Advances in Microbiology
Vol.3 No.7(2013), Article ID:40121,9 pages DOI:10.4236/aim.2013.37070
The 19 kDa Protein from Mycobacterium avium subspecies paratuberculosis Is a Glycolipoprotein
1Burnett School of Biomedical Sciences, College of Medicine, University of Central Florida, Orlando, USA
2BioSynse, Cary, US
Email: *saleh.naser@ucf.edu
Copyright © 2013 Saleh A. Naser et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received September 23, 2013; revised October 23, 2013; accepted November 30, 2013
Keywords: Mycobacterium Avium Subspecies Paratuberculosis; Glycolipoprotein; 19 KDa Protein; Post Translational Modification
ABSTRACT
This study characterizes the 19 kDa protein expressed by Mycobacterium avium subspecies paratuberculosis (MAP) as a glycolipoprotein, providing the foundation for future experiments regarding its antigenicity and role in disease pathogenicity. We have previously shown that a 4.8 kb insert from MAP will produce a 16 kDa recombinant protein when expressed in Escherichia coli and 19 kDa recombinant protein when expressed in M. smegmatis (smeg19K). The difference of 3 kDa in size of these expressed proteins may be related to post translational modifications that occur in Mycobacterium species. We hypothesized that smeg19K is a glycolipoprotein since BLAST analysis revealed approximately 76% amino acid identity between the MAP 19 kDa protein and a known lipoglycoprotein, the 19 kDa protein of M. tuberculosis. This prediction was confirmed by the following positive staining of smeg19K with Sudan Black 4B, a postelectrophoresis dye used to stain for lipids. Smeg19K has also stained positively for glycosylation with the lectin concavalin A, a highly specific stain for mannose residues. As expected, treatment with tunicamycin (an antibiotic known to inhibit N-glycosylation) and treatment with deglycosylation assay (non-specific for mannose), showed no reduction in size of 19 kDa glycolipoprotein.
1. Introduction
The genus Mycobacterium has well recognized pathogenic species. M. tuberculosis and M. leprae are the causative agents of infectious diseases, tuberculosis and leprosy, respectively [1], then M. avium and M. intracellulare, opportunistic pathogens, are associated with infection in hosts such as patients with AIDS [2,3]. The development and improvement needed for therapeutics and vaccines for these microorganisms have led to the genetic characterization of mycobacterium antigens, and to the identification of highly conserved protein families [4]. Despite decades of research, not much is known about the M. avium subsp. paratuberculosis (MAP or M. paratuberculosis) proteins. However, with the genome sequence of M. paratuberculosis identified [5], molecular biology techniques can be used to study different proteins/antigens expressed by this microorganism.
Previously in our laboratory, a genomic library of M. paratuberculosis was constructed in E. coli. Colonies were screened for gene expression using immunoblot and rabbit anti-M. paratuberculosis antibody. Clones specific to M. paratuberculosis were identified when they reacted negatively with rabbit anti-M. tuberculosis, rabbit anti-M. leprae and rabbit anti-M. bovis and rabbit anti-M. avium subspecies avium antibodies. pMptb#28 recombinant clone contained the insert of 4.8 kb and encoded for a single protein of 16 kDa. Surprisingly, M. paratuberculosis lacked a 16 kDa protein band when screened with rabbit anti-M. paratuberculosis antibodies. When the 4.8 kb BamH I fragment was cloned in M. smegmatis by using a pNEZ6.3 shuttle plasmid, the expressed product was estimated at 19 kDa protein (Smeg19k) similar to the size observed in extracts from M. paratuberculosis. This present study focused on the characterization of the 19 kDa protein expressed by M paratuberculosis regarding posttranslational modification.
Recent studies have shown some post-translational modification in mycobacterial antigens such as acylation and glycosylation [6-8], which is lacking in the majority of Gram negative bacteria such as E. coli. In acylated proteins, the lipid can attach through cysteine amino acid residues, while in glycoproteins, the carbohydrate moiety attaches via its anomeric carbon through a glycosidic link to the functional group −OH of a serine or threonine amino acid residue (referred to as O-linked), or through an N-glycosyl link to the amide nitrogen of an asparagine amino acid residue (N-linked) [9].
We propose smeg19K is a glycolipoprotein because there could be a homologue relation to the 19 kDa protein expressed by M. tuberculosis, known to be a glycolipoprotein [10]. BLAST analysis has revealed approximately 76% homology in amino acid identity between M. tuberculosis and M. paratuberculosis’s 19 kDa protein [10]. Young and Garbe, in 1991, showed through detergent phase separation and metabolic labeling that the 19 kDa of M. tuberculosis is a lipoprotein that constitutes an important target of the innate immune response [11]. Acylation is believed to occur at amino acids 19 - 24 [12]. Another study by Garbe et al., in 1993, published evidence for glycosylation in M. tuberculosis based on the ability of the 19 kDa lipoprotein to bind lectin concavalin A (ConA). ConA is highly specific for mannose residues linked via their anomeric carbon in α position [13]. Herrmann et al., in 1996, concluded that 5 threonine residues at positions 13, 14, 15, 19 and 20, of M. tuberculosis 19 kDa played a role in lectin ConA binding [8].
It is known that covalent modification of proteins with acyl or glycosyl moieties alter immunogenicity and/or pathogenicity. Some lipoproteins have been identified as major antigens from Treponema pallidum, Haemophilus influenzae and Mycoplasma hyorhinis based on their hydrophobicity during phase partitioning in the detergent Triton-Xll4 [14,15], and by metabolic labeling with fatty acid precursors [14,16,17]. In addition, glycoprotein antigens have been found in pathogenic mycobacteria [7, 18,19]. The ultimate goal of this study is to characterize the posttranslational modification of the 19 kDa M. paratuberculosis protein, which can play a role in its antigenicity. The discovery of effective vaccines to prevent Johne’s disease requires identification of antigens and their ability to modulate the immune response causing acquired protective immunity.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
E. coli strain Top 10 purchased from Invitrogen (Invitrogen Corp., Carlsbad, CA) and its recombinants were grown in 5 mL Luria Britani (LB) broth media containing 1% w/v tryptone, 0.5% w/v yeast extract and 1% w/v NaCl. This broth was supplemented with 12.5 μl ampicillin (Sigma) at a final concentration of 50 μg/mL. These cultures were incubated at 37˚C until growth was observed. Samples were aliquoted into 1 mL tubes, centrifuged for 5 min at 7000 rpm, and cell pellet was stored at −80˚C. The E. coli clone containing the plasmid pMptb#28 and E. coli pcDNAII were obtained from our genomic library as previously described [20]. M. smegmatis and M. smegmatis recombinant clone (SMEG19K) were grown in BBL MGIT Mycobacterium growth indicator tube (110 μl of fluorescent indicator and 4 mL broth: modified middlebrook 7H9 broth base, casein peptone). MGIT tube was supplemented with 5 μl kanamycin at a final concentration of 20 μg/mL. Samples were aliquoted into 1 mL tubes, centrifuged for 5 min at 7000 rpm, and cell pellet was stored at −80˚C. M. tuberculosis and M. paratuberculosis were previously grown in our lab from stock solutions and stored at −80˚C.
