American Journal of Plant Sciences
Vol.4 No.4(2013), Article ID:29690,12 pages DOI:10.4236/ajps.2013.44097
The Expressed Parasitism Genes in the Reniform Nematode (Rotylenchulus reniformis)
![]()
1Department of Biological and Environmental Sciences, Alabama A & M University, Normal, USA; 2Arthritis & Clinical Immunology Department, Oklahoma Medical Research Foundation, Oklahoma City, USA; 3The Advanced Centre for Genome Technology, University of Oklahoma, Normal, USA; 4Department of Biological Sciences, University of Alabama in Huntsville, Huntsville, USA.
Email: *seloame.nyaku@aamu.edu
Copyright © 2013 Seloame T. Nyaku et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received February 9th, 2013; revised March 11th, 2013; accepted March 30th, 2013
Keywords: Parasitism Genes; Reniform Nematode; Transcriptome
ABSTRACT
The reniform nematode (RN), Rotylenchulus reniformis, is an agriculturally important pest with a broad host range that results in a large economic impact in tropical, subtropical and in warm temperate zones. In an initial effort to understand the transcriptome and gene expression in RN, we present EST results that reveal numerous putative parasitism-related genes some of which play roles in plant cell wall modification. The characterized contigs included 8362 (40.6%) matches to unique proteins. Coding contigs predicted were 10,656 (51.7%) or 3079 (14.9%), that was similar to those identified in Brugia malayi and Caenorhabditis elegans as reference organisms respectively. Specific transcripts studied in more detail include putative plant parasitism genes, prominent among them were several plant cell wall modification genes. Contigs matching 14 parasitism genes found in sedentary endoparasitic nematodes included expansins, hexosaminidase, glycosyl hydrolases family, 14-3-3 protein, xylanases, glutathione peroxidase, pectate lyase, β-1,4-endoglucanase, major sperm protein, aminopeptidase, c-type lectin, chitin synthase, FMR famide-like peptide, and calreticulin. These genes function in suppression of host defenses and development of feeding sites.
1. Introduction
Rotylenchulus reniformis, commonly referred to as reniform nematode (RN), is a semi-endoparasitic nematode with a broad host range of over 300 plant species. Infections of RN begin when the female penetrates the root cortex via specialized nematode feeding cells that are regulated by nematode parasitism genes expressed within the esophageal glands, and delivered into the feeding cell through the stylet [1,2]. One major approach employed in expression profiling involving nematode parasitism is transcriptome sequencing [3]. These studies have enabled identification of genes such as glutathione peroxidase [4], pectate lyase [5], polygalacturonase [6], β-1,4-endoglucanase, β-1,4-endoxylanase [7], and chorismate mutase [8,9]. Other techniques employed in transcriptome sequencing include Serial Analysis of Gene Expression (SAGE) [10]. This method detects genes through sequencing of tags 14 to 25 bp in length, however, a limitation of this method is having most reads towards the 5’ end of the transcript; again full transcript copies are not obtained. Microarrays have also been applied in quantification of relative expression levels of known genes in the soybean cyst nematode [11-13]. A disadvantage of this technique, however, relates to certain genes not being detected because their sequences are unknown, also cross-hybridization of closely related gene sequences produces unreliable results [14]. These limitations have been overcome through the use of high throughput sequencing platforms such as the 454/Roche GS-FLX Pyrosequencer in transcriptome profiling, generating sufficient data for full-length transcripts assembly often greater than 5 kb [15]. The 454 sequencing technique has been extensively used in expression profiling studies and in discovery of novel genes [16]. Extensive information exists for the well-studied plant-parasitic nematode Caenorhabditis elegans, and the study of nematode biology has greatly increased our understanding of numerous members of nematode genera. Transcriptome sequencing of C. elegans in its first larval stage generated 30 Mb of sequence data and 14% of novel EST sequences (300,000) could effectively be mapped to genomic regions of C. elegans with no known annotated genes or splice variants, contributing to identification of new genetic structures [17]. Genomic resources including expression data for the RN is in its infancy, because to date only 2004 ESTs are available for this nematode in GenBank [3]. This therefore necessitates the need for further sequencing of the RN transcriptome using next-generation platforms for identification of parasitic genes. The production and availability of ESTs will also enable investigation of the evolutionary history of nematodes.
The objective of this study was to generate sufficient coverage ESTs to permit identification of genes that are expressed at elevated levels in cDNA libraries prepared from eggs and vermiform life stages of the RN to identify candidate parasitism genes.
2. Materials and Methods
2.1. Collection, Sterilization and Hatching of Reniform Nematode Eggs
Eggs of RNs cultured on roots of greenhouse-grown Micro-Tom tomato plants, were extracted from the roots and surface disinfested by immersing them in 5% bleach with shaking for 4 minutes in a beaker. The egg-containing solution was then poured through a sterilized 325-mesh sieve nested on a 500-mesh sieve autoclaved using a dry cycle (120˚C for 1 hr). The trapped eggs on the 500-mesh sieve were rinsed immediately with ~300 mL of sterilized distilled water to wash off the bleach for about 5 minutes, and then transferred into sterilized beakers containing 10 mL of sterilized distilled water. One mL of the solution containing the sterilized eggs was placed onto agar plates, these were sealed with Parafilm, and covered with aluminum foil, and placed in an incubator set at 25˚C for 2 to 4 days for the eggs to hatch into Juvenile 1 (J1), and permit growth up to Juvenile 2 (J2) stage on the 8th day.
2.2. RNA Extraction
Total RNA was extracted from eggs and vermiform stages of pooled nematodes using the PicoPure RNA kit (LifeTechnologies, Grand Island, NY) and treated with RNase-free DNase (Qiagen) following the manufacturer’s instructions. The quality and concentration of the RNA was assessed using the ExperionTM RNA StdSen Analysis Kit (Bio-Rad Laboratories, Inc., Hercules, CA), and Nanodrop 100 Spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE).
2.3. Construction of cDNA Libraries
These libraries were constructed using the CreatorTM SMARTTM cDNA library kit (Clonetech, CA, USA) using Long Distance PCR (LD PCR) according to the manufacturer’s instructions. Amplified products were purified using a Sigma Aldrich GeneEluteTM PCR CleanUp kit (Sigma-Aldrich, St. Louis, MO). Concentrations of these libraries were assessed using a Nanodrop 100 Spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE) and TKO 100 fluorometer (Hoefer Scientific Instruments, San Francisco).
