Open Journal of Medical Microbiology
Vol.05 No.03(2015), Article ID:59605,9 pages
10.4236/ojmm.2015.53015
Quantitative Detection of Helicobacter pylori by Real Time PCR in Drinking Water―Environmental and Public Health Risk Significance
Virginia Montero-Campos1, Shirley Arias-Cordero2, Benedicto Valdés-Rodríguez3, Monserrat Jarquín-Cordero2
1Research & Chemical and Microbiology Services Center―CEQIATEC, School of Chemistry, Instituto Tecnológico de Costa Rica, Cartago, Costa Rica
2Biotechnology Research Center, School of Biology, Instituto Tecnológico de Costa Rica, Cartago, Costa Rica
3Water Laboratory Services Physicochemical, University of Chiriqui, Chiriqui City, Panama
Email: vmontero@itcr.ac.cr, vmonterocampos@gmail.com
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 3 July 2015; accepted 12 September 2015; published 15 September 2015
ABSTRACT
Helicobacter pylori (H. pylori) is bacteria considered to be present in half of the population and it is a public health problem worldwide. Most patients infected with H. pylori show no clinical symptoms; nonetheless, approximately 10% to 20% of these patients will develop peptic ulcers and 1% will develop gastric cancer. The International Agency for Research on Cancer has classified H. pylori as a Group 1 carcinogen, recognized as the only bacteria capable of producing cancer. Samples of drinking water (n = 44) from aqueducts with chlorination treatment in selected areas with high prevalence of gastric cancer were analyzed in Costa Rica. Samples of drinking water from Panamá (n = 44) from aqueducts supplying untreated water for human consumption in the province of Chiriquí were also analyzed. The molecular marker of H. pylori, glmM, was used, and to optimize the Real Time PCR (qPCR) technique, annealing temperature, concentration of primers and probe were standardized; also, by analyzing different standard curves, the best reaction conditions that allowed detecting and quantifying the gene were determined. The LightCycler® 480 II (LC480II) equipment from Roche Diagnostics GmbH was used, as well as the Absolute Quantification Analysis by means of the Second Derivative Maximum Method. In the case of the samples from Costa Rica, it was determined that 79.5% were positive for H. pylori; removing outlier high average, quantification of bacteria was determined in 3.6 × 103 copies/100 mL. For Panamá it was determined that 86% of the samples were found positive for the presence of H. pylori; removing outlier high average quantification of bacteria was determined at 3.3 × 102 copies/100 mL. The difference in values between the aqueducts in both countries revealed an environmental distribution of the bacteria of epidemiological interest in each case.
Keywords:
Helicobacter pylori, Drinking Water, Real Time PCR (qPCR), Chlorination Treatment

1. Introduction
1.1. Basic Concepts
H. pylori was discovered in 1982 by doctors Barry Marshall and Robin Warren from Perth, Australia, who observed the bacterium for the first time in stomach biopsies from a patient with active chronic gastritis. In recognition for their discovery, they were awarded the 2005 Nobel Prize in Medicine and Physiology. Since 1994 and to this date, the International Agency for Research on Cancer (IARC) of the World Health Organization has declared H. pylori as a carcinogenic in humans, classified in group I (there exists enough evidence that confirms it can cause cancer in human beings); being the only bacterium capable of causing this pathology amongst all groups of toxic substances.
Between individuals, the majority of transmission routes include the gastro-oral, oral-oral and fecal-oral [1] . In recent years, other routes have been suggested for viable transmission of the bacterium, such as water and foodstuffs. Authors agree that the relative importance of these transmission routes varies among developed and developing countries [2] [3] .
The bacterium can be acquired during childhood and, by the age of 10 years, more than 50% of the children around the world have been in contact with the microorganism [4] . Nevertheless, some studies have suggested that transitory, and even very common infections, can be produced [5] [6] .