2.2. SDS-PAGE and Western Blot Analysis
Sonicated samples were boiled in the presence of SDS and β-mercaptoethanol. Electrophoresis was performed under reducing conditions by the method of Laemmli [21] with gels containing a 6% stack over a 12% resolving gel. Proteins were visualized by silver nitrate [22].
Proteins extracts, subjected to SDS-PAGE, were transferred to nitrocellulose membranes (Bio-Rad). For visualization of recombinant expression, blots were staining with rabbit hyperimmune anti-M. paratuberculosis serum (1:10,000) at 4˚C overnight (Sigma). Blot was then incubated with 1:10,000 diluted Goat anti-rabbit IgG-peroxidase conjugates for 2 hours at room temperature (Sigma). Color development on the membrane was visualized using opti-4CN diluent solution substrate (Bio-Rad).
2.3. Carbohydrate Analysis
1) The procedure for staining of nitrocellulose blots with peroxidase-conjugated concavalin A (ConA) was very similar to that used for immuno-staining [7]. Color development was visualized using Opti-4CN diluent solution substrate as described by the manufacturer (BioRad).
2) M. smegmatis recombinant clone (SMEG 19K) was subcultured into a BBL MGIT (Mycobacterium Growth Indicator Tube) and placed in non-motile incubator at 37˚C for 5 days or until visible growth. Tunicamycin powder (Sigma) was dissolved completely in 5 mL of DMSO to a stock concentration of 10 mg/mL, and working solution of 1 mg/mL tunicamycin was prepared. Different concentration of antibiotic and harvested SMEG- 19K subclone were added to new MGIT tubes and tubes were placed in rotator incubator at 37˚C until visible growth observed.
2.4. Acylation Analysis
Following electrophoresis, gels were stained in Sudan Black 4B solution (SB) (Sigma). First, gels were fixed in 40% v/v methanol, 10% v/v acetic acid, 50% v/v Millipore water for 30 min at room temperature. Then, gels were stained overnight with a 60% solution of SB in ethanol. Gels were destained in 3 changes of 30% ethanol, 15% acetic acid, and 65% Millipore water [23].
2.5. Bioinformatics Analysis
Bioinformatics software was used for protein identification and characterization, including: BLAST analysis, Expasy Proteomics, and Protein Information Resource (PIR) tools as follows: http://www.ncbi.nlm.nih.gov/BLAST/, http://www.expasy.org/tools/, http://pir.georgetown.edu/cgi-bin/multialn.pl.
3. Results
The expression of 19 kDa protein in cell extracts from M. smegmatis 19 kDa recombinant, and M. tuberculosis was confirmed by immunoblot using M. smegmatis adsorbed rabbit anti-M. paratuberculosis sera for M. tuberculosis (Figure 1, lane 3), for M. smegmatis (Figure 1, lane 5). No binding was observed against protein extracts from non-recombinant M. smegmatis (Figure 1, lane 4). Additionally, there was immunoreactivity in protein extracts from E. coli recombinant protein (Figure 1, lane 9) bound antibody at 16 kDa and not from non-recombinant E. coli (Figure 1, lane 7).
3.1. Evaluation of Smeg19K Protein for Carbohydrate Content
3.1.1. By ConA Staining
ConA is a lectin that specifically binds to α-D-glucose and α-D-mannose and identifies O-glycosylation. Samples were evaluated following staining an immunoblot with conA-peroxidase conjugated. We included, as shown in Figure 2, lane 2; protein extracts from M. tuberculosis as a positive control for glycosylation of the 19 kDa. In this study, M. smegmatis expressing recombinant 19 kDa stained positive with ConA, as seen in protein extracts from M. tuberculosis, indicating presence of carbohydrate (Figure 2, lane 4). On the other hand, M. smegmatis without 19 kDa (Figure 2, lane 3), E. coli without insert (Figure 2, lane 5) and E. coli recombinant which express 16 kDa of M. paratuberculosis 19 kDa native protein (Figure 2, lane 6) did not stain with ConA, indicating lack of carbohydrate content.
3.1.2. By Tunicamycin Treatment
Glycosylation was further evaluated with tunicamycin antibiotic, derived from the bacterium Streptomyces Iysosuperficus. Tunicamycin inhibits N-glycosylation by blocking the addition of N-acetylglucosamine to dolichol phosphate, the first step in the formation of the core oligosaccharide in first steps of N-linked glycoprotein synthesis. Protein extracts from M. smegmatis with recombinant 19 kDa proteins were harvested in the presence of tunicamycin, and evaluated following immunoblotting against M. smegmatis adsorbed rabbit anti-M. paratuberculosis sera. Recombinant protein extracts from M. smegmatis treated with tunicamycin at 0.25, 0.50, 1, 2 μg/mL, respectively expressed a protein product at 19 kDa, and had no band shift to a lower molecular weight. M. tuberculosis was used as a positive control, showing a 19 kDa protein that reacted with antibody. E. coli recombinant was included to show where the 16 kDa band is expected to be located. This experiment was repeated using higher tunicamycin concentrations. M. smegmatis recombinants treated at different tunicamycin concentrations, 2, 4, 6, 8, 10 μg/mL, respectively still expressed product at 19 kDa

Figure 1. Immunoblot analysis of 19 kDa protein. Protein extracts of M. tuberculosis, M. Smegmatis recombinants, E. coli recombinants were loaded on SDS-PAGE and immunoblotted against M. smegmatis adsorbed rabbit anti-M. paratuberculosis sera. Lanes: 1, protein size standard, Lane 2, extracts from M. paratuberculosis, Lane 3, extracts from M. tuberculosis, Lane 4, extracts from M. Smegmatis/shuttle, Lane 5, extracts from M. Smegmatis/Shuttle 19, Lane 6, Empty, Lane 7, extracts from E. coli pcDNAII, Lane 8, Empty, Lane 9, extracts from E. coli/pMptb#28, Lane 10, protein size standard.
and similar amount of protein present. M. smegmatis recombinant samples treated with tunicamycin at a concentration of 2, 6, 10 μg/mL were stained with conA-peroxidase conjugate (Figure 3, lanes 5 - 7) and showed binding to conA at 19 kDa molecular weight. M. tuberculosis (Figure 3, lane 2) and M. smegmatis recombinant without tunicamycin treatment (Figure 3, lane 4) were used as positive controls to show binding at 19 kDa. Again, M. smegmatis showed no binding to conA at 19 kDa (Figure 3, lane 3).
