Open Journal of Applied Biosensor
Vol.2 No.3(2013), Article ID:35377,11 pages DOI:10.4236/ojab.2013.23011
Technology and Applications of Microbial Biosensor
Department of Electrical & Computer Engineering, State University of New York, Binghamton, USA
Email: *sechoi@binghamton.edu
Copyright © 2013 Chunhui Dai, Seokheun Choi. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received January 28, 2013; revised March 6, 2013; accepted March 17, 2013
Keywords: Microbial Biosensor; Immobilization; Transducer
ABSTRACT
A microbial biosensor is an analytical device that immobilizes microorganisms onto a transducer for the detection of target analytes. With the development of nanotechnology, nanomaterials have been used to achieve better immobilization for developing a more reliable and selective microbial biosensor. Also, significant progress has been made in the development of transducer technology leading to higher sensitivity. Microbial biosensors have become one of the most useful means of monitoring environmental, food and clinical samples. In this review, we focus on the newly developed technologies and applications of microbial biosensors in recent years.
1. Introduction
A biosensor is an analytical device that combines the biological recognition element with a signal transducer to convert the response with analytes into a measurable signal which is proportional to the concentration of the analytes [1-4]. A microbial biosensor is a biosensor that uses microorganisms which consists of numerous enzymes as the bioelements (Figure 1). The enzymes in the living cells can produce a response to the analytes specifically and selectively, without neither the necessity of
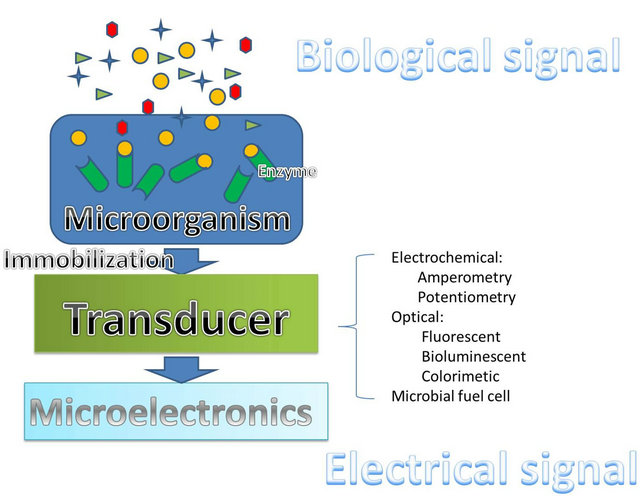
Figure 1. A schematic representation of microbial biosensor.
time-consuming and costly purification nor the negative effects of the operating environment [2,5]. In order to transfer the responses from the recognition elements to the transducers, the immobilization between the bioelements and the transducers must be intimate and stable. Integrating the microorganisms onto the transducer is the basic requirement of achieving a reliable microbial biosensor [2,3,5]. Immobilization determines not only the quality of the signal transferred from the microorganisms to the transducer but also the reusability of the microbial biosensor. Therefore, immobilization plays an important role in developing a microbial biosensor [6]. The conventional immobilization methods include adsorption, entrapment, encapsulation, covalent binding, and cross linking. However, all of these methods suffer from either poor long-term stability or negative effects from being exposed to harsh reaction conditions [3]. Advances in nanotechnology offer an alternative for better immobilization by using nanomaterial such as nanoparticles, nanotubes, and fiber optics, which promote higher reliability and stability of the bioelements [7]. The transducer is another critical part of the microbial biosensor for converting the biological response to a measurable signal [1]. Recently, microbial fuel cells (MFC) have been proposed as a new technique for microbial biosensors which relied on optical transducers as a main transducer in the past decade. With the ability to generate sustainable electricity from biodegradable organic compounds through microbial metabolism, MFCs provide high sensitivity and selective sensing capability [8,9].
With the advantages of low cost, stability and a fast response, the applications of microbial biosensor have been widely used in various fields ranging from environmental monitoring, food & fermentation industry, to clinical diagnostics. For environmental monitoring, it is necessary to find a simple, rapid, cost-effective and field portable screening method to monitor various organic and inorganic chemical contaminants which can be potential risks to human health [10-12]. The food and fermentation industries need rapid, affordable and reliable methods to ensure the quality of products and process controls [12,13]. There is also an urgent need in clinical diagnostics for accurate, fast and inexpensive devices, which can be routinely used to monitor clinically important parameters [14,15]. The conventional methods in those applications typically require analytical techniques, specialized laboratories, and practiced operators, which are costly, complicated and time consuming [14]. Microbial biosensors have significant advantages including high sensitivity, low cost, rapid response, and portability [13,10]. Several recent reviews have addressed other aspects of the microbial biosensors [16-21]. In this paper, we first briefly review the newly developed technologies in immobilization and transducers for microbial biosensors. Then, we mainly focus on the recent advances in the applications of microbial biosensor.
2. Immobilization
As the performance of the biosensor is limited by immobilization, many studies focus on improving immobilization. The nanostructures are attractive due to their small size and large surface area, resulting in enhanced surface activity and electrical conductivity [22]. Recently, nanotubes, nanoparticles and fiber optics are widely used in developing microbial biosensors [23].
Carbon nanotubes (CNT) are ideal materials for the immobilization of microbial biosensors because of CNTs’ electronic properties, large surface area, excellent electrochemical performance and good chemical stability [24]. Due to its characteristics, CNT can increase cell loading [25], catalytic activity [26] and electrical conductivity [27]. Odaci et al. constructed a microbial biosensor by entrapping bacteria cells on CNT-modified chitosan membrane, which showed good linearity and repeatability with a high operational stability [28]. Furthermore, a bacterial impedimetric biosensor for trichloroethylene (TCE) detection was developed by immobilizing Pseudomonas putida F1 strain on gold microelectrodes functionalized with single wall carbon nanotubes that were covalently linked to anti-Pseudomonas antibodies [25]. This biosensor achieved a good linearity with TCE concentration up to 150 μg∙L−1 and a low limit detection of 20 μg∙L−1. Also, anovel Nafion/bacteriadisplaying xylose dehydrogenase (XDH)/multi-walled carbon nanotubes (MWNTs) nanocomposite electrode for determination of d-xylose provided a rapid response, a low detection limit of 0.5 μM and a broad linear range from 0.6 to 100 μM [29].
Nanoparticles (NP), especially Au NPs, are also widely used to modify the surface properties of the electrodes to achieve good immobilization performances due to NPs’ high conductivity, biocompatibility and high catalytic activity [30]. Au NPs can promote the electron transfer between the microbial cell wall and the electrode surface without resulting in the denaturalization of redoxactive proteins [31]. Deng et al. constructed an Au@Pt NPs modified Silk-derived carbon fiber to detect E. coli activity, which achieved a low detection limit of 0.09 mg/L and a wide linear range from 0.5 mg/L to 36.6 mg/L [30]. Furthermore, an Au NPs modified conducting polymer which was used as a platform for immobilization for glucose analysis showed significant advantages in biocompatibility, stability, sensitivity and possibility of electrocatalysis [7].