Most patients infected with H. pylori show no clinical symptoms; across the world it is considered as the main cause of chronic gastritis and peptic ulcers, also as the main risk factor for development of gastric cancer, including gastric atrophy, intestinal metaplasia and gastric cancer as B cell MALT lymphoma [7] , as well as other extragastric pathologies: cardiovascular diseases [8] [9] , hypertension [10] and hematological disorders [11] . Moreover, an association has been observed between chronic infection by H. pylori and resistance to insulin [12] [13] ; attention has been drawn upon its presence as a possible risk cofactor in hepatocellular carcinoma [14] and there is known association between infection by H. pylori and cirrhosis in patients with hepatitis C virus [15] .
1.2. Effects of Persistence in the Stomach
The strongest risk factor for gastric cancer is H. pylori-associated chronic gastric inflammation [16] . Indeed, epidemiological studies have determined that approximately 75% of the risk for gastric cancer is attributable to infection by H. pylori [17] . Stomach cancer is the second leading cause of cancer death in both sexes worldwide (738,000 deaths, 9.7% of the total) making it currently the fourth most common malignancy in the world, behind cancers of the lung, breast and colo-rectum. More than 70% of cases (714,000 cases) occur in developing countries (467,000 in men, 247,000 in women). Estimations locate the highest mortality rates in Eastern Asia; high mortality rates are also present in both sexes in Central America [18] .
The human stomach is considered to be exclusively inhabited by H. pylori; furthermore, it lacks colonizing non-H. pylori bacterial flora, due to the acidic environment. However, a limited number of studies using molecular-based methods have recently provided a broader picture of the stomach microbiota [19] .
Persistent infection of the gastric mucosa by Helicobacter pylori can initiate an inflammatory cascade that progresses into atrophic gastritis, a condition associated with reduced capacity for secretion of gastric acid and an increased risk of developing gastric cancer. The role of H. pylori as an initiator of inflammation is evident but the mechanism for development into gastric cancer has not been proven yet. A reduced capacity for gastric acid secretion allows survival and proliferation of other microbes that are normally killed by the acidic environment. It has been postulated that some of these species may be involved in the development of gastric cancer [20] .
Recent advances in DNA sequencing technology have uncovered a complex community of non-cultivatable inhabitants of the human stomach. The interaction between these inhabitants―collectively referred to as the gastric microbiota―and H. pylori, likely impacts gastric immunobiology and possibly the sequelae of H. pylori infection. Thus, characterization of the gastric microbiota in subjects with and without H. pylori infection could provide new insight into gastric homeostasis and the pathogenesis of H. pylori-associated disease, including gastric cancer [16] . Key Message: Though H. pylori infection and the non-H. pylori microbiota play a role in gastric cancer, the properties of gastric microbiota and mechanisms by which they participate in the genesis of gastric cancer are still not clearly depicted. Moreover, how the presence of microbiota along with H. pylori infection affects the progress from gastric disease to cancer still remains to be understood [21] .
1.3. Genetic Diversity and Molecular Detection
H. pylori present a significant genetic diversity, showing variation in the genetic order from one strain to another and in the diversity of sequences within some conserved genes. This bacterium exhibits a recombinant or panmictic population structure, which indicates frequent genetic exchange among strains [22] . Between its genes 16S and 23S, there exists a extremely variable space both in sequence as well as in extension, the 16S sequences being physically removable and positionable in other places of the genome, which generates an RNAr operon of atypical structure. Specific study of this variable ribosomal region is difficult since it generates a high genetic variability within the whole Helicobacter genus. Dewhirst mentions that these modifications in the 16S RNAr sequence are due to a horizontal exchange of genes [23] . Shahamat et al. demonstrated that gene glmM was the major target for detection of H. pylori for PCR amplification [24] , and it is also considered to form part of the housekeeping genes [25] [26] ; for these reasons, the use of genes other than 16S is recommended for phylogenetic analysis and identification within the Helicobacter genus, for example RNAr 23S genes and glmM [23] .