3.1.2. By Enzymatic Deglycosylation
This enzymatic method removes carbohydrate/s from protein without protein degradation. If deglycosylation occurred, a mobility shift on an SDS-PAGE gel would be observed. E-DEGLY kit from Sigma Aldrich contains all enzymes (PNGase F, Neuraminidase, O-GlycosidaseGlycosidase, N-Acetylglucosaminidase) required to completely remove all N-linked and O-linked carbohydrates from glycoprotein. In this experiment, Bovine fetuin was used as positive control, which contains N-linked and O-linked oligosaccharides. As shown in Figure 4, lanes 1 and 2 contain Bovine fetuin glycoprotein before and after deglycosylation, respectively. Clearly, there was a shift in the size of the protein before and following the deglycosylation treatment. This observation confirms the validity of the purchased kit to be used for evaluation of glycoproteins. When the same experiment was applied on protein extracts from the recombinant M. smegmatis expressing the 19 kDa protein, there was no shift in protein size (Figure 4, lanes 3 and 4). Protein extract from the recombinant M. smegmatis was evaluated for possible inhibitors (Figure 4, lane 5). In this experiment, protein extract from the recombinant M. smegmatis was
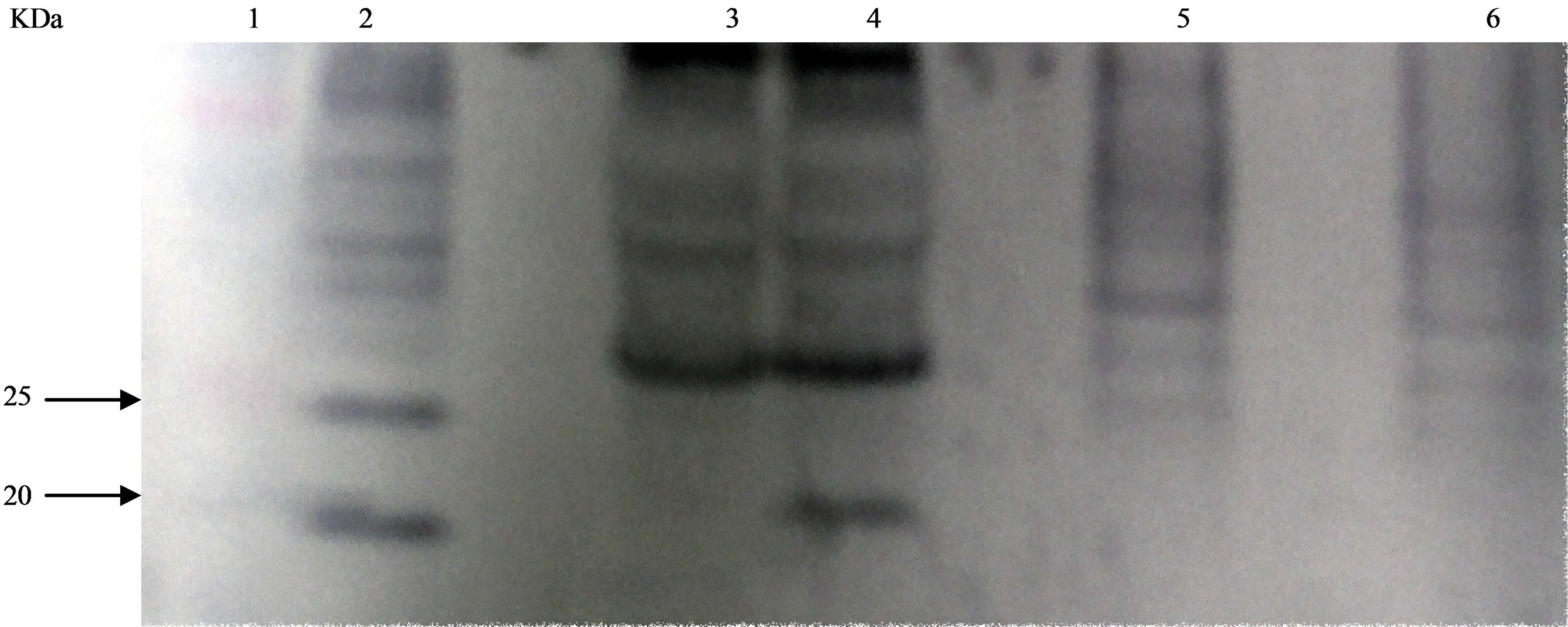
Figure 2. Evaluation of Smeg19K protein for Carbohydrate Content by ConA Staining. Protein extracts of M. tuberculosis, M. Smegmatis recombinants, E. coli recombinants were loaded on SDS-PAGE, transferred onto nitrocellulose membrane, and ConA stained. Lanes: 1, protein size standard, Lane 2, extracts from M. tuberculosis, Lane 3, extracts from M. Smegmatis/ shuttle, Lane 4, extracts from M. Smegmatis/Shuttle 19, Lane 5, extracts from E. coli pcDNAII, Lane 6, extracts from E. coli/ pMptb#28.
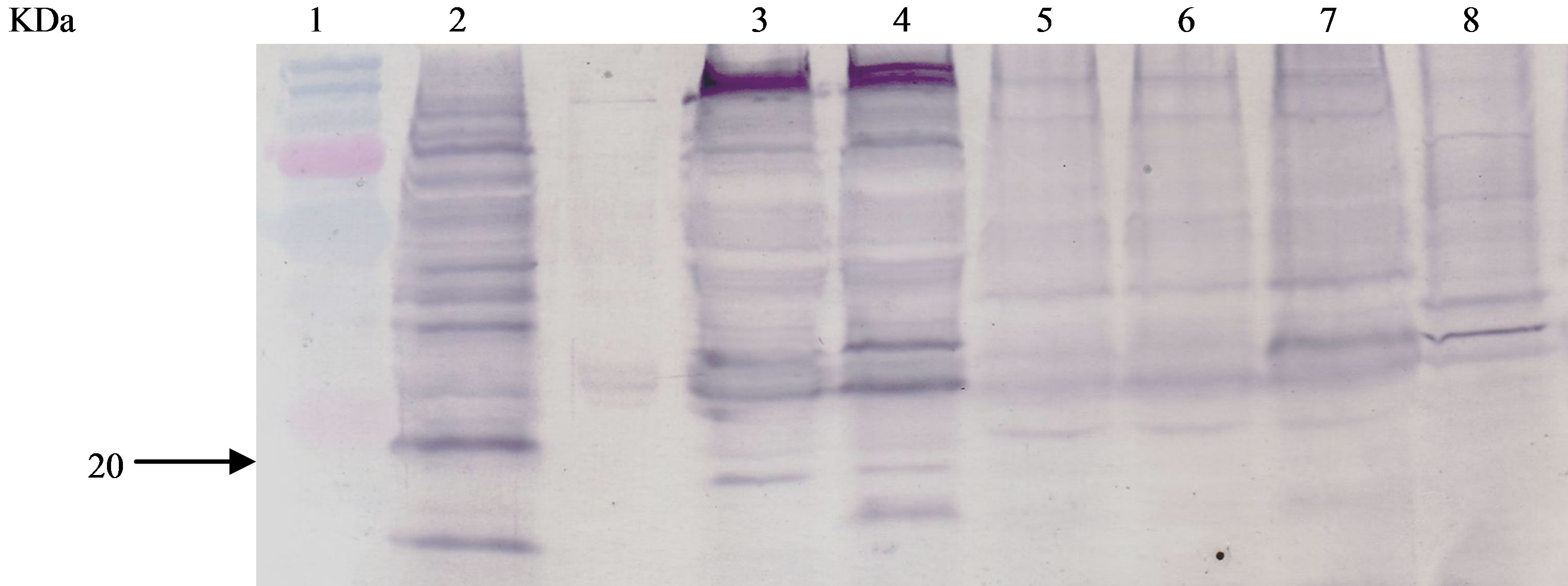
Figure 3. ConA of Tunicamycin-Treated Protein Extracts of M. smegmatis/Shuttle 19. Protein extracts of M. tuberculosis, tunicamycin-treated M. Smegmatis recombinants, M. smegmatis recombinants, E. coli recombinant were loaded on SDS-PAGE, transferred onto nitrocellulose membrane, and ConA stained. Lanes: 1, protein size standard, Lane 2, extracts from M. tuberculosis, Lane 3, extracts M. smegmatis/shuttle, Lane 4, extracts from M. smegmatis/shuttle 19, Lanes 5-7, extracts from M. smegmatis/Shuttle 19 treated at antibiotic concentrations of 2, 6, 10 μg/mL respectively, Lane 8, extracts from E. coli/pMptb- #28.
