Advances in Microbiology
Vol.05 No.03(2015), Article ID:54880,12 pages
10.4236/aim.2015.53017
Characterization of Egyptian Botrytis cinerea Isolates from Different Host Plants
Hala Abdel Wahab
Department of Plant Pathology, Faculty of Agriculture, Ain Shams University, Cairo, Egypt
Email: hala_abdelwahab@agr.asu.edu.eg
Copyright © 2015 by author and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 20 February 2015; accepted 10 March 2015; published
ABSTRACT
Gray mold causes considerable economic losses of fruit and vegetable production. The current study on Egyptian population structure of Botrytis cinerea demonstrates that this species is composed of four TE genotypes: transposa, vacuma, boty and flipper types using transposable elements and sensitivity to the hydroxyanilide fungicide, fenhexamid. The results show that transposa is the predominant isolate type (63.6%) in the sampled populations of B. cinerea. However, the four isolate types are fenhexamid-sensitive regardless of location, host plant and plant organ. Additionally, B. cinerea isolates collected from different host plants do not exhibit any host preference using artificial infection test on lettuce. Furthermore, no relation is found between isolate type and aggressiveness and no divergence event has occurred among the isolates collected from different locations and host plants. The results suggest that host specialization of B. cinerea has not been occurred in the current sampled crops.
Keywords:
Fenhexamid Resistance, Gray Mold, Molecular Characterization, Pathogenicity Variation, Transposable Elements

1. Introduction
Botrytis cinerea Pers.: Fr. (teleomorph: Botryotinia fuckeliana (de Bary) Whetzel) is a necrotrophic fungus responsible for gray mold on hundreds of dicot plants [1] and a notorious pathogen of a variety of crops which are affected in temperate and subtropical regions, causing soft rotting of all aerial plant parts, and rotting of vegetables, fruits and flowers post-harvest worldwide [2] . B. cinerea has become an important model for molecular study of necrotrophic fungi [3] and is also capable of colonizing plants internally as an endophyte without causing any disease or stress symptoms [4] . Generally, one of the fungal genomic variability sources is the existence of transposable elements (TEs) [5] [6] . Previous studies on French, Chilean populations of B. cinerea demonstrated that this species was composed of two sympatric species, transposa and vacuma [7] [8] , while three TE genotypes had been found in American populations of B. cinerea [9] . Transposa has two types of transposable elements: boty [10] and flipper [11] , whereas these two elements are absent in vacuma. Furthermore, it was shown that the transposa isolates did not differ in pathogenicity from the vacuma isolates [12] . Recently, molecular studies of different nuclear genes have suggested that B. cinerea populations are grouped into two different clades: group I and group II, phylogenetic species [13] [14] . Described group I isolates are exclusively from the vacuma transposon type, while group II can feature transposa, flipper (containing only flipper), boty (containing only boty) or vacuma genotype [8] [9] [13] [15] -[19] .
Previous studies demonstrated that some vacuma isolates were resistant to the hydroxyanilide fungicide, fenhexamid [13] [20] [21] , while other studies showed the sensitivity of vacuma isolates [19] . Furthermore, it was thought that B. cinerea had no host specificity [22] [23] as it attacked a wide range of host plants. Giraud et al. (1999) reported that the prevalence of transposa and vacuma was significantly different in B. cinerea populations collected from different host plants. In light of these recent studies, it appears that B. cinerea may have host specialization, which contradicts the traditional view [22] [23] . Currently, it is unclear whether the two sympatric species are present in B. cinerea populations in Egypt and their resistance to the more recent fungicide, fenhexamide. The objectives of the current study are to determine: 1) The species type(s) of B. cinerea existed in Egypt; 2) Pathogenic specificity in B. cinerea populations collected from different host plants; 3) Sensitivity of B. cinerea isolates to the hydroxyanilide fungicide, fenhexamid; and 4) Molecular divergence among isolates and its association with the pathogenicity of isolates.
2. Materials and Methods
2.1. Isolate Collection of B. cinerea
Thirty three isolates of B. cinerea were collected from different host plants: grape, strawberry and lettuce by using the modified selective medium, m1KERS, which was previously developed [24] [25] . The composition of such medium consists of the following components (gL−1 distilled water): glucose, 20 g; NaNO3, 1 g; KH2PO4, 1.2 g; MgSO4∙7H2O, 0.2 g; KCl, 0.15 g; agar, 25 g. This medium was autoclaved at 121˚C for 20 min. After cooling to 65˚C, the following ingredients were added: chloramphenicol, 0.05 g; tannic acid, 5 g; CuSO4, 2.2 g; Cabrio Top fungicide (Pyraclostrobin), 0.1 g. Different organs from strawberry (cv. Florida & Festival), grape (cv. Superior & Flame) and lettuce (cv. Baladi) were tested for Botrytis infection (Table 1). The plant samples were dipped separately in sterile water for 5 min, dried on paper towels, then plated onto m1KERS and incubated at 23˚C for 3 - 21 d. Table 1 shows the locations, host plants and plant organs from which the isolates were collected.
2.2. Determination of Mycelial Growth Rate
Mycelial agar plugs (6 mm diameter) were transferred from the colony margins of 3-d-old isolate culture of B. cinerea to the center of PDA petri dishes (9 cm diameter). Two replicates were done for each isolate and incubated at 23˚C. The mycelial growth rate (cm/d) was obtained daily by measuring the colony diameter during one week. This test was repeated twice.
2.3. Pathogenicity Assay
Detached leaves of lettuce plant (cv. Baladi) were used for testing the pathogenicity of B. cinerea isolates. Leaves were excised from the central part of each plant. Four leaves were selected for each replicate and placed on moist towels in a plastic tray. Mycelial agar plugs were removed from the colony margin of PDA cultures of each isolate of B. cinerea using a sterilized cork borer and placed on the leaves with the mycelial side of each plug facing the leaf surface. One or two plug(s) per leaf and four leaves for each isolate were prepared. Each tray was covered with a transparent plastic cover to maintain high humidity and incubated in a growth chamber at 20˚C under fluorescent light (12 h light/12 h dark). Lesion diameter around each mycelial agar plug on lettuce leaves was measured after incubation for 72 h. This test was repeated twice.
2.4. Fungicide Sensitivity Assay
Fenhexamid (Sigma-Aldrich) was dissolved in sterile water, adjusted to 10 mg/ml as a stock solution, and
Table 1. B.cinerea isolates collected from different organs of many host plants.