2.4. Pyrosequencing (454) and Data Analysis
Purified constructed cDNA libraries (5 µg) were used in high-throughput sequencing at the Advanced Centre for Genome Technology, University of Oklahoma. The raw reads generated were assembled using the SeqMan Lasergene software (DNASTAR Inc., Madison WI, USA) after removal of primer sequences, poly (A/T) tails, and ribosomal sequences. All EST sequences generated were submitted to GenBank at NCBI under the short read archive (SRA) with accession numbers SRX098224 and SRX098225. BLAST 2.2.21 was downloaded from NCBI ftp://ftp.ncbi.nlm.nih.gov/blast/) and used in creating a local Blast database for initially comparing the RN sequence reads to reniform ESTs available in GenBank with expected value (E) of <1.0e−10. Downloaded reniform ESTs in GenBank [3] were assembled into 107 contigs and 519 singletons, using the CLC Genomics Workbench 4.9 with the following parameters: Mismatch cost = 2, insertion cost = 3, deletion cost = 3, length fraction = 0.5, similarity = 0.9, alignment = global, conflict resolution = vote (A, C, G, and T), word size = automatic, non-specific matches = random and minimum contig length = 200. ESTs were then accessed for their coding potential using the AUGUSTUS gene prediction software [18] with B. malayi and C. elegans serving as model organisms. This tool employs a Generalized Hidden Markov Model (GHMM), defined by probability distributions for portions of genomic sequences thus identifying an optimal parse for a given genomic sequence and divides the sequence into states based on statistical models.
The universal Gene Ontology annotation, visualization, and analysis tool, (Blast2go) http://blast2go.org was employed for annotation of the functions and identification of protein-coding genes within our ESTs. The gene ontology (GO) classification scheme was used in categorizing transcripts by their putative function. These analyses were categorized into molecular function, biological process, and cellular components. Candidate parasitism genes were identified by restricting BlastX search with expected value (E) of <1.0e−10. Potential homologs in other nematodes within the reniform ESTs were through comparisons with NCBI EST others database using TBlastX. Further, comparisons of our sequences were with ESTs of C. elegans, B. malayi, Meloidogyne incognita, and Pristionchus pacificus with E-values of <1.0e−10.
3. Results and Discussion
3.1. Analysis of the Reniform Nematode Transcriptome
Over 50,000 sequence reads were generated from the RN transcriptome, resulting in more than 4 Mb (4,781,676 bases) of data that were assembled into 20,596 contigs (Table 1). Assembly of the RN raw reads downloaded from GenBank resulted in 107 contigs and 519 singletons (626 unigenes). Comparison of our ESTs to those assembled from GenBank identified 617 entries out of which 553 were unique. The alignment lengths for homology varied from 32 to 754 bp, with similarities of greater than 77%. We therefore observed 209/626 (33.4%) unique GenBank ESTs with matches to our 410/20,596 (1.9%) sequence reads.
3.2. Gene Ontology (GO) Assignments and AUGUSTUS Gene Predictions
The most highly represented activities under each of these categories were ATP binding (Figure 1), oxidation-reduction (Figure 2), and integral to membrane (Figure 3) for molecular function, biological process, and cellular components respectively. Among the several contigs generated, 10,656 (51.7%) and 3079 (14.9%) coded for genes using B. malayi and C. elegans, as reference organisms respectively. More contigs coded for genes with B. malayi serving as a reference organism probably because of the parasitic nature of B. malayi as opposed to the free living nature of C. elegans. Sixteen contigs were predicted to code for genes both by C. elegans and B. malayi (Table 2). These contigs could further be studied because they may probably be involved in
Table 1. Reniform nematode transcriptome assembly.
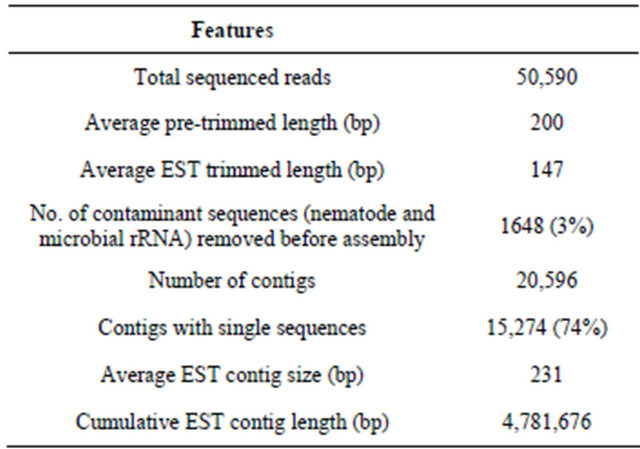
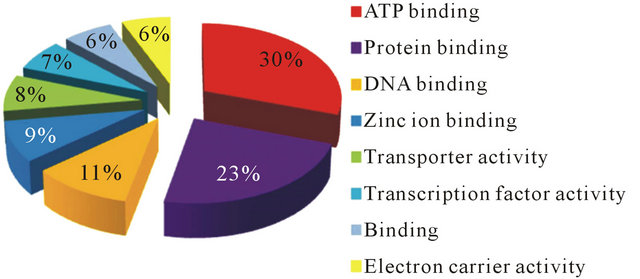
Figure 1. Gene ontology (GO) distributions after BlastX analysis for reniform nematode EST sequences as grouped by molecular function.
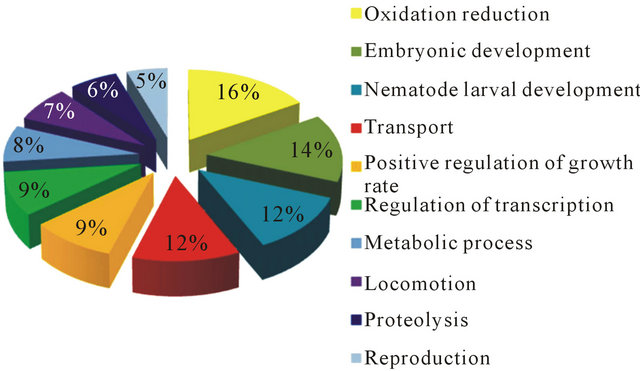
Figure 2. Gene ontology (GO) distributions after BlastX analysis for reniform nematode EST sequences as grouped by biological process.
