Plant Cell, Vol. 17, No. 3, 2005, pp. 637-690. doi:10.1105/tpc.104.027839
- S. Little, “Amplification-Refractory Mutation System (ARMS) Analysis of Point Mutations,” Current Protocols in Human Genetics, 2001. doi:10.1002/0471142905.hg0908s07
- C. R. Newton, A. Graham, L. E. Heptinstall, S. J. Powell, C. Summers, N. Kalsheker, J. C. Smith and A. F. Markham, “Analysis of any Point Mutation in DNA. The Amplification Refractory Mutation System (ARMS),” Nucleic Acids Research, Vol. 17, No. 7, 1989, pp. 2503- 2516.
- W. E. Glaab and T. R. Skopek, “A Novel Assay for Allelic Discrimination that Combines the Fluorogenic 5’ Nuclease Polymerase Chain Reaction (TaqMan) and Mismatch Amplification Mutation Assay,” Mutation Research, Vol. 430, No. 1, 1999, pp. 1-12. doi:10.1016/S0027-5107(99)00147-5
- L. Milani, M. Gupta, M. Andersen, S. Dhar, M. Fryknäs, A. Isaksson, R. Larsson and A. C. Syvänen, “Allelic Imbalance in Gene Expression as a Guide to Cis-Acting Regulatory Single Nucleotide Polymorphisms in Cancer Cells,” Nucleic Acids Research, Vol. 35, No. 5, 2007, p. e34. doi:10.1093/nar/gkl1152
- J. W. Davey and M. L. Blaxter, “RADSeq: Next-Generation Population Genetics,” Briefings in Functional Genomics, Vol. 9, No. 5-6, 2010, pp. 416-423. doi:10.1093/bfgp/elq031
- J. W. Davey, P. A. Hohenlohe, P. D. Etter, J. Q. Boone, J. M. Catchen and M. L. Blaxter, “Genome-Wide Genetic Marker Discovery and Genotyping Using Next-Generation Sequencing,” Nature Reviews Genetics, Vol. 12, No. 7, 2011, pp. 499-510.
- M. R. Miller, J. Dunham, A. Amores, W. Cresko and E. Johnson, “Rapid and Cost-Effective Polymorphism Identification and Genotyping using Restriction Site Associated DNA (RAD) Markers,” Genome Research, Vol. 17, 2007, pp. 240-248. doi:10.1101/gr.5681207
- N. A. Baird, P. D. Etter, T. S. Atwood, M. C. Currey, A. L. Shiver, Z. A. Lewis, E. U. Selker, W. A. Cresko and E. A. Johnson, “Rapid SNP Discovery and Genetic Mapping using Sequenced RAD Markers,” PLoS One, Vol. 3, No. 10, 2008, Article ID: e3376. doi:10.1371/journal.pone.0003376
- P. A. Hohenlohe, S. Bassham, P. D. Etter, N. Stiffler, E. A. Johnson and W. A. Cresko, “Population Genomics of Parallel Adaptation in Three Spine Stickleback Using Sequenced RAD Tags,” PLoS Genetics, Vol. 6, No. 2, 2010, Article ID: e1000862. doi:10.1371/journal.pgen.1000862
- D. Scaglione, A. Acquadro, E. Portis, M. Tirone, S. J. Knapp and S. Lanteri, “RAD Tag Sequencing as a Source of SNP Markers in Cynara cardunculus L.,” BMC Genomics, Vol. 13, 2012, p. 3. doi:10.1186/1471-2164-13-3.
- W. F. Pfender, M. C. Saha, E. A. Johnson and M. B. Slabaugh, “Mapping with RAD (Restriction-Site Associated DNA) Markers to Rapidly Identify QTL for Stem Rust Resistance in Lolium perenne,” Theoretical and Applied Genetics, Vol. 122, No. 8, 2011, pp. 1467-1480. doi:10.1007/s00122-011-1546-3
- H. Yang, Y. Tao, Z. Zheng, C. Li, M. Sweetingham and J. Howieson, “Application of Next Generation Sequencing for Rapid Marker Development in Molecular Plant Breeding: A Case Study on Anthracnose Disease Resistance in Lupinus angustifolius L.,” BMC Genomics, Vol. 13, 2011, p. 318. doi:10.1186/1471-2164-13-318.
- S. Deschamps, V. Llaca and G. D. May, “Genotypingby-Sequencing in Plants,” Biology, Vol. 1, No. 3, 2012, pp. 460-483. doi:10.3390/biology1030460
NOTES
*This work was supported by the Portuguese Foundation for Science and Technology with the FCT/Ciência 2008 to DB and MMR. Ecological data presented is an output from the project PTDC/AGR-AAM/ 73144/2006 funded by the Portuguese Foundation for Science and Technology. The authors would like to thank to Ezequiel Correia for assistance on georeferencing.
#Equally contributed as first authors.