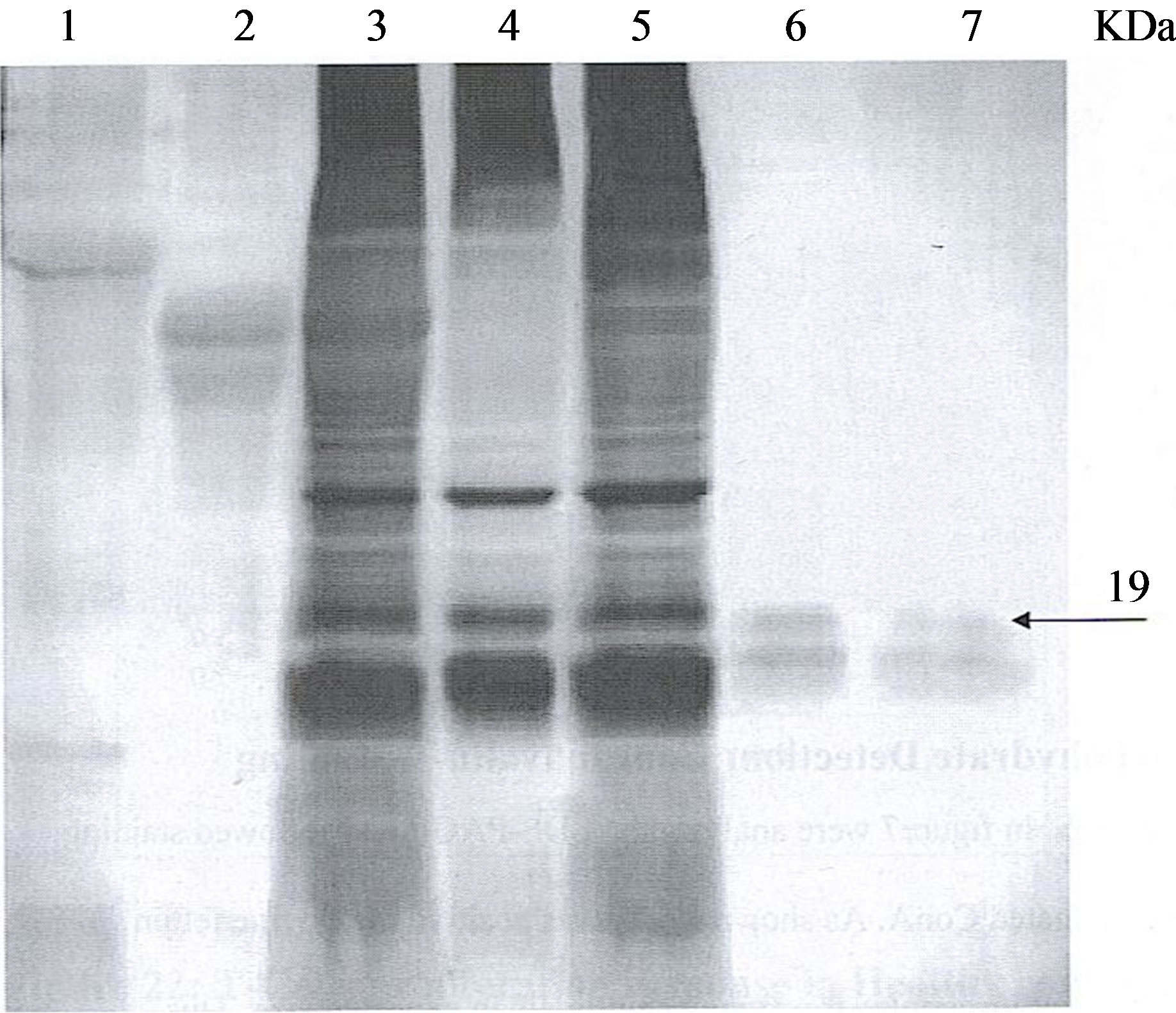
Figure 4. Evaluation of Smeg19K for Carbohydrate content. Silver Staining of samples before and after deglycosylation Bovine Fetuin as positive control and cell crude lysate from recombinant clone smeg/pShuttle 19 loaded on an SDS gel before and after applying deglygcosylation kit. Lanes: 1, 2 Bovine Fetuin before and after deglycosylation 3, 4 cell lysate from recombinant clone M. smegmatis/shuttle 19 before and after deglycosylation 5, spike (mix of positive control and cell lysate M. smeg/shuttle 19) before and after deglycosylation 6, 7, purified 19 kDa recombinant protein from smeg/shuttle 19 k (eluted from polyacrylamide gel) before and after deglycosylation.
spiked with Bovine fetuin glycoprotein control and then subject to enzymatic deglycosylation. The Fetuin glycolprotein has shifted in size as expected following the treatment illustrating absence of any inhibitors. The 19 kDa protein was not affected by the deglycosylation treatment (Figure 4, lane 5).