Figure 3. Gene ontology (GO) distributions after BlastX analysis for reniform nematode EST sequences as grouped by cellular component.
house-keeping roles. TBlastX analysis produced 11,588 matches (56.3%) with assigned functions. Majority of hits were observed with B. malayi, (620/11,588 or 5.4%). Further comparative analysis between RN ESTs predicted to code for genes by AUGUSTUS using B. malayi as the model organism (10,656 ESTs) with BlastX hits (11,588) and ESTs with GO IDs (8362) resulted in 179 and 135 matches, respectively (Figure 4). Similarly, when C. elegans was used as a model organism, 149 TBlastX hits and 99 ESTs with GO IDs matched with ESTs coding for genes (Figure 5). This analysis suggests that, majority of contigs predicted to code for genes have not been fully annotated within the reniform transcriptome. A full list of contigs with GOs and functions is available.
Table 2. Sixteen common EST contigs among C. elegans and B. malayi predicted by AUGUSTUS with annotations.
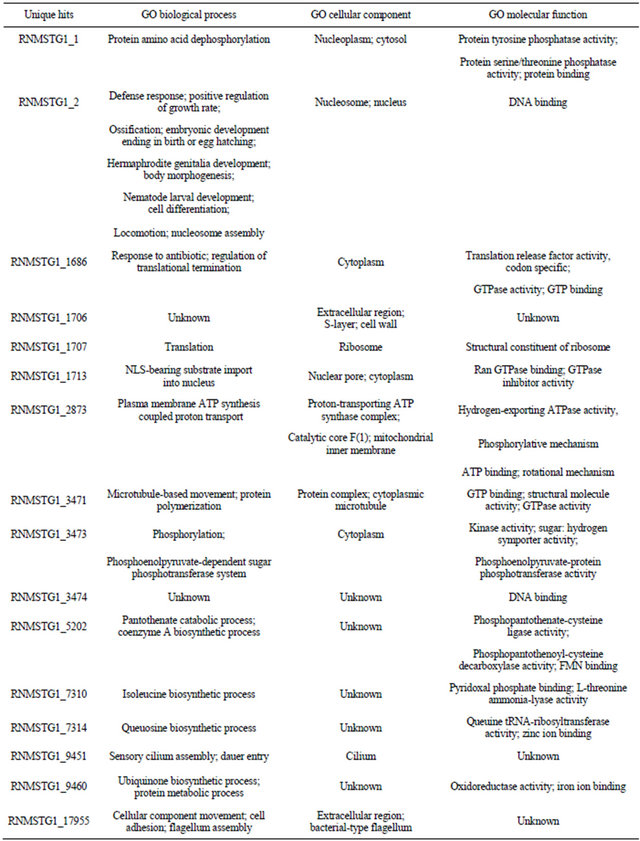
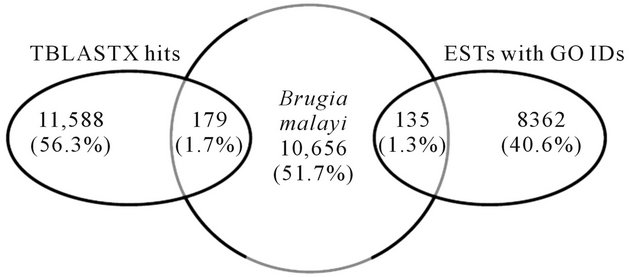
Figure 4. Comparison of ESTs predicted as genes by AUGUSTUS using Brugia malayi as a model organism with TBlastX homology and ESTs with Gene Ontology IDs.
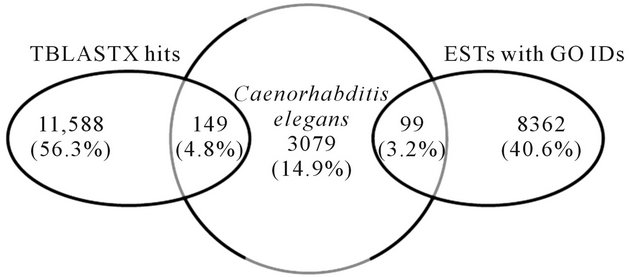
Figure 5. Comparison of ESTs predicted as genes by AUGUSTUS using Caenorhabditis elegans as a model organism with TBlastX homology and ESTs with Gene Ontology IDs.