*T: transposa; V: vacuma; B: boty; F: flipper.
added to the potato dextrose agar (PDA) (Difco, Dickinson and Company) medium after sterilization to produce the concentrations of 0.003, 0.01, 0.1, 0.5, 1, 5, 10 and 13 µg fenhexamid per ml of PDA medium. The mycelial plug (6 mm) was cut from the edge of four-d-old colony for each isolate of B. cinerea and placed on the center of a PDA dish amended with each of the fungicide concentration. Two plates for each concentration were used, and then the experiment was performed twice. Cultures were incubated at 23˚C for 3 d. For each plate, the colony diameter was measured in two perpendicular directions with the original mycelial plug diameter (6 mm) subtracted. For each isolate, the 50% effective concentration (EC50), which is the fungicide concentration that results in 50% mycelial growth inhibition, was calculated.
2.5. Transmission Electron Microscopy
The mycelial agar plugs of both hypovirulent isolate BCL29 and virulent isolate BCL11 of B. cinerea were placed on PDA in a petri dish (6 cm diameter) and incubated at 23˚C for 7 d. Small pieces (about 3 × 3 mm) of the mycelia of such two isolates were sampled from the colony margin areas and fixed in 2% of glutaraldehyde in 0.05 M sodium cacodylate buffer (pH 7.0) at 4˚C overnight. The specimen pieces were washed in 0.2 M sodium phosphate buffer (pH 7.0) three times at room temperature (25˚C), 10 min each time, and post-fixed in 2% osmium tetroxide in 0.2 M sodium phosphate buffer for 2 h, and embedded in hard resin, then cut into ultra-thin slices (100 nm) by ultramicrotome. Sections were stained with 5% of uranyl acetate and rinsed in 50% of ethanol for 1 h. Then, they were dehydrated in graded series of ethanol, stained with 5% of aqueous lead citrate and 5% of uranyl acetate, and examined with the transmission electron microscope (JEOL 1200ESII) at 80 kV.
2.6. Statistical Analysis
Regression model and subsequently the EC50 were estimated using SPSS [26] . Data scored from lesion and colony diameter for both Pathogenicity and fenhexamid sensitivity assays, respectively, and analyzed by analysis of variance (ANOVA) implemented in SPSS to determine the significant variation within and among the studied host plants. Data means were treated using the least significant difference test at P = 0.05 level.
2.7. DNA Preparation
Each isolate was grown in potato dextrose broth (Difco Laboratories, Dickinson and Company) at 23˚C for 7 d. Mycelia were harvested and washed in sterile water, frozen in liquid nitrogen, and lyophilized. Fungal genomic DNA was extracted using the GeneJET Plant GenomicDNA extraction KIT® (Thermo Scientific).
2.8. Detection of Transposable Elements (Boty and Flipper)
Boty, the long-terminal repeats (LTRs) [10] and flipper, a mobile Fot1-like transposable element [11] have been identified from B. cinerea. The PCR primer pair F300 (5’-GCA CAA AAC CTA CAG AAG A-3’) and F1550 (5’-ATT CGT TTC TTG GAC TGT A-3’) has been used to detect the flipper element [11] . To detect the boty element, a pair of primers BotyF4 (5’-CAG CTG CAG TAT ACT GGG GGA-3’) and BotyR4 (5’-GGT GCT CAA AGT GTT ACG GGA G-3’) was used to amplify the LTR according to Ma and Michalilides (2005) [9] . The PCR reaction was performed (TECHNE TC-3000) in 25 µl of 2X PCR Master Mix (Thermo Scientific) containing 1 µM of each primer, 100 ng of fungal DNA. The PCR was performed using the following parameters: an initial denaturation for 5 min at 95˚C, followed by 40 cycles of denaturation at 94˚C for 1 min, annealing at 60˚C for the primer pair F300 and F1550, or 68˚C for the primer pair BotyF4 and BotyR4, for 1 min, extension at 72˚C for 1 min (for boty primers) or 3 min (for flipper primers), and terminated with a final extension at 72˚C for ten min. PCR products were tested and verified on 1.5% agarose gels using standard protocol.
2.9. ITS and LTR Amplification and Cloning
Two primers, ITS1 (5’-TCC GTA GGT GAA CCT GCG G-3’) and ITS4 (5’-TCC TCC GCT TAT TGA TAT GC-3’) were used to amplify the internal transcribed spacer (ITS) according to White et al. (1990) [27] . While, the primer pair BotyF4 and BotyR4 was previously used to amplify the boty LTR [9] . The amplification was conducted for each region in a final volume of 50 µl containing 1 µM of each primer, 0.2 mM of each dNTP (Thermo Scientific), 1× Pfu buffer with MgSO4 and 2 U of Pfu DNA polymerase (Thermo Scientific), 100 ng of fungal DNA. For ITS amplification, reactions were performed in a TechneTM thermocycler programmed as the following steps: denaturation at 94˚C for 5 min, followed by 35 cycles, were run at 94˚C for 30 sec, 55˚C for 30 sec, and 72˚C for 1 min for denaturing, annealing and extension, respectively, and the final extension at 72˚C for 5 min. The PCR product was then kept at 4˚C. In order to study the divergence among different B. cinerea isolates, the boty element was cloned from randomly selected isolates: BCG17 (Clone 1 and 2), BCL2 (Clone 5 and 6), BCG6 (Clone 5 and 6), BCG14 (Clone 1) using Transform Aid Bacterial Transformation Kit (Thermo Scientific). Each clone was re-amplified using the previously mentioned conditions.
2.10. ITS and LTR Sequencing
Amplified fragments were purified and concentrated using Silica-column based method (GeneJET PCR Purification Kit, Thermo Scientific), and then sequenced by sequencing service (Macrogen Europe, Netherlands). Sequence chromatograms were compiled using Bioedit v3 [28] to assemble the sequences. Haplotypes sequences were submitted into the GenBank database (http://www.ncbi.nlm.nih.gov).
2.11. Phylogenetic Analysis
The aligned sequences were analyzed using Maximum Parsimony (MP) [29] . The MP analyses were run with PAUP* 4b10 [30] using the default setting and assessed using 1000 bootstrap replicates. Additionally, the data were analyzed by Bayesian inference (BI) [31] . The best models for nucleotide substitution were determined for LTR region by JModelTest [32] , a program which uses intensively PhyML [33] . Two runs were conducted with 1,000,000 generations. Trees were sampled every 1000th generation and the first 40,000 trees were discarded (burn-in) in order to exclude such trees before the chain reached the stationary phase. Burn-in validity was tested using Tracer v1.4 [34] . Trees were visualized using TreeGraph 2 [35] . A maximum likelihood (ML) analysis was carried out by MEGA5 [36] . Tree inference options were set to Nearest Neighbor Interchange. Gaps/miss- ing data were treated as partial deletions with site coverage cutoff = 95%. A bootstrap analysis with 1000 replicates was carried out in order to study the clade support values. In all methods, trees were generated in the presence of the available LTR sequences found by BLAST tool (NCBI website). The number of base substitutions per site (Genetic distance) was estimated between two groups (Egyptian isolates versus GenBank accessions) using MEGA5. Analyses were conducted using the Maximum Composite Likelihood model. The rate variation among sites was modeled with a gamma distribution (shape parameter = 0.42). The analysis involved 25 nucleotide sequences. All ambiguous positions were removed from each sequence pair.