3.2. Evaluation of Smeg19K Protein for Lipid Content
Presence of lipid was assessed by staining with Sudan Black, a lipophilic dye, post-gel electrophoresis. As shown in Figure 5, M. smegmatis (lane 7) and M. smegmatis recombinant (lane 6) stained positively with Sudan black, indicating 19 kDa lipoprotein. However, level of expression of M. smegmatis recombinant is higher, as observed in a thicker band. Silver stain of samples showed similar amount of protein present for M. smegmatis and M. smegmatis recombinant (data not shown). E. coli recombinant and non-recombinant strains stained positively with Sudan black at 19 kDa. Immunoblot against M. smegmatis adsorbed rabbit anti-M. paratuberculosis sera confirmed this 19 kDa belongs to an endogenous E. coli protein, and not to E. coli recombinant of 16 kDa (data not shown). Immunoblot also showed M. smegmatis recombinant protein present at 19 kDa and no 19 kDa was recognized with antibody for M. smegmatis (data not
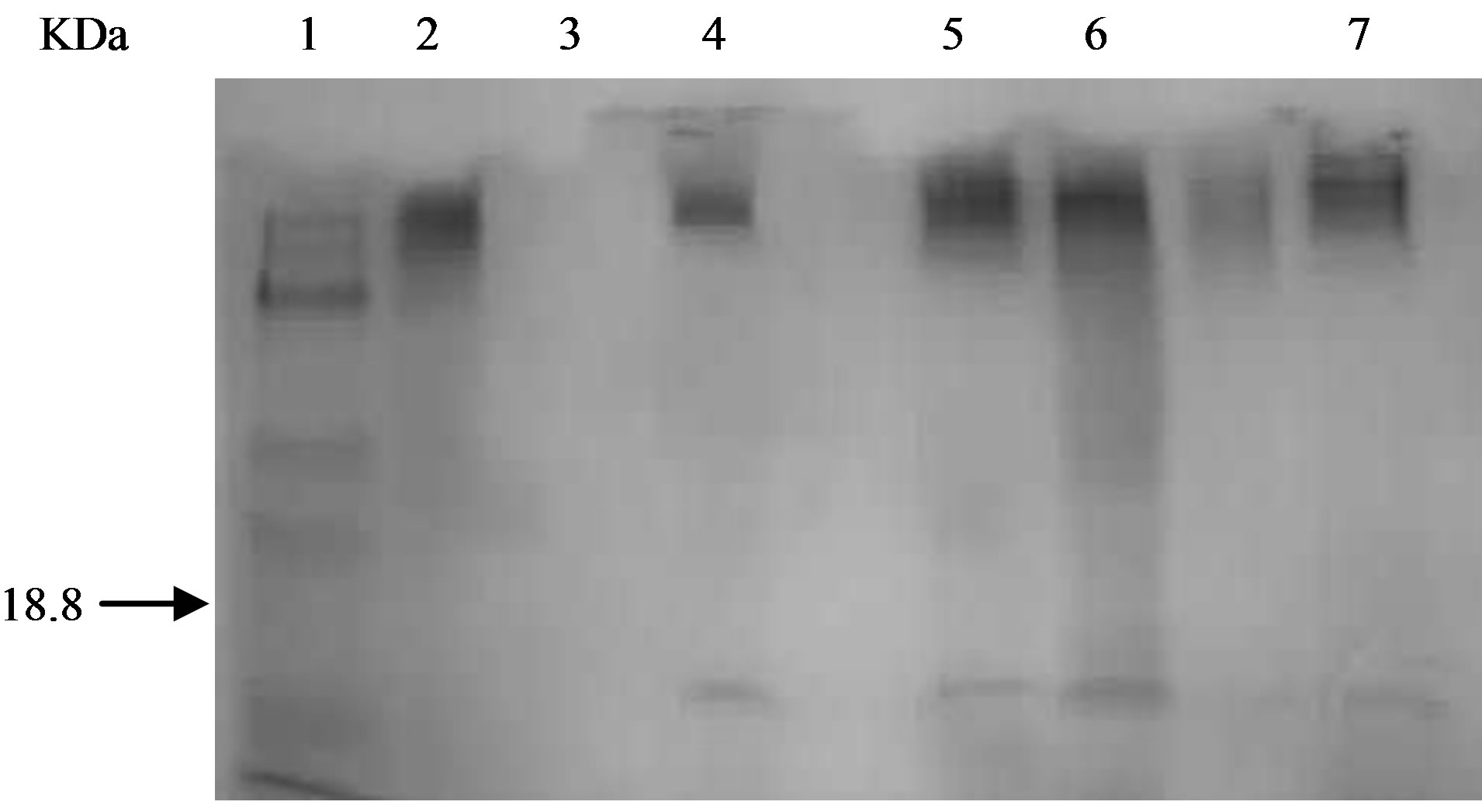
Figure 5. Evaluation of Smeg19K protein for lipid Content. A: by Sudan Black post SDS-PAGE. Protein extracts of M. tuberculosis, M. Smegmatis recombinants, E. coli recombinants were loaded on SDS-PAGE and stained with Sudan Black 4B to test for acylation. Lanes: 1, protein size standard, Lane 2, extracts from M. tuberculosis, Lane 3, extracts from M. paratuberculosis, Lane 4, extracts from E. coli pcDNAII, Lane 5, extracts from M. Smegmatis/Shuttle 19 + S, Lane 6, extracts from M. Smegmatis/shuttle 19, Lane 7, extracts from M. Smegmatis/Shuttle.
shown). M. tuberculosis 19 kDa was used as positive control for lipoprotein (Figure 5, lane 2).
In addition, M. smegmatis recombinant samples treated with tunicamycin at concentrations of 2, 4, 6, 8 μg/mL were stained with Sudan black post-electrophoresis (data not shown). These samples stained positive, indicating presence of lipid.
4. Discussion
Covalent modification in prokaryotic proteins is less common than in eukaryotic proteins but influences the overall structure thus affecting protein function. Numerous virulence factors of mycobacterial proteins have been found to be covalently modified [24]. Understanding the structure of proteins can allow for immunological characterization, which can further lead to vaccine or drug development.
The objective of this study was to characterize the 19 kDa M. paratuberculosis protein as glycosylated, acylated, or both. We chose to express the 19 kDa protein in a rapidly growing mycobacterial homologue host, M. Smegmatis, because M. paratuberculosis is a very fastidious grower and requires specific growth conditions.
BLAST analysis of amino acid sequence revealed approximately 76% homology in amino acid identity between M. tuberculosis and M. paratuberculosis’s 19 kDa protein [10]. Potential sites of glycosylation in M. paratuberculosis included a set of serine residues, found in same region, where M. tuberculosis 19 kDa protein has threonine residues implicated in O-glycosylation [8]. Unfortunately, there is no sequence motif indicative of possible O-glycosylation, as it has been found for N-glycosylation in eukaryotic and some prokaryotic proteins. This consensus motif includes Ser/Thr-Xaa-Asn, in which Xaa, with exception of proline, can be any amino acid and it has been found in M. tuberculosis and Clostridium thermocellum [25]. In addition to threonine and serine clusters involved in carbohydrate attachment, adjacent proline and alanine residues can have local conformational effects in O-glycosylation [26-29].