Comparative analysis between RN ESTs and M. incognita revealed that 10.5% of RN sequences matched those of M. incognita. The RN ESTs also had 0.8% matches to both B. malayi and C. elegans ESTs, although more EST contigs were predicted to code for genes (Figures 4 and 5). These low percentages could be as a result of phylogenetic distance of these nematodes from the RN, and also the nature of some of the ESTs which may not be of full length. The least organism having matches to the RN ESTs was P. pacificus (0.7%) (Figure 6). The complete list of Blastn analysis between RN ESTs and these nematodes is available
3.3. Identification of Nematode Parasitism Genes
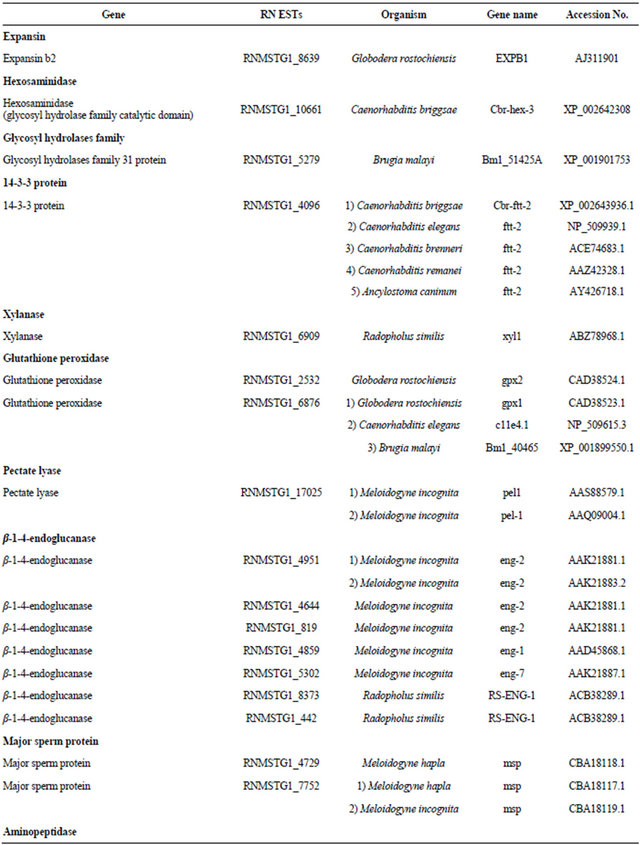
Fourteen plant parasitic nematode genes critical to modification of plant cell walls were identified within our RN ESTs (Table 3). These had expected values (E) of <1.0e−10 to regions of homology with parasitic genes in other nematodes (Table 4). The nematode parasitism genes identified include expansins, hexosaminidase, glycosyl hydrolases family, 14-3-3 protein, xylanases, glutathione peroxidase, pectate lyase, β-1,4-endoglucanase, major sperm protein, aminopeptidase, c-type lectin, chitin synthase, FMR famide-like peptide, and calreticulin. The nematode parasitome consists mostly of secreted gene products critical in plant parasitism [19]. Among these are expansin and xylanase which are cellwall modifying proteins. Expansins digest cell walls by
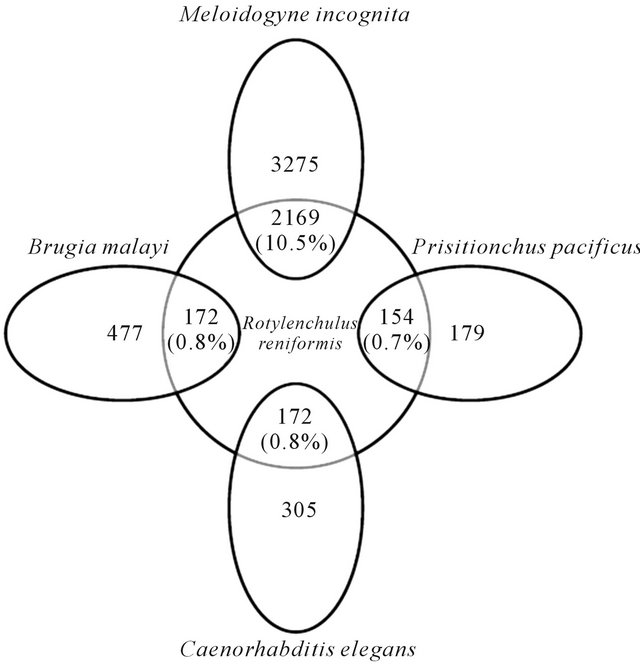
Figure 6. Comparative analysis among RN ESTs and ESTs of Brugia malayi, Meloidogyne incognita, Caenorhabditis elegans, and Pristionchus pacificus.
cleaving the non-covalent bonds and thus enhancing the activities of cell wall-degrading, carbohydrate-active enzymes (CAZymes) [20,21]. A second class of enzyme identified within the RN ESTs was hexosaminidase (glycosyl hydrolases), which is highly conserved across certain domains of bacteria and human. The β-Hexosaminidases (EC 3.2.1.52), has a specific role in removal of terminal protein glycosylation with O-linked N-acetylglucosamine (O-GlcNAc) residues found in glycoproteins and glycolipids [22]. The RNMSTG1_10661 contig had homology to the hexosaminidase gene (Cbr-hex-3) within B. malayi, suggesting the presence of this enzyme in the RN genome. Chitinases belong to glycosyl hydrolases family (EC 3.2.1.14), known to be involved in hydrolysis of β-1,4-N-acetyl-D-glucosamine linkages in chitin polymers. They catalyze reactions by hydrolyzing glycosidic bonds between two carbohydrate molecules, or a carbohydrate and a non-carbohydrate molecule, and have been identified in filarial nematodes [23,24]. The RNMSTG1_5271 contig had homology to a chitinase gene (Bm1_51425A) in B. malayi, indicating the involvement of this enzyme probably in hydrolysis of glycosidic bonds in chitin during RN parasitism. SXPRAL-2, 14-3-3, and RanBPM-like family proteins participate in regulation of cell-cycle, calcium ion binding, defense regulation mechanisms, and may act as molecular chaperones [20]. Within the RN ESTs, the ftt-2 gene belonging to 14-3-3 protein class was identified in the RNMSTG1_4096 contig, and had homologs in C. briggsae, C. elegans, C. brenneri, C. remanei, and Ancylostoma
Table 3. Candidate parasitism genes identified within the RN ESTs.