For the rapid detection of analytes, fiber-optic-based platforms have been constructed for the immobilization of the microorganisms [32]. The fiber-optic-based biosensors have advantages over other biosensors because of their fast response and stable immobilization capabilites. A flow-through fiber-optic-based real time biosensor for detection of toxicity in water was fabricated [33]. By immobilizing the bacteria stains on the fiber optic, the biosensor has advantages with the respect to regulation, as the bacteria do not leave the device with the water. Eltzov et al. also developed a fiber-optic-based biosensor for air toxicity monitoring [34].
3. Type of Microbial Biosensors (Transducers)
3.1. Optical Biosensor
An optical biosensor is a device that makes use of an optical transducer to produce changes in diverse optical properties such as adsorption, fluorescence, luminescence, or refractive index, which are proportional to the concentration of the analytes [35]. Fluorescence, bioluminescence, and colorimeter based biosensors are widely investigated due to their properties of compactness, selectivity, sensitivity, flexibility, resistance to electrical nose and small probe size [2,13].
3.1.1. Fluorescent Microbial Biosensor
Fluorescent microbial biosensors are widely used in analysis processes, which can emit fluorescent light that is directly proportional to the analytes concentration at a low level [2,3,6]. The basis of the fluorescent microbial biosensor is to fuse an inducible promoter to a reporter gene to encode a fluorescent protein which can emit detectable fluorescence in a genetically engineered microorganism [5]. Due to the advantages of stability and sensitivity, green fluorescent protein is most commonly used in fabrication of fluorescent microbial biosensors [37]. Recombinant Escherichia coli cells which are transformed with plasmids, harboring three tandem copies of the ars promoter/operator-the gene for gfp, were developed for the detection of arsenic [38]. Compared to cells that used plasmids harboring only one copy, the recombinant Escherichia coli cells doubled the signal-to-noise ratio and decreased the detection limit form 20 to 7.5 μg/L. The recombinant yeast, Green ScreenTM, has the ability to emit fluorescence by expressing green fluorescent proteins when it is exposed to genotoxins. Based on this mechanism, a microfluidic chip which retained yeast within the chip was developed for the detection of toxic compounds [39].
3.1.2. Bioluminescent Microbial Biosensor
Bioluminescence based microbial biosensors have been extensively used in environmental monitoring for detection of toxicity due to its ability to closely reflect to toxicity [11]. As a proportional response to the concentration of the analytes, the changes in the density of the bioluminescence emitted by the living cells can be measured by the bioluminescent microbial biosensor. According to the mechanism of production of bioluminescence, the method to control the expression of the lux gene can be divided into two manners: the constitutive manner and the inducible manner. In the constitutive manner, the bioluminescence caused by lux gene-coded luciferase exists constitutively as long as the organism is active. As the density of the bioluminescence can be affected by the additional compounds such as the toxicity, it can be used as a parameter to determine the additional compounds. In the inducible manner, the lux gene is fused with a promoter regulated by the concentration of the analytes. Based on this mechanism, the bioluminescence can not be detected until the concentration of the analytes approaches a critical value [2,5]. Several bioluminescent microbial biosensors have been developed in recent years. A whole-cell bioluminescent biosensor, based on genetically engineered Escherichia coli bacteria, carrying a recA::lucCDBAE promoter-reporter fusion, was developed for the detection of water toxicity [40]. Further, Kuncova et al. constructed a biosensor for the detection of water pollutions, based on Pseudomonas putida TVAS, harboring chromosomal tod-lux CDABE fusion [41]. By immobilizing bioluminescent bacteria, TV1061 strain, in wells of a microtiter plate, Eltzov et al. fabricated a microbial biosensor for air toxicity monitoring and achieved a good response to a low concentration of chloroform (6.65 ppb) [33].
3.1.3. Colorimetric Microbial Biosensor
Colorimetric microbial biosensors make use of the changes in the color of the special compound to determine the concentration of the target analytes. Methyl parathion can be hydrolyzed by bacterium into chromophoric product, p-nitrophenol (PNP), which can be measured by a colorimetric method. Based on this mechanism, colorimetric transducers have been widely used in developing microbial biosensors for the detection of methyl parathion. A colorimetric microbial biosensor based on the immobilization of Flavobacterium˚ sp. in glass fiber filter was constructed for the detection of methyl parathion with a detection limit of 0.3 μM and a linear range from 4 - 80 μM [42]. Further, Kumar et al. immobilized Sphingomonas bacteria onto the surface of the wells of polystyrene microplates (96 wells) to construct a colorimetric microbial biosensor, which had the same linear range to methyl parathion but achieved an advantage of multiple detections [43]. By immobilizing the Sphingomonas bacteria on inner epidermis of onion bulb scale, a colorimetric microbial biosensor for detection of methyl parathion was developed and achieved a stable characteristic [44].
3.2. MFC Biosensor
The ability to convert organic substrates into electricity through microbial catabolism makes it possible for microbial fuel cells (MFCs) to work as a transducer in microbial biosensors [45]. A typical two-chamber MFC consists of an anodic and a cathodic chamber which are separated by a proton exchange membrane [9]. In the anodic chamber, fuel is oxidized by microbes, generating electrons and protons which are transferred to the cathodic chamber through the external electric circuit and the membrane separately. They combine with oxygen to form water in the cathodic chamber [36]. MFCs have been widely used as biosensors, especially for measuring biochemical oxygen demand and water toxicity, because of its portability, long-term stability and fast response [46]. However, MFC biosensors suffer from low sensitivity because the power generated from MFCs is very low. We have improved an array of microliter-sized MFCs, generating 100 μW and 1.8 V output voltage, which contributes to achieve a higher sensitivity [45]. There are several new MFC biosensors shown in Table 1.
The linear relationship between the current density generated by the MFCs and the BOD concentration makes MFCs work as BOD biosensors [47]. Compared to the conventional methods for BOD analysis which take 5 or 7 days, the microbial BOD biosensors have fast response. Kumlanghan et al. showed a novel MFC BOD sensor system operated by integrating an anaerobic bioreactor for continuous supply of stable anaerobic consortium, which had a response time around 3 - 5 min without the need to wait for the metabolic recovery of anaerobic
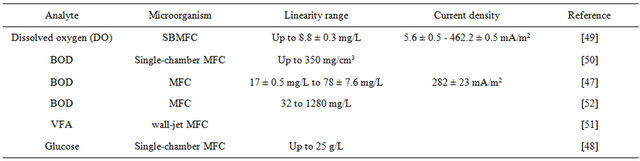
Table 1. MFC biosensors.
consortium in the anodic compartment [48]. Further, a simple method for monitoring the dissolved oxygen based on a submersible microbial fuel cell (SBMFC) assured the maximum response time of less than 4 minutes [49]. A MFC-type of BOD biosensor is advantageous over other types of BOD biosensors because they have a high reproducibility, long-term stability and wide linear range. Lorenzo used a single-chamber MFC with an air cathode to fabricate a BOD sensor which had a linear range up to 350 ppm and was still stable after 7 months with a total variation of only 15 % [50].