The expression of the recombinant 19 kDa antigen of M. paratuberculosis in M. smegmatis, is O-glycosylated, as evidenced by ConA binding (Figure 2). M. tuberculosis known 19 kDa glycolipoprotein [11-13] was used as positive control. The 19 kDa recombinant protein expressed in E. coli system showed no evidence of glycosylation; similar to what was observed when the recombinant 19 kDa protein of M. tuberculosis was cloned in E. coli [7]. Since lectin conA is highly specific for α-mannose residues [8], mannose could be attached to serine residues through an O-glycosidic bond; similar to mannose residues attached to threonine residues in 19 kDa of M. tuberculosis [8]. To confirm the presence of 19 kDa M. paratuberculosis recombinant protein, samples were immunoblotted against M. smegmatis adsorbed rabbit anti-M. paratuberculosis sera. Results showed the presence of 19 kDa protein from M. tuberculosis and 19 kDa recombinant protein in M. smegmatis. No binding between antibody and M. smegmatis was observed at 19 kDa. As expected, the E. coli recombinant protein bound antibody at 16 kDa while E. coli had no binding at 16 kDa.
Treatment of M. smegmatis recombinant proteins with tunicamycin supports that 19 kDa M. paratuberculosis is O-glycosylated, as evidence by conA staining. M. smegmatis recombinant proteins showed no band shift from 19 kDa to a lower molecular weight (Figure 3) when subjected to different concentration of tunicamycin (0.25, 0.50, 1, and 2 μg/mL) [30]. The experiment was repeated at higher concentrations up to 10 ug/mL to rule out the possibility the resistance to lower antibiotic concentrations. Results showed no band shift at 19 kDa for the treated M. smegmatis recombinants. Additionally, high concentrations of antibiotic affected cell metabolism, as observed with decreased thickness of bands. Silver stain of SDS-gels containing these samples, showed similar amount of protein content present for all of the treated M. smegmatis recombinants. Furthermore, M. smegmatis recombinant samples treated with tunicamycin, at a concentration of 2, 6, 10 μg/mL, were stained with conAperoxidase conjugate and showed binding to conA at 19 kDa molecular weight (Figure 3). M. tuberculosis and M. smegmatis recombinant without tunicamycin treatment were used as positive controls to show binding to conA of 19 kDa protein. Again, M. smegmatis showed no binding to conA at 19 kDa.
Previously in our lab, an enzymatic deglycosylation kit was performed on the M. smegmatis recombinant protein. E-DEGLY kit (Sigma Aldrich) used contained all enzymes (PNGase F, Neuraminidase, O-Glycosidase, Glycosidase, N-Acetylglucosaminidase) required to completely remove all N-linked and O-linked carbohydrates from glycoprotein. If deglycosylation occurred, a mobility shift on an SDS-PAGE gel would be observed. After deglycosylation treatment, a lower molecular weight in the recombinant M. smegmatis 19 kDa protein was not observed (Figure 4). Mobility shift was observed on positive control, Bovine fetuin. Results showed Bovine fetuin had a lower molecular weight, but no band shift was detected for recombinant protein. A plausible reason may be that, mannose directly O-linked to proteins cannot be removed enzymatically [6]. Mannose is the sugar O-glycosylated in the 19 kDa lipoprotein of M. tuberculosis [8]. Thus, tunicamycin treatment and enzymatic deglycosylation kit suggested O-glycosylation with man-nose sugar directly linked to protein, further confirming results obtained from conA binding.
The second objective of this study was to characterize if acylation occurred. Comparison of amino acid sequence between 19 kDa of M. paratuberculosis and M. tuberculosis, showed conservation of a cysteine residue at amino acid 22 across both species. Presence of highly conserved six-residue consensus sequences for acylation in M. tuberculosis, 19-LSGCSS-24, and in M. avium subsp. Intracellulare, 19-ISGSG-24, located adjacent to hydrophobic signal peptide, and biochemical evidence [29,31], suggest that the 19 kDa protein from M. paratuberculosis, 19-VSGCSS-24, undergoes acylation and subsequent cleavage of signal peptide prior to secretion of the mature lipoprotein across cell membrane. Confirmation that 19 kDa M. paratuberculosis is lipoprotein was defined by staining with Sudan black dye post-gel electrophoresis (Figure 5). Both, M. smegmatis and M. smegmatis recombinant, stained positive with lipophilic dye. However, level of expression of M. smegmatis recombinant is higher, as observed in a thicker band. Silver stain of samples, showed similar amount of protein present for both protein extracts. BLAST analysis comparing 19 kDa sequence of M. paratuberculosis to M. smegmatis resulted in 48% homology in amino acid identity and discovery that M. smegmatis has endogenous lipoprotein. E. coli recombinant and E. coli stained positively with Sudan black at 19 kDa weight. Immunoblot against M. smegmatis adsorbed rabbit anti-M. paratuberculosis sera confirmed this 19 kDa band belongs to an endogenous E. coli protein, and not to E. coli recombinant of 16 kDa (data not shown). Immunoblot also showed M. smegmatis recombinant protein present at 19 kDa and no 19 kDa protein was recognized with antibody for M. smegmatis. M. tuberculosis 19 kDa was used as positive control for lipoprotein.
In this study, the data illustrates that the 19 kDa M. paratuberculosis is lipoglycosylated. Confirmation that 19 kDa protein is glycosylated will require structural evidence of covalent sugar-protein linkage. Future studies on the 19 kDa protein of M. paratuberculosis should exploit the availability of rapidly growing homologue mycobacterial hosts to study biological function and immunological significance. Furthermore, to confirm if the serine residues of the amino acid sequence play a role in O-glycosylation, site-directed mutagenesis should be performed. Substitution of serine residues with nonpolar amino acid residues such as valine or alanine, which cannot support glycosylation, will show which serine residues are involved with ConA binding. Herrmann et al., in 1996, studied the 19 kDa protein expressed by M. tuberculosis. Site-directed mutagenesis substituted suspected threonine residues near N-terminus with valine residues, and showed that these amino acid residues played a role in conA binding [8]. Since the recombinant 19 kDa antigen from M. tuberculosis is expressed at 16 kDa in E. coli in comparison to M. smegmatis recombinant [7]; threonine substitutions had a very slight effect on molecular size of 16 kDa in E. coli [8]. Also, these recombinants showed no detectable conA binding [8]. To study acylation, another homologue host without endogenous 19 kDa should be used.
The importance of this study was to characterize the possible posttranslational modification of the 19 kDa MAP protein which can play a role in its antigenicity. Studies on M. tuberculosis 19 kDa glycolipoprotein shows this antigen stimulates both a T and B cell response as well as induces a number of TH1 cytokines. It has been shown to be immunodominant in both mice [12, 32,33], and in humans [34,35]. It stimulates a CD4+ T cell proliferation and the release of IL-2, IFN-Y, and IL-12 [11,36]. Glycosylation modification inhibits innate immune response, such as the release of TNF-α, IL-6, and IL-10 from macrophages, while not inhibiting antibody binding [7,19,23]. More importantly, experimental infection and staining of macrophages have shown that this 19 kDa protein is secreted by live M. tuberculosis residing within phagolysosomal compartment [37-40].