Continued
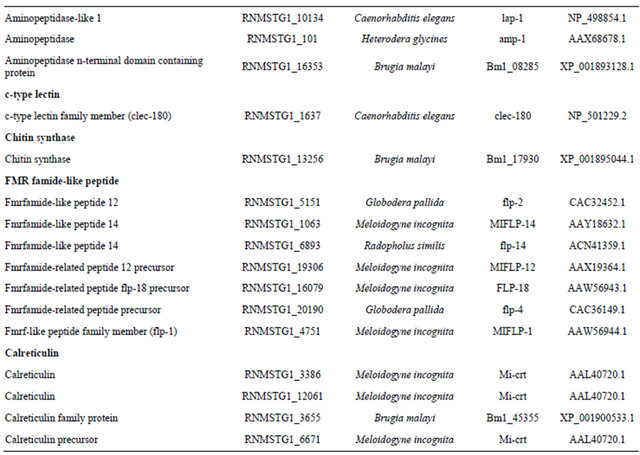
caninum. Xylanase is a component of hemicellulose, which functions in cleaving β-1,4-linkage of xylopyranose subunits [25]. The presence of xylanase in the RN ESTs, specifically in the RNMSTG1_6909 contig with homology to RS-xyl1 gene in Radopholus similis may suggest the importance of this enzyme in RN genome for the breakdown of xylan during parasitism. Glutathione peroxidases (GpX) plays roles in protecting animal parasitic nematodes through removal of hydrogen peroxide defensively released by plants in response to nematode attack [26]. This enzyme complex also catalyzes the conversion of peroxidised fatty acids to alcohols. The RNMSTG1_2532 contig had matches to gxp2 gene in G. rostochiensis while RNMSTG1_6876 contig had significant identity to gxp1, clle4.1, and Bm1_40465 genes in G. rostochiensis, C. elegans, and B. malayi, respectively, suggesting the importance of glutathione peroxidase in the RN for parasitism. The RNMSTG1_17025 contig encoded a pectate lyase gene (pel-1) gene previously cloned in M. incognita. Pectate lyases are commonly associated with proteomes of bacterial and fungal pathogens [27], and disrupt the glycosidic bonds in the primary cell wall and in middle lamella [28]. These enzymes act as pathogenicity factors, because of their disruptive nature to host cell walls [29]. A qPCR and RNAi analysis in H. glycines targeting pectate lyase revealed an increase in male: female ratios after J2 nematodes were soaked in a solution containing double-stranded (ds) RNA [30]. In another study involving M. incognita, two cDNAs (Mi-pel-1 and Mi-pel-2) encoding pectate lyase have been isolated from the esophageal gland-cell through subtractive cDNA libraries [31]. The β-1,4-endoglucanase gene identified in our RN ESTs, showed high homology to the endoglucanases of M. incognita and R. similis respectively. Cellulases belong to the glycosyl hydrolase family 5 and similarity of these genes to bacterial homologs suggests that, they may have been acquired through horizontal gene transfer (HGT) [1]. These genes have also been identified in H. glycines, G. rostochiensis, M. incognita, B. xylophilus, P. penetrans, and Heterodera spp. Respectively [32-40]. In invertebrates including nematodes, sperms serve as signals involved in meiosis during the arrested oocytes developmental stage [41]. The MSP has been identified as a 14.1-kD peptide using MALDI-TOF mass spectroscopy [42], and known to suppress Heterodera glycines reproduction in transgenic soybeans plants expressing specific siRNAs to the MSP gene [43]. The RNMSTG1_4729 and RNMSTG1_7752
Table 4. Candidate parasitism genes identified within the RN ESTs and their E-values.

contigs encoded MSP genes suggesting the importance of the MSP in RN reproduction. Aminopeptidases are known to mediate processes such as neuropeptide and signal transduction [44], embryo nourishment and digestion [45], molting [46], and reproduction [47]. These enzymes can serve as targets for nematode control [48]. The RNMSTG1_10134, RNMSTG1_101, and RNMSTG1_16353 contigs had matches to lap-1, amp-1, and Bm1_08285 genes in C. elegans, H. glycines, and B. malayi, respectively. C-type lectin, a pathogen recognition protein was identified within the RN dataset. The RNMSTG1_1637 contig showed high homology to the clec-180 gene. In C. elegans silencing of this gene through RNAi, resulted in decrease in cyst nematode numbers [49]. The RNMSTG1_13256 contig encoded for the chitin synthase gene which has been previously silenced in M. artiellia hindering the formation and survival of the eggs in this nematode [50]. Chitin is an important component in the fungal cell wall, cuticle of insects [51], and nematodes [52]. This structure is composed of β-1,4 linked N-acetylglucosamine residue chains. FMRFamide-like peptides or FLPs identified in our ESTs are a class of neuropeptides with specific roles in sensory and motor activities in nematodes, and also serve as neurotransmitters or neuromodulators within the nervous system. Recently, two of these genes flp-12 and flp-16 have been cloned and characterized in H. avenae [53]. These genes have been proposed to serve as critical targets for plant parasitic nematode management. The C-terminal signature (Arg-Phe-NH2) of these peptides is highly conserved across C. elegans, A. suum, and M. incognita. Three of the reniform contigs coded for this class of genes. Calreticulins are a class of calcium-binding proteins, which are conserved in both plants and animals. These proteins are localized in the endoplasmic reticulum and primarily act as chaperones [54]. Other functions of calreticulins include transportation of proteins from the nucleus [54], mRNA degradation [55], cell adhesion [56], calcium regulation, cell cycle, endocytosis, exocytosis, secretion, and cell growth and differentiation [57]. Calreticulins are abundant in the esophageal secretions of nematodes especially at the nematode feeding sites (NFS). An example is Mi-CRT gene synthesized in the esophageal glands of M. incognita [58]. Three RN contigs were homologous with calreticulins of M. incognita and B. malayi respectively.
3.4. Conclusion
This study has generated a number of novel transcripts with hitherto unknown function that will need further characterization. The nematode parasitism gene identified here could serve as RNAi targets for investigating plant resistance against RNs and be further explored through functional analysis for effective design of management strategies
4. Acknowledgements
This work was supported by grants to RVK and/or to GCS: USDA-CSREES Grant # 2004-38814-15160, USDA ALAX-011-706 and NSF/PGRP award #DBI 0703470 and Agricultural Experiment Station. This is Journal article # 656 of Alabama A & M Agricultural Experiment Station.