The toxicity in the water caused by pesticides and the waste water from the industry has been a big risk to human health. Conventional methods for detection of toxic compounds have a lot of limitations such as time consumption and high cost. A silicon-based MFC utilized as a toxicity biosensor was validated to minimize the time and the cost [46].
VFA, particularly acetate and propionate, as the important inter mediator of anaerobic digestion (AD), can be used as a process indicator. Liu et al. developed a wall-jet MFC biosensor to reflect the real time microbial activity by the detection of acetate [51].
4. Applications
4.1. Environmental Monitoring
Pollutants in the environment are great risks for the health of human beings. Several microbial biosensors for detection of organic and inorganic toxicities are shown in Table 2. Being extensively used in industry, heavy metal becomes a main toxicant in waste water. The non-biodegradability of metal ions results in its accumulation in living organisms and causes various diseases [53]. A low cost, specific, simple and quick tool is needed for monitoring heavy metals. The microbial biosensor provides an opportunity to solve this problem. The constitutive manner (light-off) and the inducible manner (light-on) are two general strategies for developing a microbial biosensor for monitoring heavy metal toxicity [54]. In the constitutive manner, the lux gene exists constitutively. The presence of the toxic heavy metal affects the expression of the lux gene and reduces the light density [5]. As it can respond to any substance that is toxic to the microbe, this microbial biosensor is nonspecific [55]. Specific biosensors, which are based on inducible promoters fused to reporter genes, are more sophisticated and sensitive [36]. Only the specific biosensor can be used for in situ measurement of contaminants. Heavy metal ions can act as an acute enzyme inhibitor and then cause some changes that can be used as the signal for detecting heavy metal ions. As mercury can inhibit the activity of alkaline phosphate enzymes present in the cell wall of Chlorella sp., Singh et al. developed a biosensor for determination of mercury by immobilizing Chlorella sp. on a glassy carbon surface [56]. The use of genetically engineered bacteria, which can produce measurable signals when contacted with bio-components, is the best approach for detecting heavy metal [57]. Ravikumarzra et al. constructed a biosensor for detecting zinc and copper based on engineered bacteria, where P and cusC promoters were fused to a dual-labeling reporter protein as an interactive biocomponent for zinc and copper to generate a signal from the constructed biosensor [58]. A promoterless enhanced green fluorescent protein (egfp) gene was fused with the czcR3 promoter, which could respond quantitatively to zinc, for specific detection of zinc [59].
Using dead biomass to uptake heavy metals passively is a more efficient, economical and easier way for detecting them. Compared to living cells, dead biomass requires no nutrients, is easy to handle and store, and has high tolerance to toxic harsh reaction environments [60]. Pseudomonas aeruginosa were used in a heat dried form to construct a microbial biosensor for the detection of heavy metal Pb (II), which achieved a linear response to Pb (II) from 1.0 μM to 2.0 μM and a detection limit of 0.6 μM [61].
Organic toxicity is another main pollutant in the environment which is harmful to human beings. A rapid, low-cost, and specific method for detection of various organic toxicities is needed. Microbial biosensors provide an alternative to solve this problem. Methyl parathion which has been widely used in agriculture as Organophosphorus (OP) pesticides are not only harmful to
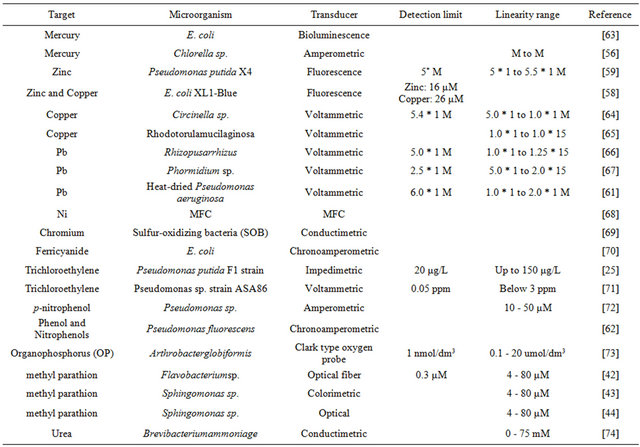
Table 2. Microbial biosensors for detection of organic and inorganic toxicities.
insect, but to human beings as well. Based on the princeple that methyl parathion can be hydrolyzed into electrochemical or colorimetric detectable product p-nitrophenol (PNP) by organophosephorus hydrolase (OPH), an optical microbial biosensor that used Sphingomonas sp. immobilized on the bottom surface of the wells of the microplate was developed for the detection of methyl parathion pesticide [43]. Trichloroethylene (TCE), another main organic pollutant which has been used as an organic solvent and degreasing agent in industry, can cause impairment to the central nervous system. TCE can be degraded into the conductometric measurable products glyoxylate and formate ions by the toluene dioxygenase enzyme in toluene-grown Pseudomonas putida F1 (PpF1). Based on this reaction mechanism, an impedimetric microbial biosensor based on the immobilization of PpF1 strain on gold microelectrode was developed for the detection of trichloroethylene [25]. Furthermore, Liu et al. presented a microbial biosensor based on Pseudomonas fluorescens for detection of phenol and nitrophenols [62].
4.2. Food and Fermentation
As the quality of the products is required by both the customers and the government, rapid and affordable methods to assure the quality of products and process controls are needed [75]. Table 3 shows the recent developed microbial biosensors used in food and fermentation. Fermentation is widely used for the production of foodstuffs and drinks, which requires a carefully performed fermentation system operation [76]. Microbial biosensors are used to monitor the materials in order to control the fermentation process. Because ethanol is very important and necessary in different fermentation process, microbial biosensors have been used for sensitive determination of ethanol in order to monitor the fermentation process. An amperometric biosensor based on Candida tropicalis cells immobilized in gelatin by using glutaraldehyde was developed for the determination of ethanol in the range from 0.5 mM to 7.5 mM [77]. Further, Valach et al. constructed a new microbial amperometric biosensor for the measurement of ethanol in flow injection analysis, which achieved a linear response to ethanol in the range from 10 μM to 1.5 mM in 3 minutes [78]. Furthermore, based on the immobilization of Methylobacterium organophilium on eggshell membrane and an oxygen electrode, an ethanol biosensor got a linear range of 0.050 - 7.5 mmol/L and a detection limit of 0.035 mmol/L to ethanol [79].