3. Results
3.1. Isolation of B. cinerea Populations Using m1KERS Medium
Different organs from the host plants: lettuce, strawberry and grape were screened to detect B. cinerea after incubation on m1KERS medium during 3 - 21 d. The appearance of brown halo formation surrounding the infected plant samples was observed at the third day of incubation indicating the presence of B. cinerea in the plant samples. All B. cinerea isolates were then identified morphologically using light microscope, and conserved in paraffin oil at 4˚C.
3.2. Determination of Isolate Type Using Transposable Elements
The PCR results showed that the primer pair F300 & F1550 has amplified the expected 1250 bp PCR fragment from 24 out of 33 isolates, and the primer pair BotyF4 & BotyR4 has generated a 510 bp PCR fragment from 28 out of 33 isolates (Data not shown). While, two isolates have neither of these two elements (Table 1). Among 33 isolates tested, 21 have two transposable elements, boty and flipper (transposa type), which indicated that transposa isolates were predominant (63.6%) in almost all of B. cinerea populations isolated from different host plants and plant organs. While, seven have the boty element (boty type, 21.2%), three have the flipper element (flipper type, 9.1%) and two have neither of these two elements (vacuma type, 6.1%; Table 1).
3.3. Pathogenicity of B. cinerea Isolates
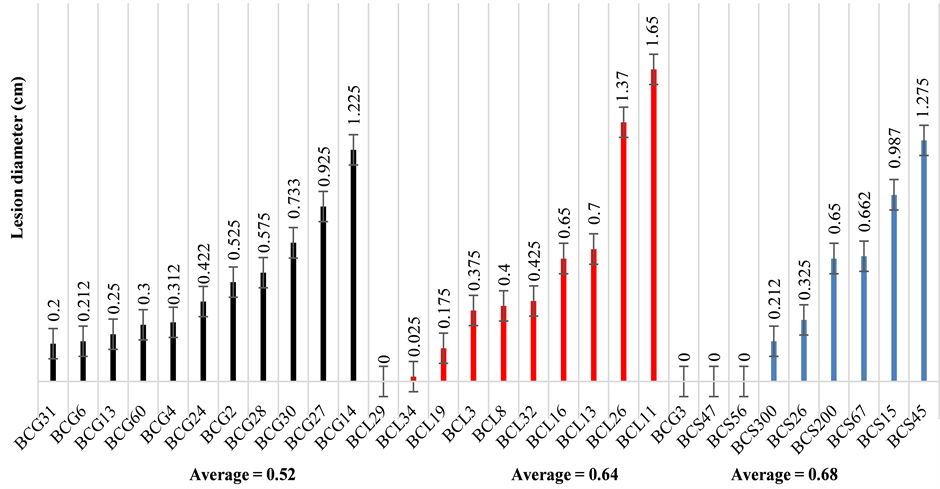
Statistical analysis of the pathogenicity test of 30 isolates of B. cinerea, collected from different host plants, on lettuce leaf showed a significant difference among isolates depending on their aggressiveness regardless of location and host plant (Figure 1). After incubation at 20˚C for 3 d, four isolates (BCG3, BCS47, BCS56, BCL29) out of 30, obtained from different plant species, have shown their hypovirulence on lettuce leaves (Table 1 and Figure 1), whereas BCL11 isolate, collected from lettuce stem, caused no infection on lettuce leaf (Figure 1 and Figure 2). The lesion diameter varied among the virulent isolates of B. cinerea, ranging from 0.02 to 1.65 cm (Figure 1), whereas the hypovirulent isolate BCL29, collected from lettuce stem, caused no infection on lettuce leaves (Figure 1 and Figure 2). Moreover, there was no significant difference regarding pathogenicity between all isolate types, as has been found in previous reports [37] .
All B. cinerea isolates, but one, BCL29 (Table 2), grew rapidly on PDA at 23˚C with average mycelial radial growth rate (MGR) ranging from 2 to 3.6 cm/d (Table 2). As shown in Table 2, the MGR of the virulent isolate BCL11 (3.6 cm/d) is higher than that of the hypovirulent isolate BCL29 (1.2 cm/d). Many isolates developed and covered the entire dish (9 cm diameter) within 3 - 4 d. Aerial mycelia, conidia, and/or sclerotia were formed after 15-d-old cultures of many isolates such as BCL11 (data not shown). In contrast, some of them like the hypovirulent islolate BCL29 often produced sparse conidia and did not produce any sclerotia on the colonies after incubation for 15 d (data not shown). These results confirmed a clear variation between such two isolates, BCL11 and BCL29, at both morphological and pathogenicity levels.
3.4. Sensitivity of B. cinerea Isolates to Fenhexamid
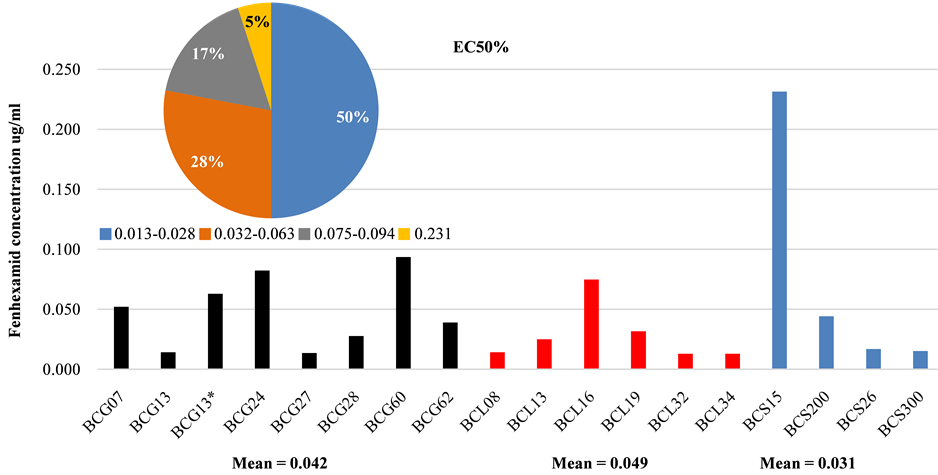
The EC50 values of all B.cinerea isolates were lower than 1 µg/ml (Figure 3) indicating that these isolates were
Figure 1. Pathogenicity of the B. cinerea isolates collected from three host plants: grape, strawberry and lettuce. Results were expressed as mean lesion diameter (cm) ± standard error.