REFERENCES
- E. L. Davis, R. S. Hussey, T. J. Baum, J. Bakker and A. Schots, “Nematode Parasitism Genes,” Annual Review of Phytopathology, Vol. 38, 2000, pp. 365-396. doi:10.1146/annurev.phyto.38.1.365
- E. L. Davis, R. S. Hussey and T. J. Baum, “Getting to the Roots of Parasitism by Nematodes,” Trends in Parasitology, Vol. 20, No. 3, 2004, pp. 134-141. doi:10.1016/j.pt.2004.01.005
- M. J. Wubben, F. E. Callahan and B. S. Scheffler, “Transcript Analysis of Parasitic Females of the Sedentary Semi-Endoparasitic Nematode Rotylenchulus reniformis,” Molecular and Biochemical Parasitology, Vol. 172, No. 1, 2010, pp. 31-40. doi:10.1016/j.molbiopara.2010.03.011
- J. T. Jones, B. Reavy, G. Smant and A. E. Prior, “Glutathione Peroxidases of the Potato Cyst Nematode Globodera rostochiensis,” Gene, Vol. 324, 2004, pp. 47-54. doi:10.1016/j.gene.2003.09.051
- H. Popeijus, H. Overmars, J. Jones, V. Blok, A. Goverse, J. Helder, A. Schots, J. Bakker and G. Smant, “Degradation of Plant Cell Walls by a Nematode,” Nature, Vol. 406, 2000, pp. 36-37. doi:10.1038/35017641
- S. Jaubert, J. B. Laffaire, P. Abad and M. N. Rosso, “A Polygalacturonase of Animal Origin Isolated from the Root-Knot Nematode Meloidogyne incognita,” FEBS Letters, Vol. 522, No. 1-3, 2002, pp. 109-112. doi:10.1016/S0014-5793(02)02906-X
- M. Dautova, N. Rosso, P. Abad, F. J. Gommers, J. Bakker and G. Smant, “Single Pass cDNA Sequencing—A Powerful Tool to Analyse Gene Expression in Pre-Parasitic Juveniles of the Southern Root-Knot Nematode, Meloidogyne incognita,” Nematology, Vol. 3, No. 2, 2001, pp. 29-139. doi:10.1163/156854101750236259
- K. N. Lambert, K. D. Allen and I. M. Sussex, “Cloning and Characterization of an Esophageal-Gland-Specific Chorismate Mutase from the Phytoparasitic Nematode Meloidogyne javanica,” Molecular Plant-Microbe Interactions, Vol. 18, No. 6, 1999, pp. 593-601. doi:10.1094/MPMI-18-0593
- J. T. Jones, C. Furlanetto, E. Bakker, B. Banks, V. Blok, Q. Chen, M. Phillips and A. Prior, “Characterization of a Chorismate Mutase from the Potato Cyst Nematode Globodera pallida,” Molecular Plant Pathology, Vol. 4, No. 1, 2003, pp. 43-50. doi:10.1046/j.1364-3703.2003.00140.x
- V. E. Velculescu, L. Zhang, B. Vogelstein and K. W. Kinzler, “Serial Analysis of Gene Expression,” Science, Vol. 270, No. 5235, 1995, pp. 484-487. doi:10.1126/science.270.5235.484
- D. P. Puthoff, D. Nettleton, S. R. Rodermel and T. J. Baum, “Arabidopsis Gene Expression Changes during Cyst Nematode Parasitism Revealed by Statistical Analyses of Microarray Expression Profiles,” Plant Journal, Vol. 33, No. 5, 2003, pp. 911-921. doi:10.1046/j.1365-313X.2003.01677.x
- R. Khan, N. Alkharouf, H. Beard, M. MacDonald, I. Chouikha, S. Meyer, J. Grefenstette, H. Knap and B. Matthews, “Microarray Analysis of Gene Expression in Soybean Roots Susceptible to the Soybean Cyst Nematode Two Days Post Invasion,” Journal of Nematology, Vol. 36, No. 3, 2004, pp. 241-248.
- J. M. de Boer, J. P. McDermott, X. Wang, T. Maier, F. Qui, R. S. Hussey, E. L. Davis and T. J. Baum, “The Use of DNA Microarrays for the Developmental Expression Analysis of cDNAs from the Oesophageal Gland Cell Region of Heterodera glycines,” Molecular Plant Pathology, Vol. 3, No. 4, 2002, pp. 261-270. doi:10.1046/j.1364-3703.2002.00122.x
- R. Kothapalli, S. J. Yoder, S. Mane and T. P. Loughran Jr., “Microarray Results: How Accurate Are They?” BMC Bioinformatics, Vol. 3, 2002, p. 22. doi:10.1186/1471-2105-3-22
- J. H. Leamon, M. S. Braverman and J. M. Rothberg, “High-Throughput, Massively Parallel DNA Sequencing Technology for the Era of Personalized Medicine,” Gene Therapy and Regulation, Vol. 3, No. 1, 2007, pp. 15-31. doi:10.1142/S1568558607000046
- T. T. Torres, M. Metta, B. Ottenwalder and C. Schlotterer, “Gene Expression Profiling by Massively Parallel Sequencing,” Genome Research, Vol. 18, 2008, pp. 172-177. doi:10.1101/gr.6984908
- H. Shin, M. Hirst, M. N. Bainbridge, V. Magrini, E. Mardis, D. G. Moerman, M. A. Marra, D. L. Baillie and S. J. M. Jones, “Transcriptome Analysis for Caenorhabditis elegans Based on Novel Expressed Sequence Tags,” BMC Biology, Vol. 6, 2008, p. 14. doi:10.1186/1741-7007-6-30
- M. Stanke, M. Diekhans, R. Baertsch and D. Haussler, “Using Native and Syntenically Mapped cDNA Alignments to Improve de Novo Dene Finding,” Bioinformatics, Vol. 24, No. 5, 2008, pp. 637-644. doi:10.1093/bioinformatics/btn013
- R. S. Hussey, E. L. Davis and T. J. Baum, “Secrets in Secretions: Genes That Control Nematode Parasitism in Plants,” Brazilian Journal of Plant Physiology, Vol. 14, No. 3, 2002, pp. 183-194. doi:10.1590/S1677-04202002000300002