The control of food quality and freshness is of growing interest for both the consumer and the food industry [75]. The demand for quick and specific analytical tools is needed for monitoring nutritional parameters and food contaminants [3]. Microbial biosensors work as a rapid and affordable method to assure the quality of products. As an index in the determination of the quality of coffee, caffeine needs to be detected sensitively and rapidly. Babu et al. developed an amperometric biosensor for the determination of caffeine by immobilizing Pseudomonas alcaligenes MTCC 5264 on a cellophane membrane, which responded linearly to caffeine over a range of 0.1 - 1 mg/mL within 3 minutes [80]. D-glucose and D-xylose are the two ideal sweeteners and nutritional agents which are widely used in food. Based on the co-immobilization of glucose oxidase and xylose dehydrogenase displayed XDH-bacteria on multiwalled CNT nanocomposite films modified electrode, a voltametric biosensor was developed for detection of D-glucose and D-xylose [81]. Contaminants also should be carefully detected in order to assure the quality of the products. Zearalenone family mycotoxins are common contaminants in milk, which canlead to mycotoxicoses [82]. In order to assure the quality of milk, genetically modified Saccharomyces cerevisiae strain were used as the bioelement of the microbial biosensor for the detection of zearalenone family mycotoxins in milk [83].
4.3. Clinical Diagnostics
Conventional techniques for the diagnosis of various diseases suffer from slow response time, time consumption, and complicated process, which make critical care during emergencies difficult [14]. Compared to enzyme based biosensors, microbial biosensors require no purifycation which is time consuming and expensive. Microbial biosensors provides a rapid, accurate and inexpensive way for diagnosis of hormones, pathogens and DNA, which are important parameters of a living individual. Akyilmaz et al. fabricated a novel microbial biosensor for the determination of epinephrine by immobilizing white rot fungi (Phanerochaete chrysosporium ME446) in gelatin using glutaraldehyde crosslinking agent on a Ptelectrode, which achieved a linear range of 5 - 100 μM and a detection limit of 1.04 μM [86]. In this biosensor, epinephrine was turned into epinephrine quinone through a redox activity catalyzed by lactase in the fungal cells, causing an increase in the current. As a cause of virus diseases, the detection of pathogens plays an important role in clinical diagnostics. Rat basophilic leukemia (RBL) mast cells which could produce a dramatic exocytotic response within minutes of antigen addition were used to fabricate a microbial biosensor for the detection of pathogens [87]. DNA damage which can affect DNA replication, repair and gene expression can lead to many diseases including cancer. An E. coli SOS-EGFP based on SOS response was constructed for detection of DNA damage. The SOS response could be triggered by harmful chemicals for DNA to produce fluorescent protein controlled by recAgene promoter [88].
5. Future Directions
Microbial biosensors have been widely used in the environmental, food and diagnostics industry due to its advantages of low cost, stability and fast response. Compared to enzymes, the microorganisms that are used as bioelements can make use of the enzyme to specifically respond to the analytes without time consuming and expensive purification. Based on its attractive properties, several directions for the development of the microbial biosensors have shown great promise.
Lab-on-a-chip is one direction that attracts a lot of researches’ focus [90]. The biotic-micro electrochemical system developed by the integration of a microbe onto the microfluidic chip offers promising characteristics of a fast response and small size. Lab-on-a-chip technology
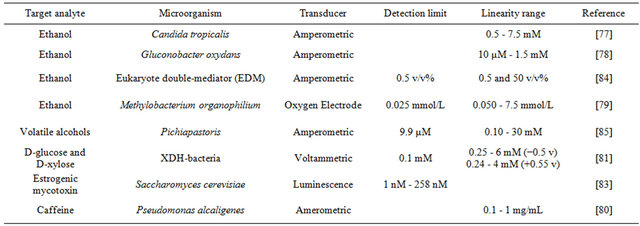
Table 3. Microbial biosensor for detection in food and fermentation industry.
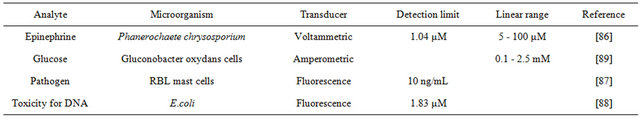
Table 4. Microbial biosensors for clinical diagnostics.
are widely used in fabricating microfluidic devices [29], which enable the detection of analytes at ultra-low concentrations by actively transporting target analytes to the surface of the microbial biosensors [91,92]. Lab-ona-chip shows great promise for the development of microbial biosensors.
Another promising direction microbial biosensors is forensic identification. Forensic identification is the recognition of the fine physical features of an object, which are specifically distinguished from other objects of the same kind [93,94]. Body fluidic traces recovered at crime scenes, which contain valuable DNA and miRNA evidence, are mostly used for forensic identification [95,96]. However, the conventional methods for forensic identification are complicated and time-consuming, which bring many difficulties towards solving a criminal case. Since the microbial biosensor has the ability to analyze DNA, microbial biosensors may be used for forensic identification.
Finally, a portable microbial biosensor array system will be promising in minimizing the time, the cost and the personnel required for detecting the toxicity of water while intensive industrialization and farming have led to the release of many toxic compounds in the environment, causing an important pollution of aquatic ecosystems.
REFERENCES
- S. P. Mohanty and E. Kougianos, “Biosensosrs: A Tutorial Review,” IEEE Potentials, Vol. 25, No. 2, 2006, pp. 35-40. doi:10.1109/MP.2006.1649009
- Y. Lei, W. Chen and A. Mulchandani, “Microbial Biosensors,” Analytica Chimica Acta, Vol. 568, No. 1-2, 2006, pp. 200-210. doi:10.1016/j.aca.2005.11.065
- S. F. D’Souza, “Microbial Biosensors,” Biosensor and Bioelectronics, Vol. 16, No. 6, 2001, pp. 337-353. doi:10.1016/S0956-5663(01)00125-7
- S. Chauhan, V. Rai and H. B. Singh, “Biosensors,” Biosensor, 2004, pp. 33-44.
- L. Su, W. Jia, C. Hou and Y. Lei, “Microbial Biosensors: A Review,” Biosensors and Bioelectronics, Vol. 26, No. 5, 2011, pp. 1788-1799. doi:10.1016/j.bios.2010.09.005
- S. F. D’Souza, “Immobilization and Stabilization of Biomaterials for Biosensor Applications,” Applied Biochemistry and Biotechnology, Vol. 96, No. 1-3, 2001, pp. 225- 238. doi:10.1385/ABAB:96:1-3:225
- S. Tuncagil, C. Ozdemir, D. Demirkol, S. Timur and L. Toppare, “Gold Nanoparticle Modified Conducting Polymer of 4-(2,5-di(thiophen-2-yl)-1H-pyrrole-1-l) Benzenamine for Potential Use as a Biosensing Material,” Food Chmeistry, Vol. 127, No. 3, 2011, pp. 1317-1322. doi:10.1016/j.foodchem.2011.01.089
- S. Choi and J. Chae, “Optimal Biofilm Formation and Power Generation in a Micro-Sized Microbial Fuel Cell (MFC),” Sensors & Actuators A: Physical, Vol. 195, 2012, pp. 206-212.