Figure 2. The virulence of BCL11 (right) and the hypovirulence of BCL29 (left) on lettuce leaf.
Figure 3. Sensitivity of B. cinerea isolates to fenhexamid. Additional pie chart shows the percentage of EC50 concentration ranges for all isolates regardless of their host plants.
Table 2. Determination of mycelial growth rate (MGR) of different B. cinerea isolates.
sensitive to fenhexamid according to the previous studies [38] -[40] . In addition, the EC50 values were within the range of 0.013 - 0.23 µg/ml fenhexamid (Figure 3) for B.cinerea isolates which were collected from different host plants and showed all TE genotypes (transposa, vacuma, boty and flipper) suggesting that no relation was evident between isolate type and resistance to fenhexamid. This case is in accordance with numerous studies but in contrast with some other studies [7] [9] . Among all isolates tested, only one isolate has shown that EC50 value is greater than 0.2 µg/ml (Figure 3). The minimum inhibitory concentrations (MICs) of the majority of isolates are less than 13 µg/ml. Thus, these isolates are considered to be fenhexamid-sensitive based on previous studies on the sensitivity of B. cinerea to fenhexamid [38] [39] [41] -[44] .
3.5. Hyphal Cell Malformation of BCL29
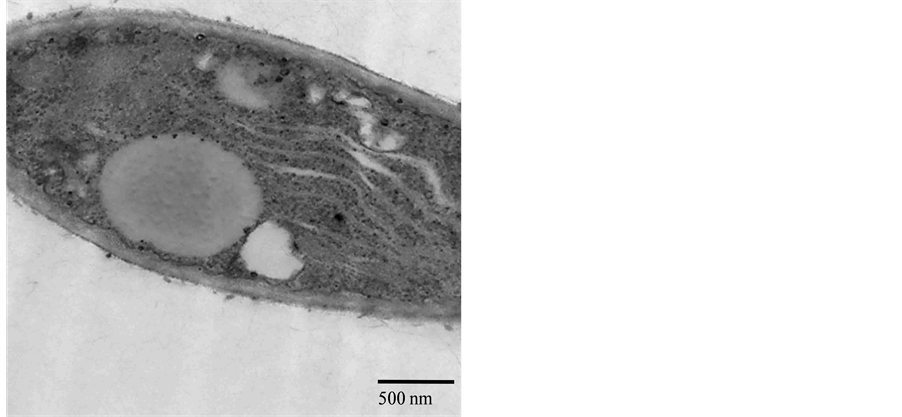
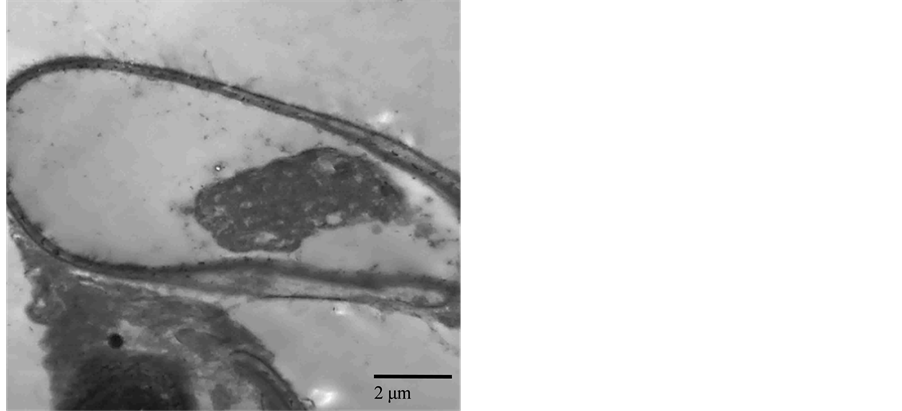
TEM observation of both the virulent BCL11 and the hypovirulent BCL29 showed that the hyphal cell of BCL11 had a dense and distributed cytoplasm and formed numerous mitochondria with normal cristae inside each mitochondrion. In contrast, the hyphal cell of BCL29 produced an accumulated cytoplasm and showed a cellular degeneration (Figure 4). This result indicated that the variation between these two isolates existed also at cellular level.
3.6. Sequences and Phylogenetic Analysis
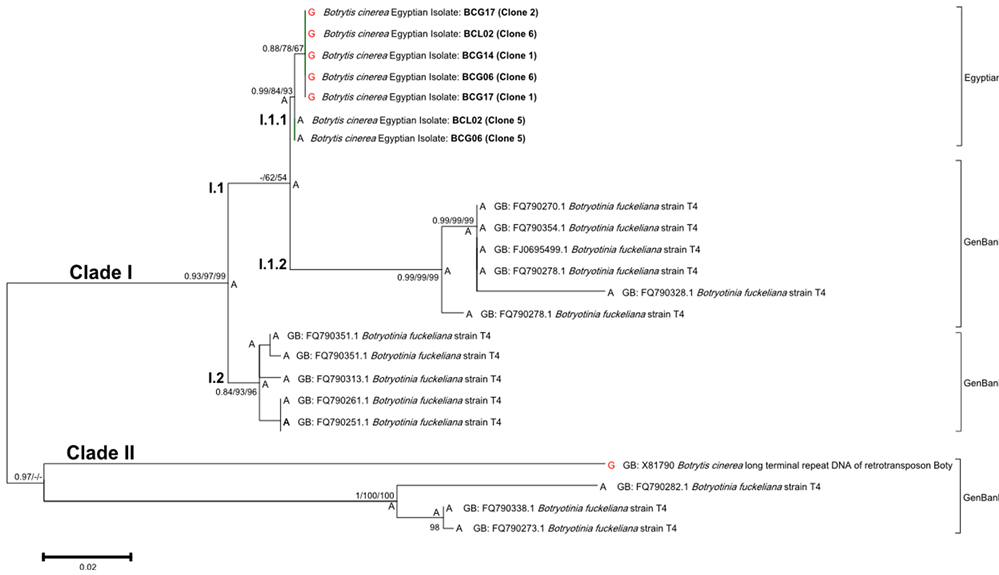
The primer combination ITS1 and ITS4 produced a fixed region length of 509 bp for four isolates of B. cinerea. By comparing their sequences with the NCBI database through Blast search, a complete query of exact matching and coverage were found to Botryotinia fuckeliana (GenBank accession no. X81790). Only one haplotype (accession no. KM276776) was found as no point mutation was detected for the sequenced isolates and marked no differences with the GenBank accession. The primer combination BotyF4 and BotyR4 produced a fixed region length of 510 bp for seven sequences from four isolates of B. cinerea. BLAST search demonstrated that the maximum score of alignment was to the long terminal repeat of the retrotransposon boty (GenBank accession no. X81790). Two haplotypes (accession no. KM276777 and KM276778) were found, with a unique transition point mutation (A > G) in the nucleotide site no. 139 of the 510 bp amplified sequence (Figure 5). Estimated genetic distance was 0.121 ± 0.031 between the GenBank accessions and 0.01 ± 0.01 between the Egyptian isolates, while the genetic distance was 0.085 ± 0.022 between the two groups.