- M. G. Mitchum, R. S. Hussey, E. L. Davis and T. J. Baum, “Application of Biotechnology to Understand Pathogenesis of Nematode Plant Pathogens,” In: Z. K. Punja, S. DeBoer and H. Sanfacon, Eds., Biotechnology & Plant Disease Management, CABI International, 2007, pp. 58-86. doi:10.1079/9781845932886.0058
- L. Qin, U. Kudla, E. H. A. Roze, A. Goverse, H. Popeijus, H. O. Nieuwland, J. T. Jones, A. Schots, G. Smant, J. Bakker and J. Helder, “A Nematode Expansin Actingon Plants,” Nature, Vol. 427, 2004, p. 30. doi:10.1038/427030a
- M. Gutternigg, D. Rendić, R. Voglauer, T. Iskratsch and I. B. H. Wilson, “Mammalian Cells Contain a Second Nucleocytoplasmic Hexosaminudase,” Biochemical Journal, Vol. 419, 2009, pp. 83-90. doi:10.1042/BJ20081630
- J. Flach, P. E. Pilet and P. Jollés, “What’s New in Chitinase Research,” Experientia, Vol. 48, No. 8, 1992, pp. 701-716. doi:10.1007/BF02124285
- J. A. Fuhrman, W. S. Lane, R. F. Smith, W. F. Piessens and F. B. Perler, “Transmission-Blocking Antibodies Recognize Microfilarial Chitinase in Brugian Lymphatic Filariasis,” Proceedings of National Academy of Sciences of United States, Vol. 89, No. 5, 1992, pp. 1548-1552. doi:10.1073/pnas.89.5.1548
- T. Collins, C. Gerday and G. Feller, “Xylanases, Xylanase Families and Extremophilic Xylanases,” FEMS Microbiology Reviews, Vol. 29, No. 1, 2005, pp. 3-23. doi:10.1016/j.femsre.2004.06.005
- G. H. Waetzig, M. Sobczak and F. M. W. Grundler, “Localization of Hydrogen Peroxide during the Defence Response of Arabidopsis thaliana Against the Plant Parasitic Nematode Heterodera glycines,” Nematology, Vol. 1, No. 7-8, 1999, pp. 681-686. doi:10.1163/156854199508702
- S. R. Herron, J. A. E. Benen, R. D. Scavetta, J. Visser and F. Jurnak, “Structure and Function of Pectic Enzymes: Virulence Factors of Plant Pathogens,” Proceedings of the National Academy of Sciences of United States, Vol. 97, No. 16, 2000, pp. 8762-8769. doi:10.1073/pnas.97.16.8762
- M. D. Yoder, N. T. Keen and F. Jurnak, “New Domain Motif: The Structure of Pectate Lyase C, a Secreted Plant Virulence Factor,” Science, Vol. 260, No. 5113, 1993, pp. 1503-1507. doi:10.1126/science.8502994
- P. Alghisi and F. Favaron, “Pectin-Degrading Enzyme and Plant-Parasite Interactions,” European Journal of Plant Pathology, Vol. 101, No. 4, 1995, pp. 365-375. doi:10.1007/BF01874850
- M. Bakhetia, P. E. Urwin and H. J. Atkinson, “qPCR Analysis and RNAi Define Pharyngeal Gland Cell-Expressed Genes of Heterodera glycines Required for Initial Interactions with the Host,” Molecular Plant-Microbe Interaction, Vol. 20, No. 3, 2007, pp. 306-312. doi:10.1094/MPMI-20-3-0306
- G. Huang, R. Dong, R. Allen, E. L. Davis, T. J. Baum and R. S. Hussey, “Developmental Expression and Molecular Analysis of Two Meloidogyne incognita Pectate Lyase Genes,” International Journal for Parasitology, Vol. 35, No. 6, 2005, pp. 685-692. doi:10.1016/j.ijpara.2005.01.006
- G. Smant, J. P. W. G. Stokkermans, Y. T. Yan, J. M. de Boer, T. J. Baum, X. H. Wang, R. S. Hussey, F. J. Gommers, B. Henrissat, E. L. Davis, J. Helder, A. Schots and J. Bakker, “Endogenous Cellulases in Animals: Isolation of Beta-1,4-Endoglucanase Genes from Two Species of Plant-Parasitic Cyst Nematodes,” Proceedings of the National Academy of Sciences of United States, Vol. 95, No. 9, 1998, pp. 4906-4911. doi:10.1073/pnas.95.9.4906
- Y. Yan, G. Smant, J. Stokkermans, L. Qin, J. Helder, T. J. Baum, A. Schots and E. L. Davis, “Genomic Organization of Four β-1,4-Endoglucanase Genes in Plant-Parasitic Cyst Nematodes and Its Evolutionary Implications,” Gene, Vol. 220, No. 1-2, 1998, pp. 61-70. doi:10.1016/S0378-1119(98)00413-2
- J. de Meutter, T. Tytgat, E. Van der Schueren, G. Smant, A. Schots, A. Coomans, M. Van Montagu and G. Gheysen, “Cloning of Two Endoglucanase Genes from Heterodera schachtii,” Mededelingen Faculteit Landbouwkundige en Toegepaste Biologische Wetenschappen, Universiteit Gent, Vol. 63, 1998, pp. 619-623.
- B. Gao, R. Allen, T. Maier, E. L. Davis, T. J. Baum and R. S. Hussey, “Defining a Plant-Parasitic Nematode: A Profile of Putative Parasitism Genes Expressed in the Esophageal Gland Cells of Heterodera glycines,” Nematology, Vol. 4, 2002, p. 218.
- M. Goellner, G. Smant, J. M. De Boer, T. J. Baum and E. L. Davis, “Isolation of Beta-1,4-Endoglucanase Genes from Globodera tabacum and Their Expression during Parasitism,” Journal of Nematology, Vol. 32, No. 2, 2000, pp. 154-165.