- Z. W. Du, H. R. Li and T. Y. Gu, “A State of the Art Review on Microbial Fuel Cells: A Promising Technology for Wastewater Treatment and Bioenergy,” Biotechnology Advances, Vol. 25, No. 5, 2007, pp. 464-482. doi:10.1016/j.biotechadv.2007.05.004
- K. R. Rogers, “Recent Advances in Biosensor Techniques for Environmental Monitoring,” Analytica Chimica Acta, Vol. 568, No. 1-2, 2006, pp. 222-231. doi:10.1016/j.aca.2005.12.067
- S. M. Steinberg, E. J. Poziomek, W. H. Engelmann and K. R. Rogers, “A Review of Environmental Applications of Bioluminescence Measurements,” Chemosphere, Vol. 30, No. 11, 1995, pp. 2155-2197. doi:10.1016/0045-6535(95)00087-O
- S. Belkin, “Microbial Whole-Cell Sensing Systems of Environmental Pollutants,” Current Opinion in Microbiology, Vol. 6, No. 3, 2003, pp. 206-212. doi:10.1016/S1369-5274(03)00059-6
- P. Arora, A. Sindhu, N. Dilbaghi and A. Chaudhury, “Biosensors as Innovative Tools for the Detection of Food Borne Pathogens,” Biosensors and Bioelectronics, Vol. 28, No. 1, 2011, pp. 1-12. doi:10.1016/j.bios.2011.06.002
- B. D. Malhotra and A. Chaubey, “Biosensors for Clinical Diagnostics Industry,” Sensors & Actuators, Vol. 91, No. 1-3, 2003, pp. 117-127. doi:10.1016/S0925-4005(03)00075-3
- D. Ivnitski, I. Abdel-Hamid, P. Atanasov and E. Wilkins, “Biosensors Detection of Pathogenic Bacteria,” Biosensors and Bioelectronics, Vol. 14, No. 7, 1999, pp. 599- 624. doi:10.1016/S0956-5663(99)00039-1
- H. J. Shin, “Genetically Engineered Microbial Biosensors for in Situ Monitoring of Environmental Pollution,” Applied Microbiology and Biotechnology, Vol. 89, No. 4, 2011, pp. 867-877. doi:10.1007/s00253-010-2990-8
- F. Zhang and J. Keasling, “Biosensors and Their Applications in Microbial Metabolic Engineering,” Trends in Microbiology, Vol. 19, No. 7, 2011, pp. 323-329. doi:10.1016/j.tim.2011.05.003
- B. Pérez-López and A. Merkoci, “Nanomaterials Based Biosensors for Food Analysis Applications,” Trends in Food Science & Technology, Vol. 22, No. 11, 2011, pp. 625-639. doi:10.1016/j.tifs.2011.04.001
- P. D’Orazio, “Biosensors in Clinical Chemistry—2011 update,” Clinica Chimica Acta, Vol. 412, No. 19-20, 2011, pp. 1749-1761. doi:10.1016/j.cca.2011.06.025
- L. Ding, D. Du, X. J. Zhang and H. X. Ju, “Trends in Cell-Based Electrochemical Biosensors,” Current Medicinal Chemistry, Vol. 15, No. 30, 2008, pp. 3160-3170. doi:10.2174/092986708786848514
- S. Dhanekar and S. Jain, “Porous Silicon Biosensor: Current Status,” Biosensors and Bioelectronics, Vol. 41, No. 15, 2013, pp. 54-64. doi:10.1016/j.bios.2012.09.045
- N. L. Rosi and C. A. Mirkin, “Nanostructures in Biodiagnostics,” Chemical Reviews, Vol. 105, No. 4, 2005, pp. 1547-1562. doi:10.1021/cr030067f
- C. Kaittanis, S. Santra and J. M. Perez, “Emerging Nanotechnology-Based Strategies for the Identification of Microbial Pathogenesis,” Advanced Drug Delivery Reviews, Vol. 62, No. 4, 2010, pp. 408-423. doi:10.1016/j.addr.2009.11.013
- R. Andrews, D. Jacques, D. L. Qian and T. Rantell, “Multiwall Carbon Nanotubes: Synthesis and Application,” Accounts of Chemical Research, Vol. 35, No. 12, 2002, pp. 1008-1017. doi:10.1021/ar010151m
- M. Hnaien, S. Bourigua, F. Besseueille, J. Bausells, A. Errachid, F. Lagarde and N. Jaffrezic-Renault, “Impedimetric Microbial Biosensor Based on Single Wall Carbon Nanotube Modified Microelectrodes for Trichloroethylene Detection,” Electrochimica Acta, Vol. 56, No. 28, 2011, pp. 10353-10358. doi:10.1016/j.electacta.2011.04.041
- C. E. Banks, R. R. Moore, T. J. Davies and R. G. Compton, “Investigation of Modified Basal Plane Pyrolytic Graphite Electrodes: Definitive Evidence for the Electrocatalytic Properties of the Ends of Carbon Nanotubes,” Chemical Communications, Vol. 21, No. 16, 2004, pp. 1804-1805.
- A. Merkoci, M. Pumera, X. Llopis, B. Perez, M. Valle and S. Alegret, “New Materials for Electrochemical Sensing VI: Carbon Nanotubes,” TrAC Trends in Analytical Chemistry, Vol. 24, No. 24, 2005, pp. 826-838. doi:10.1016/j.trac.2005.03.019
- D. Odaci, S. Timur and A. Telefoncu, “Bacterial Sensors Based on Chitosan Matrices,” Sensors & Actuators B: Chemical, Vol. 134, No. 1, 2008, pp. 89-94. doi:10.1016/j.snb.2008.04.013
- P. Li, N. Lei, D. A. Sheadel, J. Xu and W. Xue, “Integration of Nanosensors into a Sealed Microchannel in a Hybrid Lab-on-a-Chip Device,” Sensors and Actuators B: Chemical, Vol. 166, No. 1, 2012, pp. 870-877. doi:10.1016/j.snb.2012.02.047
- L. Deng, S. Guo, M. Zhou, L. Liu, C. Liu and S. Dong, “A Silk Derived Carbon Fiber Mat Modified with Au@Pt Urchilike Nanoparticles: A New Platform as Electrochemical Microbial Biosensor,” Biosensors and Bioelectronics, Vol. 25, No. 10, 2010, pp. 2189-2193. doi:10.1016/j.bios.2010.02.005
- Q. Sheng, K. Luo, L. Li and J. Zheng, “Direct Electrochemistry of Glucose Oxidase Immobilized on NdPO4 Nanoparticles/Chitosan Composite Film on Glassy Carbon Electrodes and Its Biosensing Application,” Bioelectrochemistry, Vol. 74, No. 2, 2009, pp. 246-253. doi:10.1016/j.bioelechem.2008.08.007
- D. R. DeMarco, E. W. Saaski, D. A. McCrae and D. V. Lim, “Rapid Detection of Escherichia coli O157: H7 in Ground Beef Using a Fiber-Optic Biosensor,” Journal of Food Protection, Vol. 62, No. 7, 1999, pp. 711-716.