5. Conclusion
Collectively, our study clearly illustrates for the first time that M. paratuberculosis 19 kDa expressed protein is experimentally confirmed to be a lipoglycosylated protein. Furthermore, this posttranslational modification includes O-glycosylation and mannose as possible sugar subunit. The data confirm observation reported in 19 kDa from M. tuberculosis. The significance of this study may aid in investigating the pathogenicity of M. paratuberculosis infection in animals and humans.
6. Acknowledgements
This study was supported, in part, by a grant from the Eli Broad Foundation.
REFERENCES
- C. M. G. Ranes, J. Rauzier, M. Lagraderie, M. Gheorghiu and B. Gicquel, “Functional Analysis of pAL5000, a Plasmid from Mycobacterium Fortuitum: Construction of a “Mini” Mycobacterium-Escherichia coli Shuttle Vector,” Journal of Bacteriology, Vol. 172, No. 5, 1990, pp. 2793- 2797.
- J. O. Falkinham, “Epidemiology of Infection by Nontuberculosis Mycobacteria,” Clinical Microbiology Reviews, Vol. 9, No. 2, 1996, pp. 177-215.
- C. B. Inderleid, C. A. Kemper and L. E. M. Bermudez, “The Mycobacterium Avium Complex,” Clinical Microbiology Reviews, Vol. 6, No. 3, 1993, pp. 266-310.
- E. Mahenthiralingam, B. I. Marklund, L. A. Brooks, D. A. Smith, G. L. Bancroft and R. W. Stokes, “Site-Directed Mutagenesis of the 19-Kilodalton Lipoprotein Antigen Reveals No Essential Role for the Protein in the Growth and Virulence of Mycobacterium Intracellulare,” Infection and Immunity, Vol. 66, No. 8, 1998, pp. 3626-3634.
- J. P. Bannantine, R. G. Barletta, J. R. Stabel, M. L. Paustian and V. Kapur, “Application of the Genome Sequence to Address Concerns That Mycobacterium Avium Subspecies Paratuberculosis Might Be a Foodborne Pathogen,” Foodborne Pathogens and Disease, Vol. 1, No. 1, 2004, pp. 3-15. http://dx.doi.org/10.1089/153531404772914419
- A. Chiba, et al., “Structures of Sialylated O-Linked Oligosaccharides of Bovine Peripheral Nerve α-Dystroglycan. The Role of Novel O-Mannosyl-Type Oligosaccharide in the Birding of α-Dystroglycan with Laminin,” The Journal of Biological Chemistry, Vol. 272, No. 4, 1997, pp. 2156-2162. http://dx.doi.org/10.1074/jbc.272.4.2156
- T. Garbe, D. Harris, M. Vordermeier, R. Lathigra, J. Ivanyi and D. Young, “Expression of the Mycobacterium Tuberculosis 19-Kilodalton Antigen in Mycobacterium Smegmatis: Immunological Analysis and Evidence of Glycosylation,” Infection and Immunity, Vol. 61, No. 1, 1993, pp. 260-267.
- J. L. Herrmann, P. O’Gaora, A. Gallagher, J. E. R. Thole and D. Young, “Bacterial Glycoproteins: A Link between Glycosylation and Proteolytic Cleavage of a 19 kDa Antigen from Mycobacterium Tuberculosis,” EMBO Journal, Vol. 15, No. 14, 1996, pp. 3547-3554.
- D. L. Nelson and M. M. Cox, “Lehninger Principles of Biochemistry,” 4th Edition, United States of America, Freeman, 2005, p. 256.
- J. Huntley, J. R. Stabel and J. P. Bannantine, “Immunoreactivity of the Mycobacterium Avium subsp. Paratuberculosis 19-kDa Lipoprotein,” BMC Microbiology, Vol. 5, No. 1, 2005, p. 3. http://dx.doi.org/10.1186/1471-2180-5-3
- H. D. Brightbill, D. H. Libraty, S. R. Krutzik, R. B. Yang, J. T. Belisle, J. R. Bleharski, M. Maitland, M. V. Norgard, S. E. Plevy, S. T. Smale, P. J. Brennan, B. R. Bloom, P. J. Godowski and R. L. Modlin, “Host Defense Mechanisms Triggered by Microbial Lipoproteins through Toll-Like Receptors,” Science, Vol. 285, No. 5428, 1999, pp. 732- 736. http://dx.doi.org/10.1126/science.285.5428.732
- D. P. Harris, H. M. Vordermeier, S. J. Brett, G. Pasvol, C. Moreno and J. Ivanyi, “Epitope Specificity and Isoforms of the Mycobacterial 19-Kilodalton Antigen,” Infection and Immunity, Vol. 62, No. 7, 1994, pp. 2963-2972.
- D. L. Nelson and M. M. Cox, “Lehninger Principles of Biochemistry,” 4th Edition, United States of America, Freeman, 2005, p. 262.
- T. M. Bricker, M. J. Boyer, J. Keith, R. Watson-McKown and K. S. Wise, “Association of Lipids with Integral Membrane Surface Proteins of Mycoplasma Hyorhinis,” Infection and Immunity, Vol. 56, No. 2, 1988, pp. 295-301.
- WebMD, “Definition of lipoprotein,” 2008. http://www.webmd.com/cholesterol-management/tc/lipid-panel-topic-overview
- N. R. Chamberlain, M. E. Brandt, A. L. Erwin, J. D. Radolf and M. V. Norgard, “Major Integral Membrane Protein Immunogens of Treponema Pall’Dum Are Proteolipids,” Infection and Immunity, Vol. 57, No. 9, 1989, pp. 2872-2877.
- H. C. Wu and M. Tokunaga, “Biogenesis of Lipoproteins in Bacteria,” Current Topics in Microbiology and Immunology, Vol. 125, No. 1, 1986, pp. 127-157. http://dx.doi.org/10.1007/978-3-642-71251-7_9
- C. Espitia and R. Mancilla, “Identification, Isolation, and Partial Characterization of Mycobacterium Tuberculosis 50- to 55-Kilodalton and Mycobacterium Bovis BCG 45- to 47-Kilodalton Antigens,” Infection and Immunity, Vol. 63, No. 3, 1989, pp. 580-584.
- T. Fifis, C. Costopoulos, A. J. Radford, A. Bacic and P. R. Wood, “Purification and Characterization of Major Antigens from a Mycobacterium Bovis Culture Filtrate,” Infection and Immunity, Vol. 59, No. 3, 1991, pp. 800-807.