The phylogenetic tree of LTR sequences was generated using BI and confirmed by both MP and ML methods (Figure 5). The tree was formed from seven LTR sequences amplified from the Egyptian isolates along with 15 GenBank accessions that showed a similar coverage range. Two main clades could be clearly distinguished. Clade I was supported with 0.93 of BI posterior probability, 97 bootstrap values of MP method and 99 bootstrap values of ML method. Sub-clade I.1 was divided into two partitions and the partition I.1.1 was formed by the Egyptian isolates sequences BCG17 (Clone 1 and 2), BCL2 (Clone 6), BCG6 (Clone 6), BCG14 (Clone 1) which possessed the G base in the dimorphic site along with BCL2 (Clone 5) and BCG6 (Clone 5). Such partition was supported with 0.99 of BI posterior probability, 84 bootstrap values of MP method and 93 bootstrap values of ML method. While the partition I.1.2, Sub-clade I.2 and Clade II were formed by the accessions obtained from the GenBank database.
4. Discussion
In the current study, four TE genotypes of B. cinerea isolates: transposa, boty, flipper and vacuma, were described from different Egyptian locations and collected from numerous symptomless plant organs. The results showed that transposa was the predominant isolate type (63.6%) in the sampled populations of B. cinerea. The
Figure 4. Transmission electron micrographs showing the cellular difference between two B. cinerea isolates (virulent: BCL11, left and hypovirulent: BCL29, right). Bar represents 0.5 - 2 μm.
Figure 5. Rooted tree for LTR sequences based on Bayesian inference method. Clades I and II are indicated. For each clade, BI posterior probability values and bootstrap support values above 50% for MP and ML are written, respectively. The tree is drawn to show the ancestral state of the dimorphic nucleotide site no. 139, where G and A are written in red and black, respectively.
other isolate types found in our samples were: boty isolates (21.2%), flipper isolates (9.1%), and vacuma isolates (6.1%). This result was in agreement with previous studies that had demonstrated the significant predominance of transposa type [19] [45] -[47] . Otherwise, the vacuma isolates had been previously reported as a predominant type in other host plants [37] . Additionally, we have found that all four isolate types, collected from different host plants, plant organs and locations, are fenhexamid-sensitive populations with a concentration between 0.013 - 0.23 µg/ml indicating a fenhexamid sensitivity [48] . Thus, such fungicide concentration would be recommended as an effective fungicide against B. cinerea infections. Interestingly, none of the isolate tested was phenotyped as HydR1, which corresponded to the natural resistance exhibited only by B. cinerea group I [13] . Meanwhile, these current isolates involved in B. cinerea group as they did not have any naturally resistant species as B. pseudocinerea recently described [49] . Indeed, these isolates could be related to B. cinerea group II [21] [50] [51] , but would be further confirmed at a molecular level. However, the observed absence of resistant isolates could be due either to the sampling strategy or to a real absence of these isolates in Egypt, so that it remained to be further investigated. Moreover, no clear relationship was found between sensitivity to fenhexamid and the TE genotype in B. cinerea. Otherwise, previous studies had shown that some vacuma isolates were resistant to the fungicide, fenhexamid [20] [21] . In fact, the hydroxyanilide fungicide, fenhexamid is a recently introduced fungicide with a high preventive activity against gray mold on various crops in Egypt (Prof. Dr. El-Shimy H., ARC, personal communication). Although newly introduced botryticides face the possibility of resistance development [21] , other studies have shown resistance to such fungicide in some areas where it have never been used. In addition, the results demonstrated, consistently with other studies, that resistance to fenhexamid had not also been found from boty isolates [7] [13] . Contrarily, it was reported that all boty isolate types were fenhexamid-resistant populations [9] . However, fenhexamid resistance alone was not a reliable character to classify B. cinerea [19] .
Differentiation in pathogenicity was observed among isolates of B. cinerea depending on isolate aggressiveness regardless of the host plant. The majority of B. cinerea isolates (86.7%) were virulent, whereas 13.3% of them, collected from different host plants, caused no infection on lettuce leaves. In fact, B. cinerea can infect a wide range of host plants and plant organs under laboratory conditions, and it had long been thought that B. cinerea had no host specificity [22] [23] , while other studies had shown the occurrence of host specificity [52] . In addition, the Egyptian B. cinerea populations did not demonstrate any relation between TE genotype and isolate virulence, consistently with the previous results of Greek isolates [37] . Otherwise, this relation had been positively documented [12] [45] . Recent studies on genetic population of B. cinerea had shown that there were significant genetic differentiations among isolates collected from different host plants in France [10] [53] and in Chile [8] . In addition, the results here showed variation of mycelial growth and sclerotia production among all isolates tested. One of them, BCL29, grew slowly on PDA with the average radial growth rate 1.2 cm/d. The TEM test investigates a cellular degeneration in the hypovirulent isolate BCL29 compared with the virulent isolate BCL11 of B. cinerea. This may be caused by one of many factors such as mycoviruses cellular association as previously reported [54] -[56] .