- E. Eltzov, V. Pavluchkov, M. Burstin and R. S. Marks, “Creation of a Fiber Optic Based Biosensor for Air Toxicity Monitoring,” Sensors and Actuators B: Chemical, Vol. 155, No. 2, 2011, pp. 859-867. doi:10.1016/j.snb.2011.01.062
- E. Eltzov, R. S. Marks, S. V. Bart, A. Wullings and M. B. Heringa, “Flow-Through Real Time Bacterial Biosensor for Toxic Compounds in Water,” Sensors and Actuators B: Chemical, Vol. 142, No. 1, 2009, pp. 11-18. doi:10.1016/j.snb.2009.08.024
- M. N. Velasco-Garcia, “Optical Biosensors for Probing at the Cellular Level: A Review of Recent Progress and Future Prospects,” Seminars in Cell & Developmental Biology, Vol. 20, No. 1, 2009, pp. 27-33. doi:10.1016/j.semcdb.2009.01.013
- X. Xu and Y. Ying, “Microbial Biosensors for Environmental Monitoring and Food Analysis,” Food Reviews International, Vol. 27, No. 3, 2011, pp. 300-329.
- J. C. Pickup, F. Hussain, N. D. Evans, O. J. Rolinski and D. J. S. Birch, “In vivo Glucose Monitoring: The Clinical Reality and the Promise,” Biosensors and Bioelectronics, Vol. 20, No. 10, 2005, pp. 2555-2565. doi:10.1016/j.bios.2004.10.002
- C. Tani, K. Inoue, Y. Tani, M. Harun-ur-Rashid, N. Azuma, S. Ueda, K. Yoshida and I. Maeda, “Sensitive Fluorescent Microplate Bioassay Using Recombinant Escherichia coli with Multiple Promoter-Reporter Units in Tandem for Detection of Arsenic,” Journal of Bioscience and Bioengineering, Vol. 108, No. 5, 2009, pp. 414- 420. doi:10.1016/j.jbiosc.2009.05.014
- J. Garcia-Alonso, G. M. Greenway, J. D. Hardege and S. J. Haswell, “A Prototype Microfluidic Chip Using Fluorescent Yeast for Detection of Toxic Compounds,” Bionsensor and Bioelectronics, Vol. 24, No. 5, 2009, pp. 1508-1511. doi:10.1016/j.bios.2008.07.074
- R. Daniel, R. Almog, A. Ron, S. Belkin and Y. S. Dianmand, “Modeling and Measurement of a Whole-Cell Bioluminescent Biosensor Based on a Single Photon Avalanche Diode,” Biosensors and Bioelectronics, Vol. 24, No. 4, 2008, pp. 882-887. doi:10.1016/j.bios.2008.07.026
- G. Kuncova, J. Pazlarova, A. Hlavata, S. Ripp and G. S. Sayler, “Bioluminescent Bioreporter Pseudomonas putida TVA8 as a Detector of Water Pollution. Operational Conditions and Selectivity of Free Cells Sensor,” Ecological Indicators, Vol. 11, No. 3, 2011, pp. 882-887. doi:10.1016/j.ecolind.2010.12.001
- J. Kumar, S. K. Jhaadn and S. F. D’Souza, “Optical Microbial Biosensor for Detection of Methyl Parathion Pesticide Using Flavobacterium sp. Whole Cells Adsorbed on Glass Fiber Filters as Disposable Biocomponent,” Biosensors and Bioelectronics, Vol. 21, No. 11, 2006, pp. 2100-2105. doi:10.1016/j.bios.2005.10.012
- J. Kumar and S. F. D’Souza, “An Optical Microbial Biosensor for Detection of Methyl Parathion Using Sphingomonas sp. Immobilized on Microplate as a Reusable Biocomponent,” Biosensors and Bioelectronics, Vol. 26, No. 4, 2010, pp. 1292-1296. doi:10.1016/j.bios.2010.07.016
- J. Kumar and S. F. D’Souza, “Immobilization of Microbial Cells on Inner Epidermis of Onion Bulb Scale for Biosensor Application,” Biosensors and Bioelectronics, Vol. 26, No. 11, 2011, pp. 4399-4404. doi:10.1016/j.bios.2011.04.049
- S. Choi and J. Chae, “An Array of Microliter-Sized Microbial Fuel Cells Generating 100 μW of Power,” Sensors & Actuators A: Physical, Vol. 177, No. 7, 2012, pp. 10-15. doi:10.1016/j.sna.2011.07.020
- D. Dávila, J. P. Esquivel, N. Sabate and J. Mas, “Silicon-Based Microfabricated Microbial Fuel Cell Toxicity Sensor,” Biosensors and Bioelectronics, Vol. 26, No. 5, 2011, pp. 2426-2430. doi:10.1016/j.bios.2010.10.025
- L. Peixoto, B. Min, G. Martins, A. G. Brito, P. Kroff, P. Parpot, I. Angelidaki and R. Nogueira, “In Situ Microbial Fuel Cell-Based Biosensor for Organic Carbon,” Bioelectrochemistry, Vol. 81, No. 2, 2011, pp. 99-103. doi:10.1016/j.bioelechem.2011.02.002
- A. Kumlanghan, J. Liu, P. Thavarungkul, P. Kanatharana and B. Mattiasson, “Microbial Fuel Cell-Based Biosensor for Fast Analysis of Biodegradable Organic Matter,” Biosensors and Bioelectronics, Vol. 22, No. 12, 2007, pp. 2939-2944. doi:10.1016/j.bios.2006.12.014
- Y. Zhang and I. Angelidaki, “A Simple and Rapid Method for Monitoring Dissolved Oxygen in Water with a Submersible Microbial Fuel Cell (SBMFC),” Biosensors and Bioelectronics, Vol. 38, No. 1, 2012, pp. 189-194. doi:10.1016/j.bios.2012.05.032
- M. Di Lorenzo, T. P. Curtis, I. M. Head and K. Scott, “A Single-Chamber Microbial Fuel Cell as a Biosensor for Wastewaters,” Water Research, Vol. 43, No. 13, 2009, pp. 3145-3154. doi:10.1016/j.watres.2009.01.005
- Z. Liu, J. Liu, S. Zhang, X. Xing and Z. Su, “Microbial Fuel Cell Based Biosensor for in situ Monitoring of Anaerobic Digestion Process,” Bioresource Technology, Vol. 102, No. 22, 2011, pp. 10221-10229. doi:10.1016/j.biortech.2011.08.053
- O. Modin and B. Wilen, “A Novel Bioelectrochemical BOD Sensor Operating with Voltage Input,” Water Research, Vol. 46, No. 18, 2012, pp. 6113-6120. doi:10.1016/j.watres.2012.08.042
- I. Gammoudi, H. Tarbague, A. Othmane, D. Moynet, D. Rebiere, R. Kalfat and C. Dejous, “Love-Wave BacteriaBased Sensor for the Detection of Heavy Metal Toxicity in Liquid Medium,” Biosensors and Bioelectronics, Vol. 26, No. 4, 2010, pp. 1723-1726. doi:10.1016/j.bios.2010.07.118