- F. A. K. El-Zaatari, S. A. Naser, L. Engstrand, C. Y. Hachem and D. Y. Graham, “Identification and Characterization of Mycobacterium Paratuberculosis Recombinant Proteins Expressed in E. coli,” Current Microbiology, Vol. 29, No. 3, 1994, pp. 177-184. http://dx.doi.org/10.1007/BF01570760
- U. K. Laemmli, “Cleavage of Structural Proteins during the Assembly of the Head Bacteriophage T4,” Nature (London), Vol. 45, No. 5259, 1970, pp. 680-685. http://dx.doi.org/10.1038/227680a0
- J. H. Morrisey, “Silver Stain for Proteins in Polyacrylamide Gels: A Modified Procedure with Enhanced Uniform Sensitivity,” Analytical Biochemistry, Vol. 117, No. 2, 1981, pp. 307-310. http://dx.doi.org/10.1016/0003-2697(81)90783-1
- A. T. Andrews, “Electrophoresis: Theory, Techniques, Biochemical and Clinical Applications,” 2nd Edition, Oxford Science Publications, New York, 1981, p. 37
- S. M. Kamper and A. F. Barbet, “Surface Epitope Variation via Mosaic Gene Formation Is Potential Key to Long Term Survival of Trypanosoma Brucei,” Molecular and Biochemical Parasitology, Vol. 53, No. 1-2, 1992, pp. 33- 44. http://dx.doi.org/10.1016/0166-6851(92)90004-4
- S. Moens and J. Vanderleyen, “Glycoproteins in Prokaryotes,” Archieves of Microbiology, Vol. 168, No. 3, 1997, pp. 69-175. http://dx.doi.org/10.1007/s002030050484
- A. A. Gooley, B. J. Classon, R. Marschalek and K. L Williams, “Glycosylation Sites Identified by Detection of Glycosylated Amino Acids Released from Edman Degradation: The Identification of Xaa-Proo-Xaa-Xaa as a Motif for Thr-O Glycosylation. Biochem. Biophys,” Research Communications, Vol. 178, No. 3, 1991, pp. 1994-1201. http://dx.doi.org/10.1016/0006-291X(91)91019-9
- B. C. O’Connel, L. A. Tabak and N. Ramasubbu, “The Influence of Flanking Sequences on O-Glycosylation. Biochem. Biophys,” Research Communications, Vol. 180, No. 2, 1991, pp. 1024-1030. http://dx.doi.org/10.1016/S0006-291X(05)81168-4
- B. C. O’Connel, F. K. Hagan and L. A. Tabak, “The Influence of Flanking Sequence on the O-Glycosylation of Threonine in Vitro,” Journal of Biological Chemistry, Vol. 267, No. 35, 1992, pp. 25010-25018.
- I. B. H. Wilson, Y. Gavel and G. von Heijne, “Amino Acids Distribution around O-Linked Glycosylation Sites,” Biochemistry Journal, Vol. 275, No. 2, 1991, pp. 529- 534.
- N. Jun-ichi, M. Akiyoshi, S. Hirata and T. Fukuda, “Saccharomyces Cerevisiae IRE2/HAC1 Is Involved in IRE1- Mediated KAR2 Expression,” Oxford University Press, Oxford, Vol. 24, No. 21, 1996, pp. 4222-4226.
- P. S. Jackett, G. Bothamley, H. V. Batra, A. Mistry, D. B. Young and J. Inayi, “Specificity of Antibodies to Immunodominant Mycobacterial Antigens in Pulmonary Tuberculosis,” Journal of Clinical Microbiology, Vol. 26, No. 11, 1998, pp. 2313-2318.
- K. J. Erb, J. Kirman, L. Woodfield, T. Wilson, D. M. Collins, J. D. Watson and G. LeGros, “Identification of Potential CD8+ T-Cell Epitopes of the 19 kDa and AhpC Proteins from Mycobacterium Tuberculosis. No Evidence for CD8+ T-Cell Priming against the Identified Peptides after DNA-Vaccination of Mice,” Vaccine, Vol. 16, No. 7, 1998, pp. 692-697. http://dx.doi.org/10.1016/S0264-410X(97)00253-3
- D. P. Fonseca, D. Joosten, H. Snippe and A. F. Verheul, “Evaluation of T-Cell Responses to Peptides and Lipopeptides with MHC Class I Binding Motifs Derived from the Amino Acid Sequence of the 19-kDa Lipoprotein of Mycobacterium Tuberculosis,” Molecular Immunology, Vol. 37, No. 8, 2000, pp. 413-422. http://dx.doi.org/10.1016/S0161-5890(00)00066-3
- D. P. Harris, H. M. Vordermeier, G. Friscia, E. Roman, H. M. Surcel, G. Pasvol, C. Moreno and J. Ivanyi, “Genetically Permissive Recognition of Adjacent Epitopes from the 19-kDa Antigen of Mycobacterium Tuberculosis by Human and Murine T Cells,” Journal of Immunology, Vol. 150, No. 11, 1993, pp. 5041-5050.
- H. Hohn, C. Kortsik, K. Nilges, A. Necker, K. Freitag, G. Tully, C. Neukirch and M. J. Maeurer, “Human Leucocyte Antigen-A2 Restricted and Mycobacterium Tuberculosis 19-kDa Antigen-Specific CD8+ T-Cell Responses Are Oligoclonal and Exhibit a T-Cell Cytotoxic Type 2 Response Cytokine-Secretion Pattern,” Immunology, Vol. 104, No. 3, 2001, pp. 278-288. http://dx.doi.org/10.1046/j.1365-2567.2001.01307.x
- W. H. Boom, R. N. Husson, R. A. Young, J. R. David and W. F. Piessens, “In Vivo and in Vitro Characterization of Murine T-Cell Clones Reactive to Mycobacterium Tuberculosis,” Infection and Immunity, Vol. 55, No. 9, 1987, pp. 2223-2229.
- O. Neyrolles, K. Gould, M. P. Gares, S. Brett, R. Janssen, P. O’Gaora, J. L. Herrmann, M. C. Prevost, E, Perret, J. E. Thole and D. Young, “Lipoprotein Access to MHC Class I Presentation during Infection of Murine Macrophages with Live Mycobacteria,” Journal of Immunology, Vol. 166, No. 1, 2001, pp. 447-457.
- G. S. Getz, P. A. Vanderlaan and C. A. Reardon, “Natural Killer T Cells in Lipoprotein Metabolism and Atherosclerosis,” Thrombosis and Haemostasis, Vol. 106, No. 5, 2011, pp. 814-819. http://dx.doi.org/10.1160/TH11-05-0336
- Y. Zheng, M. T. Stephan, S. A. Gai, W. Abraham, A. Shearer and D. J. Irvine, “In Vivo Targeting of Adoptively Transferred T-Cells with Antibodyand Cytokine-Conjugated Liposomes,” Journal of Controlled Release: Official Journal of the Controlled Release Society, 2013.
- J. S. Chauhan, A. H. Bhat, G. P. Raghava and A. Rao, “GlycoPP: A Webserver for Prediction of Nand O-Glycosites in Prokaryotic Protein Sequences,” PloS One, Vol. 7, No. 7, 2012, Article ID: e40155. http://dx.doi.org/10.1371/journal.pone.0040155
NOTES
*Corresponding author.