- T. Kikuchi, J. T. Jones, T. Aikawa, H. Kosaka and N. Ogura, “A Family of Glycosyl Hydrolase Family 45 Cellulases from the Pine Wood Nematode Bursaphelenchus xylophilus,” FEBS Letters, Vol. 572, No. 1-3, 2004, pp. 201-205. doi:10.1016/j.febslet.2004.07.039
- M. N. Rosso, B. Favery, C. Piotte, L. Arthaud, J. M. De Boer, R. S. Hussey, J. Bakker, T. J. Baum and P. Abad, “Isolation of a cDNA Encoding a Beta-1,4-Endoglucanase in the Root-Knot Nematode Meloidogyne incognita and Expression Analysis during Plant Parasitism,” Molecular Plant-Microbe Interactions, Vol. 12, No. 7, 1999, pp. 585-591. doi:10.1094/MPMI.1999.12.7.585
- T. Uehara, A. Kushida and Y. Momota, “PCR-Based Cloning of Two β-1,4-Endoglucanases from the RootLesion Nematode Pratylenchus penetrans,” Nematology, Vol. 3, No. 4, 2001, pp. 335-341. doi:10.1163/156854101317020259
- Y. Yan, G. Smant and E. L. Davis, “Functional Screening Yields a New Beta-1,4-Endoglucanase Gene from Heterodera glycines That May Be the Product of Recent Gene Duplication,” Molecular Plant-Microbe Interactions, Vol. 14, No. 1, 2001, pp. 63-71. doi:10.1094/MPMI.2001.14.1.63
- Y. Masui, “Meiotic Arrest in Animal Oocytes,” In: A. Monroy and C. B. Metz, Eds., Biology of Fertilization, Vol. 1, Academic Press, Orlando, 1985, pp. 189-219. doi:10.1016/B978-0-12-492601-1.50014-5
- M. A. Miller, V. Q. Nguyen, M. H. Lee, M. Kosinski, T. Schedl, R. M. Caprioli and D. Greenstein, “A Sperm Cytoskeletal Protein That Signals Oocyte Meiotic Maturation and Ovulation,” Science, Vol. 291, No. 5511, 2001, pp. 2144-2147. doi:10.1126/science.1057586
- R. M. Steeves, T. C. Todd, J. S. Essig and H. N. Trick, “Transgenic Soybeans Expressing siRNAs Specific to a Major Sperm Protein Gene Suppress Heterodera glycines Reproduction,” Functional Plant Biology, Vol. 33, No. 11, 2006, pp. 991-999. doi:10.1071/FP06130
- E. P. Masler, “Aminopeptidases in Caenorhabditis elegans and Panagrellus redivivus: Detection Using Peptide and Non-Peptide Substrates,” Journal of Helminthology, Vol. 76, No. 1, 2002, pp. 45-52. doi:10.1079/JOH200193
- D. R. Brooks, N. M. Hooper and R. E. Isaac, “The Caenorhabditis elegans Orthologue of Mammalian Puromycin-Sensitive Aminopeptidase has Roles in Embryogenesis and Reproduction,” Journal of Biological Chemistry, Vol. 278, 2003, pp. 42795-47801. doi:10.1074/jbc.M306216200
- X. Hong and J. Bouvier, “Brugia pahangi: Identification and Characterization of an Aminopeptidase Associated with Larval Molting,” Experimental Parasitology, Vol. 76, No. 2, 1993, pp. 127-133. doi:10.1006/expr.1993.1015
- C. J. Lilley, S. A. Goodchild, H. J. Atkinson and P. E. Urwin, “Cloning and Characterization of a Heterodera glycines Aminopeptidase cDNA,” International Journal for Parasitology, Vol. 35, No. 14, 2005, pp. 1577-1585. doi:10.1016/j.ijpara.2005.07.017
- A. G. Maule, A. Mousley, N. J. Marks, T. A. Day, D. P. Thompson, T. G. Geary and D. W. Halton, “Neuropeptide Signaling Systems-Potential Drug Targets for Parasite and Pest Control,” Current Topics in Medicinal Chemistry, Vol. 2, No. 7, 2002, pp. 733-758. doi:10.2174/1568026023393697
- P. E. Urwin, C. J. Lilley and H. J. Atkinson, “Ingestion of Double-Stranded RNA by Pre-Parasitic Juvenile Cyst Nematodes Leads to RNA Interference,” Molecular PlantMicrobe Interactions, Vol. 15, No. 8, 2002, pp. 747-752. doi:10.1094/MPMI.2002.15.8.747
- E. Fanelli, M. Di Vito, J. T. Jones and C. De Giorgi, “Analysis of Chitin Synthase Function in a Plant Parasitic Nematode, Meloidogyne artiellia, Using RNAi,” Gene, Vol. 349, 2005, pp. 87-95. doi:10.1016/j.gene.2004.11.045
- A. F. Bird and P. G. Self, “Chitin in Meloidogyne javanica,” Fundamental & Applied Nematology, Vol. 18, No. 3, 1995, pp. 235-239.
- M. T. Harris and J. A. Fuhrman, “Structure and Expression of Chitin Synthase in the Parasitic Nematode Dirofilaria immitis,” Molecular and Biochemical Parasitology, Vol. 122, No. 2, 2002, pp. 231-234. doi:10.1016/S0166-6851(02)00102-0
- P. Thakur, A. Sharma, S. B. Rao, M. Kumar, N. G. Prasad, N. Tyagi, D. Kamaraju, P. Papolu, P. Banakar and U. Rao, “Cloning and Characterization of Two Neuropeptide Genes from Cereal Cyst Nematode, Heterodera avenae from India,” Bioinformation, Vol. 8, No. 13, 2012, pp. 617-621. doi:10.6026/97320630008617
- J. M. Holaska, B. E. Black, F. Rastinejad and B. M. Paschal, “Ca2+-Dependent Nuclear Export Mediated by Calreticulin,” Molecular and Cellular Biology, Vol. 22, No. 17, 2002, pp. 6286-6297. doi:10.1128/MCB.22.17.6286-6297.2002
- G. Nickenig, F. Michaelsen, C. Muller, A. Berger, T. Vogel, A. Sachinidis, H. Vetter and M. Bohm, “Destabilization of AT(1) Receptor mRNA by Calreticulin,” Circulation Research, Vol. 90, 2002, pp. 53-58. doi:10.1161/hh0102.102503
- S. Goicoechea, M. A. Pallero, P. Eggleton, M. Michalak and J. E. Murphy-Ullrich, “The Anti-Adhesive Activity of Thrombospondin Is Mediated by the N-Terminal Domain of Cell Surface Calreticulin,” Journal of Biological Chemistry, Vol. 277, 2002, pp. 37219-37228. doi:10.1074/jbc.M202200200
- M. Michalak, J. M. R. Parker and M. Opas, “Ca2+ Signaling and Calcium Binding Chaperones of the Endoplasmic Reticulum,” Cell Calcium, Vol. 32, No. 5-6, 2002, pp. 269-278. doi:10.1016/S0143416002001884
- S. Jaubert, A. L. Milac, A. J. Petrescu, J. Almeida-Engler, P. Abad and M. Rosso, “In Planta Secretion of a Calreticulin by Migratory and Sedentary Stages of Root-Knot Nematode,” Molecular Plant Microbe-Interactions, Vol. 18, No. 12, 2005, pp. 1277-1284. doi:10.1094/MPMI-18-1277
Abbreviations
ATP: Adenosine Triphosphate;
EST: Expressed Sequence Tag;
GO: Gene Ontology;
LD PCR: Long Distance Polymerase Chain Reaction;
MSP: Major Sperm Protein;
NFS: Nematode Feeding Site;
SRA: Short Read Achieve.
NOTES
*Corresponding author.