- A. Ivask, T. Rolova and A. Kahru, “A Suite of Recombinant Luminescent Bacterial Strains for the Quantification of Bioavailable Heavy Metals and Toxicity Testing,” BMC Biotechnology, Vol. 9, 2009, pp. 41-55. doi:10.1186/1472-6750-9-41
- S. Ramanathan, M. Ensor and S. Daunert, “Bacterial Biosensors for Monitoring Toxic Metals,” Trends in Biotechnology, Vol. 15, No. 12, 1997, pp. 500-506. doi:10.1016/S0167-7799(97)01120-7
- J. Singh and S. K. Mittal, “Chlorella sp. Based Biosensor for Selective Determination of Mercury in Presence of Silver Ions,” Sensors and Actuators B: Chemical, Vol. 165, No. 1, 2012, pp. 48-52. doi:10.1016/j.snb.2012.02.009
- Y. Yong and J. Zhong, “A Genetically Engineered Whole-Cell Pigment-Based Bacterial Biosensing System for Quantification of N-Butyryl Homoserine Lactone Quorum Sensing Signal,” Biosensors and Bioelectronics, Vol. 25, No. 1, 2009, pp. 41-47. doi:10.1016/j.bios.2009.06.010
- S. Ravikumar, I. Ganesh, I. Yoo and S. Hong, “Construction of a Bacterial Biosensor for Zinc and Copper and Its Application to the Development of Multifunctional Heavy Metal Adsorption Bacteria,” Process Biochemistry, Vol. 47, No. 5, 2012, pp. 758-765. doi:10.1016/j.procbio.2012.02.007
- P. Liu, Q. Huang and W. Chen, “Construction and Application of a Zinc-Specific Biosensor for Assessing the Immobilization and Bioavailability of Zinc in Different Soils,” Environmental Pollution, Vol. 164, 2012, pp. 66- 72. doi:10.1016/j.envpol.2012.01.023
- [61] K. Vijayaraghavan and Y.-S. Yun, “Bacterial Biosorbents and Biosorption,” Biotechnology Advances, Vol. 26, No. 3, 2008, pp. 266-291. doi:10.1016/j.biotechadv.2008.02.002
- [62] M. Yüce, H. Nazir and G. Dönmez, “Utilization of HeatDried Pseudomonas aeruginosa Biomass for Voltammetric Determination of Pb(II),” New Biotechnology, Vol. 28, No. 4, 2011, pp. 356-361. doi:10.1016/j.nbt.2010.11.005
- [63] C. Liu, D. Yong, D. Yu and S. Dong, “Cell-Based Biosensor for Measurement of Phenol and Nitrophenols Toxicity,” Talanta, Vol. 84, No. 3, 2011, pp. 766-770. doi:10.1016/j.talanta.2011.02.006
- [64] P. R. G. Barrocas, W. M. Landing and R. J. M. Hudson, “Assessment of Mercury(II) Bioavailability Using a Bioluminescent Bacterial Biosensor: Practical and Theoretical Challenges,” Journal of Environmental Sciences, Vol. 22, No. 8, 2010, pp. 1137-1143. doi:10.1016/S1001-0742(09)60229-1
- [65] S. Alpat, S. K. Aplat, B. H. Cadirci, I. Yasa and A. Telefoncu, “A Novel Microbial Biosensor Based on Circinella sp. Modified Carbon Paste Electrode and Its Voltammetric Application,” Sensor & Actuators B: Chemical, Vol. 134, No. 1, 2008, pp. 175-181. doi:10.1016/j.snb.2008.04.044
- [66] M. Yüce, H. Nazir and G. Dönmez, “A Voltammetric Rhodotorula mucilaginosa Modified Microbial Biosensor for Cu(II) Determination,” Bioelectrochemistry, Vol. 79, No. 1, 2010, pp. 66-70. doi:10.1016/j.bioelechem.2009.11.003
- [67] M. Yüce, H. Nazir and G. Dönmez, “Using of Rhizopus arrhizus as a Sensor Modifying Component for Determination of Pb(II) in Aqueous Media by Voltammetry,” Bioresource Technology, Vol. 101, No. 19, 2010, pp. 7551-7555. doi:10.1016/j.biortech.2010.04.099
- [68] M. Yüce, H. Nazir and G. Dönmez, “An Advanced Investigation on a New Algal Sensor Determining Pb(II) Ions from Aqueous Media,” Biosensor and Bioelectronics, Vol. 26, No. 2, 2010, pp. 321-326. doi:10.1016/j.bios.2010.08.022
- [69] N. Stein, H. V. M. Hamelers and C. N. J. Buisman, “On-Line Detection of Toxic Components Using a Microbial Fuel Cell-Based Biosensor,” Sensors and Actuators B: Chemical, Vol. 22, No. 9, 2012, pp. 1663, 1-7.
- [70] A. Gurung, S. Oh, K. D. Kim and B. Shin, “Semi-Continuous Detection of Toxic Hexavalent Chromium Using a Sulfur-Oxidizing Bacteria Biosensor,” Journal of Environmental Management, Vol. 106, No. 15, 2012, pp. 110- 112. doi:10.1016/j.jenvman.2012.04.010
- [71] D. Yong, C. Liu, D. Yu and S. Dong, “A Sensitive, Rapid and Inexpensive Way to Assay Pesticide Toxicity Based on Electrochemical Biosensor,” Talanta, Vol. 84, No. 1, 2011, pp. 7-12. doi:10.1016/j.talanta.2010.11.012
- [72] G. Chee, “Biodegradation Analyses of Trichloroethylene (TCE) by Bacteria and Its Use for Biosensing of TCE,” Talanta, Vol. 85, No. 4, 2011, pp. 1778-1782. doi:10.1016/j.talanta.2011.07.002
- [73] R. M. Banik, Mayank, R. Prakash and S. N. Upadhyay, “Microbial Biosensor Based on Whole Cell of Pseudomonas sp. for Online Measurement of p-Nitrophenol,” Sensors and Actuators B: Chemical, Vol. 131, No. 1, 2008, pp. 295-300. doi:10.1016/j.snb.2007.11.022
- [74] M. Stoytcheva, R. Zlatev, Z. Velkova, B. Valdex, M. Ovalle and L. Petkov, “Hybrid Electrochemical Biosensor for Organophosphorus Pesticides Quantification,” Electrochemica Acta, Vol. 54, No. 6, 2009, pp. 1721-1727. doi:10.1016/j.electacta.2008.09.063
- [75] S. K. Jha, M. Kanungo, A. Nath and S. F. D’Souza, “Entrapment of Live Microbial Cells in Electropolymerized Polyaniline and Their Use as Urea Biosensor,” Biosensors and Bioelectronics, Vol. 24, No. 8, 2009, pp. 2637- 2642. doi:10.1016/j.bios.2009.01.024
- [76] L. D. Mello and L. T. Kubota, “Review of the Use of Biosensors as Analytical Tools in the Food and Drink Industries,” Food Chemistry, Vol. 77, No. 2, 2002, pp. 237-256. doi:10.1016/S0308-8146(02)00104-8