On the other hand, the ITS sequence didn’t show any polymorphism level that would resolve the reported issue of the cryptic speciation known in B. cinerea [57] . The LTR sequences showed the presence of two abundant copies in the Egyptian isolates in which a unique transition point mutation was occurred and caused the presence of two different partitions in clade I.1.1 (revise Figure 5): Partition I contained the LTR sequences from the five clones of Egyptian isolates, while partition II contained two other amplified clones of the same isolates grouped in partition I. The presence of the two copies in the same individual impedes the possibility to correlate such single nucleotide polymorphism (SNP) to the virulence action. Nevertheless, the detected genetic distance between the Egyptian isolates and the GenBank accessions reflect the high polymorphism level within the boty mobile element. While the phylogenetic analysis shows the Egyptian isolates as a separate highly supported divergent group from the others. Combining the current ITS analysis and LTR sequences, it would confirm that no divergence events have occurred among the Egyptian isolates even if the samples are isolated from different host plants and different locations. The results suggest that the host specialization of B. cinerea does not occur in the sampled crops (grape, strawberry and lettuce). Even though, Isenegger et al. (2008) have found that the genetic differentiation, possibly as a result of geographic isolation, is likely to have occurred recently in isolates from Australia and South Asia. Otherwise, geography does not differentiate the Egyptian B. cinerea isolates under study, which reflects a high spore dispersal ability among the studied locations [58] .
The detection of the cryptic speciation of B. cinerea could have important implications in population structure analysis and thus integrated disease management strategies in Egypt and surrounding regions, it would be proposed the application of a genome-scan approach (e.g. AFLP [59] [60] ) or the next generation sequencing (NGS) technology [61] to efficiently measure the speciation level and genetic diversity within B. cinerea isolates of Egypt and its association with the isolate pathogenicity.
5. Conclusion
The Egyptian B. cinerea populations are composed of four TE genotypes that demonstrate fenhexamid sensitivity at low concentration level. Such fungicide is recommended at 0.013 - 0.23 µg/ml as an effective concentration against B. cinerea. The results show variation among B. cinerea isolates at morphological and cellular level. However, the current results demonstrate no relation between isolate type, aggressiveness, fenhexamid resistance and host preference.
Acknowledgements
This research was supported financially by the Science and Technology Development Fund (STDF), Egypt, Grant No. 2131. I Thank Dr. Magdy Mahmoud, Department of Genetics, Faculty of Agriculture, for his technical assistance in the current study.
References
- Elad, Y., Williamson, B., Tudzynski, P. and Delen, N. (2007) Botrytis spp. and Diseases They Cause in Agricultural Systems―An Introduction. In: Elad, Y., Williamson, B., Tudzynski, P. and Delen, N., Eds., Botrytis: Biology, Pathology and Control. Springer, The Netherlands, 1-8.
- Williamson, B., Tudzynski, B., Tudzynski, P. and Van Kan, J.A. (2007) Botrytis cinerea: The Cause of Grey Mould Disease. Molecular Plant Pathology, 8, 561-580. http://dx.doi.org/10.1111/j.1364-3703.2007.00417.x
- Dean, R., Van Kan, J.A., Pretorius, Z.A., Hammond-Kosack, K.E., Di Pietro, A., Spanu, P.D., Rudd, J.J., Dickman, M., Kahmann, R., Ellis, J. and Foster, G.D. (2012) The Top 10 Fungal Pathogens in Molecular Plant Pathology. Molecular Plant Pathology, 13, 414-430. http://dx.doi.org/10.1111/j.1364-3703.2011.00783.x
- Van Kan, J.A., Shaw, M.W. and Grant-Downton, R.T. (2014) Botrytis Species: Relentless Necrotrophic Thugs or Endophytes Gone Rogue? Molecular Plant Pathology, 15, 957-961. http://dx.doi.org/10.1111/mpp.12148
- Dufresne, M., Hua-Van, A., Abdel Wahab, H., Ben M’Barek, S., Vasnier, C., Teysset, L., Kema, G.H.J. and Daboussi, M.-J. (2007) Transposition of a Fungal Miniature Inverted-Repeat Transposable Element through the Action of a Tc1-Like Transposase. Genetics, 175, 441-452. http://dx.doi.org/10.1534/genetics.106.064360
- López-Berges, M.S., Di Pietro, A., Daboussi, M.-J., Abdel Wahab, H., Vasnier, C., Roncero, G., Dufresne, M. and Hera, C. (2009) Identification of Virulence Genes in Fusarium oxysporum f. sp. lycopersici by Large-Scale Transposon Tagging. Molecular Plant Pathology, 10, 95-107. http://dx.doi.org/10.1111/j.1364-3703.2008.00512.x
- Giraud, T., Fortini, D., Levis, C., Lamarque, C., Leroux, P., LoBuglio, K. and Brygoo, Y. (1999) Two Sibling Species of the Botrytis cinerea Complex, transposa and vacuma, Are Found in Sympatry on Numerous Host Plants. Phytopathology, 89, 967-973. http://dx.doi.org/10.1094/PHYTO.1999.89.10.967
- Muñoz, G., Hinrichsen, P., Brygoo, Y. and Giraud, T. (2002) Genetic Characterisation of Botrytis cinerea Populations in Chile. Mycological Research, 106, 594-601. http://dx.doi.org/10.1017/S0953756202005981
- Ma, Z.H. and Michailides, T.J. (2005) Genetic Structure of Botrytis cinerea Populations from Different Host Plants in California. Plant Disease, 89, 1083-1089. http://dx.doi.org/10.1094/PD-89-1083
- Diolez, A., Marches, F., Fortini, D. and Brygoo, Y. (1995) Boty, a Long-Terminal-Repeat Retroelement in the Phytopathogenic Fungus Botrytis cinerea. Applied and Environmental Microbiology, 61, 103-108.
- Levis, C., Fortini, D. and Brygoo, Y. (1997) Flipper, a Mobile Fot1-Like Transposable Element in Botrytis cinerea. Molecular and General Genetics, 254, 674-680. http://dx.doi.org/10.1007/s004380050465
- Martinez, F., Blancard, D., Lecomte, P., Levis, C., Dubos, B. and Fermaud, M. (2003) Phenotypic Differences between vacuma and transposa Subpopulations of Botrytis cinerea. European Journal of Plant Pathology, 109, 479-488. http://dx.doi.org/10.1023/A:1024222206991
- Albertini, C., Thebaud, G., Fournier, E. and Leroux, P. (2002) Eburicol 14α-Demethylase Gene (CYP51) Polymorphism and Speciation in Botrytis cinerea. Mycological Research, 106, 1171-1178. http://dx.doi.org/10.1017/S0953756202006561
- Fournier, E., Levis, C., Fortini, D., Leroux, P., Giraud, T. and Brygoo, Y. (2003) Characterization of Bc-hch, the Botrytis cinerea Homolog of the Neurospora crassahet-c Vegetative Incompatibility Locus, and Its Use as a Population Marker. Mycologia, 95, 251-261. http://dx.doi.org/10.2307/3762036
- ben Ahmed, D. and Hamada, W. (2005) Genetic Diversity of Some Tunisian Botrytis cinerea Isolates Using Molecular Markers. Phytopathologia Mediterranea, 44, 300-306.
- Milicevic, T., Topolovec-Pintaric, S., Cvjetkovic, B., Ivic, D. and Duralija, B. (2006) Sympatric Subpopulations of Botrytis cinerea on Strawberries Based on the Content of Transposable Elements and Their Connection with Resistance to Botryticides. Acta Horticulturae, 708, 115-118.
- Isenegger, D., Ades, P., Ford, R. and Taylor, P. (2008) Status of the Botrytis cinerea Species Complex and Microsatellite Analysis of Transposon Types in South Asia and Australia. Fungal Diversity, 29, 17-26.
- Rajaguru, B. and Shaw, M.W. (2010) Genetic Differentiation between Hosts and Locations in Populations of Latent Botrytis cinerea in Southern England. Plant Pathology, 59, 1081-1090. http://dx.doi.org/10.1111/j.1365-3059.2010.02346.x
- Esterio, M., Muñoz, G., Ramos, C., Cofré, G., Estévez, R., Salinas, A. and Auger, J. (2011) Characterization of Botrytis cinerea Isolates Present in Thompson Seedless Table Grapes in the Central Valley of Chile. Plant Disease, 95, 683-690. http://dx.doi.org/10.1094/PDIS-04-10-0298
- Fournier, E., Giraud, T., Loiseau, A., Vautrin, D., Estoup, A., Solignac, M., Cornuet, J.M. and Byrgoo, Y. (2002) Characterization of Nine Polymorphic Microsatellite Loci in the Fungus Botrytis cinerea (Ascomycota). Molecular Ecology Notes, 2, 253-255. http://dx.doi.org/10.1046/j.1471-8286.2002.00207.x
- Leroux, P., Fritz, R., Debieu, D., Albertini, C., Lanen, C., Bach, J., Gredt, M. and Chapeland, F. (2002) Mechanisms of Resistance to Fungicides in Field Strains of Botrytis cinerea. Pest Management Science, 58, 876-888. http://dx.doi.org/10.1002/ps.566
- Jarvis, W.R. (1980) Epidemiology. In: Coley-Smith, J.R., Verhoeff, K. and Jarvis, W.R., Eds., The Biology of Botrytis, Academic Press, London, 219-250.
- Lorbeer, J.W. (1980) Variation in Botrytis and Botryotinia. In: Coley-Smith, J.R., Verhoeff, K. and Jarvis, W.R., Eds., The Biology of Botrytis, Academic Press, London, 19-40.
- Abdel Wahab, H. and Younis, R.A. (2012) Early Detection of Gray Mold in Grape Using Conventional and Molecular Methods. African Journal of Biotechnology, 11, 15251-15257.
- Abdel Wahab, H. and Helal, N.S. (2013) Evaluation of Pre-Harvest Bioagent Applications for both Production and Biological Control of Onion and Strawberry Plants under Natural Botrytis Infections. African Journal of Plant Science and Biotechnology, 7, 64-69.
- IBM (2011) IBM SPSS Statistics for Windows, Version 20. IBM Corp., Armonk.
- White, T.J., Bruns, T., Lee, S. and Taylor, J. (1990) Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. and White, T.J., Eds., PCR Protocols: A Guide to Methods and Applications, Academic Press, San Diego, 315-322. http://dx.doi.org/10.1016/B978-0-12-372180-8.50042-1
- Hall, T.A. (1999) BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95-98.
- Fitch, W. (1971) Rate of Change of Concomitantly Variable Codons. Journal of Molecular Evolution, 1, 84-96. http://dx.doi.org/10.1007/BF01659396
- Swofford, D. (2002) PAUP* Version 4.0. Phylogenetic Analysis Using Parsimony (and Other Methods). Sinauer Associates, Inc., Sunderland.
- Ronquist, F. and Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics, 19, 1572-1574. http://dx.doi.org/10.1093/bioinformatics/btg180
- Posada, D. (2008) jModelTest: Phylogenetic Model Averaging. Molecular Biology and Evolution, 25, 1253-1256. http://dx.doi.org/10.1093/molbev/msn083
- Guindon, S. and Gascuel, O. (2003) A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Systematic Biology, 52, 696-704. http://dx.doi.org/10.1080/10635150390235520
- Rambaut, A. and Drummond, A. (2007) Tracer v1.4 (Internet). http://tree.bio.ed.ac.uk/software/tracer/
- Stöver, B.C. and Müller, K.F. (2010) TreeGraph 2: Combining and Visualizing Evidence from Different Phylogenetic Analyses. BMC Bioinformatics, 11, 7. http://dx.doi.org/10.1186/1471-2105-11-7
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. and Kumar, S. (2011) MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution, 28, 2731-2739. http://dx.doi.org/10.1093/molbev/msr121
- Samuel, S., Veloukas, T., Papavasileiou, A. and Karaoglanidis, G.S. (2012) Differences in Frequency of Transposable Elements Presence in Botrytis cinerea Populations from Several Hosts in Greece. Plant Disease, 96, 1286-1290. http://dx.doi.org/10.1094/PDIS-01-12-0103-RE
- Leroux, P., Chapeland, F., Desbrosses, D. and Gredt, M. (1999) Patterns of Cross-Resistance to Fungicides in Botryotinia fuckeliana (Botrytis cinerea) Isolates from French Vineyards. Crop Protection, 18, 687-697. http://dx.doi.org/10.1016/S0261-2194(99)00074-5
- Ziogas, B., Markoglou, A. and Malandrakis, A. (2003) Studies on the Inherent Resistance Risk to Fenhexamid in Botrytis cinerea. European Journal of Plant Pathology, 109, 311-317. http://dx.doi.org/10.1023/A:1023522213675
- Fekete, É., Fekete, E., Irinyi, L., Karaffa, L., Árnyasi, M., Asadollahi, M. and Sándor, E. (2012) Genetic Diversity of a Botrytis cinerea Cryptic Species Complex in Hungary. Microbiological Research, 167, 283-291. http://dx.doi.org/10.1016/j.micres.2011.10.006
- Fillinger, S., Leroux, P., Auclair, C., Barreau, C., Al Hajj, C. and Debieu, D. (2008) Genetic Analysis of Fenhexamid- Resistant Field Isolates of the Phytopathogenic Fungus Botrytis cinerea. Antimicrobial Agents and Chemotherapy, 52, 3933-3940. http://dx.doi.org/10.1128/AAC.00615-08
- Kretschmer, M., Leroch, M., Mosbach, A., Walker, A.S., Fillinger, S., Mernke, D., Schoonbeek, H.J., Pradier, J.M., Leroux, P., De Waard, M.A. and Hahn, M. (2009) Fungicide-Driven Evolution and Molecular Basis of Multidrug Resistance in Field Populations of the Grey Mould Fungus Botrytis cinerea. PLoS Pathogens, 5, e1000696. http://dx.doi.org/10.1371/journal.ppat.1000696
- Amiri, A., Heath, S. and Peres, N. (2013) Phenotypic Characterization of Multifungicide Resistance in Botrytis cinerea Isolates from Strawberry Fields in Florida. Plant Disease, 97, 393-401. http://dx.doi.org/10.1094/PDIS-08-12-0748-RE
- Schumacher, J., Gautier, A., Morgant, G., Studt, L., Ducrot, P.H., Pecheur, P.L., Azeddine, S., Fillinger, S., Leroux, P., Tudzynski, B. and Viaud, M. (2013) A Functional Bikaverin Biosynthesis Gene Cluster in Rare Strains of Botrytis cinerea Is Positively Controlled by VELVET. PLoS ONE, 8, e53729. http://dx.doi.org/10.1371/journal.pone.0053729
- Martinez, F., Dubos, B. and Fermaud, M. (2005) The Role of Saprotrophy and Virulence in the Population Dynamics of Botrytis cinerea in Vineyards. Phytopathology, 95, 692-700. http://dx.doi.org/10.1094/PHYTO-95-0692
- Martinez, F., Corio-Costet, M.F., Levis, C., Coarer, M. and Fermaud, M. (2008) New PCR Primers Applied to Characterize Distribution of Botrytis cinerea Populations in French Vineyards. Vitis, 47, 217-226.