- [77] Y. Kim, J. Park and H. Jung, “An Impedimetric Biosensor for Real-Time Monitoring of Bacterial Growth in a Microbial Fermentor,” Sensor & Actuators B: Chemical, Vol. 138, No. 1, 2009, pp. 270-277. doi:10.1016/j.snb.2009.01.034
- [78] E. Akyilmaz and E. Dinckaya, “An Amperometric Microbial Biosensor Development Based Candida tropicalis Yeast Cells for Sensitive Determination of Ethanol,” Biosensors and Bioelectronics, Vol. 20, No. 7, 2005, pp. 1263-1269. doi:10.1016/j.bios.2004.04.010
- [79] M. Valach, J. Katrlik, E. Sturdik and P. Gemeiner, “Ethanol Gluconobacter Biosensor Designed for Flow Injection Analysis: Application in Ethanol Fermentation Off-Line Monitoring,” Sensors & Actuators B: Chemical, Vol. 138, No. 2, 2009, pp. 581-586. doi:10.1016/j.snb.2009.02.017
- [80] G. M. Wen, S. M. Shuang, C. Dong and M. F. Choi, “An Ethanol Biosensor Based on a Bacterial Cell-Immobilized Eggshell Membrane,” Chinese Chemical Letter, Vol. 23, No. 4, 2012, pp. 481-483. doi:10.1016/j.cclet.2012.01.026
- [81] V. R. S. Babu, S. Patra, N. G. Karanth, M. A. Kumar and M. S. Thakur, “Development of a Biosensor for Caffeine,” Analytica Chimica Acta, Vol. 582, No. 2, 2007, pp. 329-334. doi:10.1016/j.aca.2006.09.017
- [82] L. Li, B. Liang, F. Li, J. Shi, M. Mascini, Q. Lang and A. Liu, “Co-Immobilization of Glucose Oxidase and Xylose Dehydrogenase Displayed Whole Cell on Multiwalled Carbon Nanotube Nanocomposite Films Modified Electrode for Simultaneous Voltammetric Detection of d-Glucose and d-Xylose,” Biosensors and Bioelectronics, Vol. 42, No. 12, 2013, pp. 156-162. doi:10.1016/j.bios.2012.10.062
- [83] W. L. Bryden, “Mycotoxins in the Food Chain: Human Health Implications,” Asia Pacific Journal of Clinical Nutrition, Vol. 16, Suppl. 1, 2007, pp. 95-101.
- [84] A. Valimaa, A. T. Kivisto, P. I. Leskinen and M. T. Karp, “A Novel Biosensor for the Detection of Zearalenone Family Mycotoxins in Milk,” Journal of Microbiological Methods, Vol. 80, No. 1, 2010, pp. 44-48. doi:10.1016/j.mimet.2009.10.017
- [85] H. Nakamura, R. Tanaka, K. Suzuki, M. Yataka and Y. Mogi, “A Direct Determination Method for Ethanol Concentrations in Alcoholic Beverages Employing a Eukaryote Double-Mediator System,” Food Chemistry, Vol. 117, No. 3, 2009, pp. 509-513. doi:10.1016/j.foodchem.2009.04.026
- [86] M. Hammerle, K. Hilgert, A. H. Marcus and M. Ralf, “Analysis of Volatile Alcohols in Apple Juices by an Electrochemical Biosensor Measuring in the Headspace Above the Liquid,” Sensor & Actuators B: Chemical, Vol. 158, No. 1, 2011, pp. 313-318. doi:10.1016/j.snb.2011.06.026
- [87] E. Akyilmaz, M. Turemis and I. Yasa, “Voltammetric Determination of Epinephrine by White Rot Fungi (Phanerochaete chrysosporium ME446) Cells Based Microbial Biosensor,” Biosensors an Bioelectronics, Vol. 26, No. 5, 2011, pp. 2590-2594. doi:10.1016/j.bios.2010.11.012
- [88] T. Curtis, R. M. Z. G. Naal, C. Batt, J. Tabb and D. Holowka, “Development of a Mast Cell-Based Biosensor,” Biosensors and Bioelectronics, Vol. 23, No.7, 2008, pp. 1024-1031. doi:10.1016/j.bios.2007.10.007
- [89] Z. Chen, M. Lu, D. Zou and H. Wang, “An E. coli SOSEGFP Biosensor for Fast and Sensitive Detection of DNA Damaging Agents,” Journal of Environmental Science, Vol. 24, No. 3, 2012, pp. 541-549. doi:10.1016/S1001-0742(11)60722-5
- [90] S. Tuncagil, D. Odaci, E. Yildiz, S. Timur and L. Toppare, “Design of a Microbial Sensor Using Conducting Polymer of 4-(2,5-di(thiophen-2-yl)-1H-pyrrole-1-l) Benzenamine,” Sensors &Acturators B: Chemical, Vol. 137, No. 1, 2009, pp. 42-47. doi:10.1016/j.snb.2008.10.067
- [91] E. D. Leo, L. Galluccio, A. Lombardo and G. Morabito, “Networked Labs-on-a-Chip (NLoC): Introducing Networking Technologies in Microfluidic Systems,” Nano Communication Networks, Vol. 3, No. 4, 2012, pp. 217- 228. doi:10.1016/j.nancom.2012.09.007
- [92] G. Zheng, F. Patolsky, Y. Cui, W. U. Wang and C. M. Lieber, “Multiplexed Electrical Detection of Cancer Markers with Nanowire Sensor Arrays,” Nature Biotechnology, Vol. 23, 2005, pp. 1294-1301. doi:10.1038/nbt1138
- [93] L. Yang, M. Li, Y. Qu, Z. Dong and W. J. Li, “Carbon Nanotube-Sensor-Integrated Microfluidic Platform for Real-Time Chemical Concentration Detection,” Electrophoresis, Vol. 30, No. 18, 2009, pp. 3198-3205. doi:10.1002/elps.200900126
- [94] A. Biedermann, S. Bozza and F. Taroni, “Decision Theoretic Properties of Forensic Identification: Underlying Logic and Argumentative Implications,” Forensic Science International, Vol. 177, No. 2-3, 2008, pp. 120-132. doi:10.1016/j.forsciint.2007.11.008
- [95] M. J. Saks, “Forensic Identification: From a Faith-Based ‘Science’ to a Scientific Science,” Forensic Science International, Vol. 201, No. 1-3, 2010, pp. 14-17. doi:10.1016/j.forsciint.2010.03.014
- [96] K. Virkler and I. K. Lednev, “Raman Spectroscopic Signature of Semen and Its Potential Application to Forensic Body Fluid Identification,” Forensic Science International, Vol. 193, No. 1-3, 2009, pp. 56-62. doi:10.1016/j.forsciint.2009.09.005
- [97] Z. Wang, J. Zhang, H. Luo, Y. Ye, J. Yan and Y. Hou, “Screening and Confirmation of MicroRNA Markers for Forensic Body Fluid Identification,” Forensic Science International: Genetics, Vol. 7, No. 1, 2013, pp. 116-123. doi:10.1016/j.fsigen.2012.07.006
NOTES
*Corresponding author.