- Váczy, K.Z., Sándor, E., Karaffa, L., Fekete, E., Fekete, É., Árnyasi, M., Czeglédi, L., Kövics, G.J., Druzhinina, I.S. and Kubicek, C.P. (2008) Sexual Recombination in the Botrytis cinerea Populations in Hungarian Vineyards. Phyto- pathology, 98, 1312-1319. http://dx.doi.org/10.1094/PHYTO-98-12-1312
- Rodríguez, A., Acosta, A. and Rodríguez, C. (2014) Fungicide Resistance of Botrytis cinerea in Tomato Greenhouses in the Canary Islands and Effectiveness of Non-Chemical Treatments against Gray Mold. World Journal of Microbiology and Biotechnology, 30, 2397-2406. http://dx.doi.org/10.1007/s11274-014-1665-5
- Walker, A.S., Gautier, A.L., Confais, J., Martinho, D., Viaud, M., Pecheur, P.L., Dupont, J. and Fournier, E. (2011) Botrytis pseudocinerea, a New Cryptic Species Causing Gray Mold in French Vineyards in Sympatry with Botrytis cinerea. Phytopathology, 101, 1433-1445. http://dx.doi.org/10.1094/PHYTO-04-11-0104
- Albertini, C. and Leroux, P. (2004) A Botrytis cinerea Putative 3-keto Reductase Gene (ERG27) that Is Homologous to the Mammalian 17β-Hydroxysteroid Dehydrogenase Type 7 Gene (17β-HSD7). European Journal of Plant Pathology, 110, 723-733. http://dx.doi.org/10.1023/B:EJPP.0000041567.94140.05
- Leroux, P., Gredt, M., Leroch, M. and Walker, A.S. (2010) Exploring Mechanisms of Resistance to Respiratory Inhibitors in Field Strains of Botrytis cinerea, the Causal Agent of Gray Mold. Applied and Environmental Microbiology, 76, 6615-6630. http://dx.doi.org/10.1128/AEM.00931-10
- Asadollahi, M., Fekete, E., Karaffa, L., Flipphi, M., Árnyasi, M., Esmaeili, M., Váczy, K.Z. and Sándor, E. (2013) Comparison of Botrytis cinerea Populations Isolated from Two Open-Field Cultivated Host Plants. Microbiological Research, 168, 379-388. http://dx.doi.org/10.1016/j.micres.2012.12.008
- Giraud, T., Fortini, D., Levis, C., Leroux, P. and Brygoo, Y. (1997) RFLP Markers Show Genetic Recombination in Botryotinia fuckeliana (Botrytis cinerea) and Transposable Elements Reveal Two Sympatric Species. Molecular Biology and Evolution, 14, 1177-1185. http://dx.doi.org/10.1093/oxfordjournals.molbev.a025727
- Wu, M., Zhang, L., Li, G., Jiang, D., Hou, M. and Huang, H.C. (2007) Hypovirulence and Double-Stranded RNA in Botrytis cinerea. Phytopathology, 97, 1590-1599. http://dx.doi.org/10.1094/PHYTO-97-12-1590
- Pearson, M.N. and Bailey, A.M. (2013) Viruses of botrytis. Advances in Virus Research, 86, 249-272. http://dx.doi.org/10.1016/B978-0-12-394315-6.00009-X
- Kecskeméti, E., Brathuhn, A., Kogel, K.H., Berkelmann-Löhnertz, B. and Reineke, A. (2014) Presence of Transposons and Mycoviruses in Botrytis cinerea Isolates Collected from a German Grapevine Growing Region. Journal of Phyto- pathology, 162, 582-595. http://dx.doi.org/10.1111/jph.12230
- McDonald, B.A. and Linde, C. (2002) Pathogen Population Genetics, Evolutionary Potential, and Durable Resistance. Annual Review of Phytopathology, 40, 349-379. http://dx.doi.org/10.1146/annurev.phyto.40.120501.101443
- Fournier, E. and Giraud, T. (2008) Sympatric Genetic Differentiation of a Generalist Pathogenic Fungus, Botrytis cinerea, on Two Different Host Plants, Grapevine and Bramble. Journal of Evolutionary Biology, 21, 122-132.
- Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M. and Zabeau, M. (1995) AFLP: A New Technique for DNA Fingerprinting. Nucleic Acids Research, 23, 4407-4414. http://dx.doi.org/10.1093/nar/23.21.4407
- Majer, D., Mithen, R., Lewis, B.G., Vos, P. and Oliver, R.P. (1996) The Use of AFLP Fingerprinting for the Detection of Genetic Variation in Fungi. Mycological Research, 100, 1107-1111. http://dx.doi.org/10.1016/S0953-7562(96)80222-X
- Narum, S.R., Buerkle, C.A., Davey, J.W., Miller, M.R. and Hohenlohe, P.A. (2013) Genotyping-by-Sequencing in Ecological and Conservation Genomics. Molecular Ecology, 22, 2841-2847. http://dx.doi.org/10.1111/mec.12350