1.4. Presence in the Environment
It is known that H. pylori can operate in the environment under a biofilm structure. It is also known nowadays that, depending on the conditions, a bacterium can grow sessile adhered to the surface or develop in a planktonic manner, swimming freely in the liquid medium. In planktonic biofilms, bacteria can be found in a reversible “viable but not cultivable” (VBNC) state and this is considered the main reason for the low rate of microorganism detection in biofilms by routine means of cultivation [27] . This ability to aggregate in biofilms is shown by H. pylori. It has been demonstrated that sessile bacteria generally conserve the capacity to retain their spiral form for longer in comparison with planktonic forms, which transform into coccoid sooner (VBNC) [28] . Furthermore, it has been demonstrated that coccoid forms of H. pylori are capable of colonizing gastric mucosa and cause gastritis in mice [29] [30] .
Currently, some researches worldwide have quantified the presence of H. pylori naturally in the environment, with diverse water sources being important reservoirs, such as rivers, lakes, drinking water, municipal and residual waters, among others [21] . Quantification has been carried out by means of the Real Time PCR, using the cag E gene as a reference [31] . The bacterium has also been recently found in water with residual chlorine intended for human consumption [32] . An investigation determined the presence of H. pylori in soil samples from public playing areas of Spanish parks, with contamination levels ranging from 104 cells/g to 107 cells/g [33] .
1.5. Real Time PCR Technique Analysis
Real Time PCR is a variant of the conventional technique that combines amplification and detection of PCR products in one step by correlating the product in each one of the cycles with a fluorescence intensity signal. It allows for high specificity and ample range of detection as it is capable of quantifying and detecting concentrations of nucleic acids which could not be studied otherwise. It also makes it possible to obtain results faster as it requires no posterior electrophoreses; captures fluorescence in each cycle and generates amplification curves through the software of the equipment. These amplification curves possess four regions: base fluorescence (baseline or background), the exponential or logarithmic region, the linear phase and the termination or plateau. At the beginning of the logarithmic phase there is a cycle in which fluorescence stops being basal and becomes greater. This cycle is known as Cycle threshold (Ct) or Crossing point (Cp).
Fluorescence is produced by means of a hydrolysis probe, which consists of oligonucleotides specific to a sequence in a determined gene, marked with a reporter at the 5’ end and a quencher at the 3’ end. The probe employs the FRET principle for emission of fluorescence [34] and its base pair size is equal to the distance between the two fluorophores. Once the primers have been aligned to the sequence, the probe hybridizes and, in the course of the extension cycle, polymerase hydrolyzes the probe, causing separation of the reporter and the quencher, thus generating an increase in fluorescence proportional to the increase of the PCR product. By having two quenchers, it produces a decrease in the background of the amplification curves.
1.6. Elaboration of the Standard Curve
In Real Time PCR tests, elaboration of a standard curve is a critical step for optimization of the reaction, since the data for efficiency of the reaction, reproducibility or error and the reaction gradient or kinetics are extracted from this curve. To elaborate a standard curve, a minimum of five to six 1:10 dilutions of the primer (genomic DNA, DNAc, linearized plasmids, among others) nucleic acid (NA) must be performed. Dilutions by replication must be analyzed in one run (above 3 are recommended) and a minimum of 5 logarithms reduction in the NA concentration. This is required in order to achieve a more exact quantification and to determine efficiency of the reaction [34] .
The standard curve is built by plotting the logarithm of the initial primer NA concentration against the Ct or Cp values obtained during each amplification cycle. Most of the software in existing equipment generates a linear regression with an R2 value, which is employed to assess optimization of the PCR reaction [35] .
2. Methods
For each aqueduct, a sample of 250 mL of drinking water was collected, transported and stored in cold until processed in the laboratory. If the water had received treatment, residual chlorine was determined by means of the DPD reagent addition technique and subsequent comparison of the color generated. Then, the sample was filtered in the laboratory using the membrane filtration system Biosart® 100 (0.45 µm filter), from Sartorius (Weender, Goettingen, Germany). The Rapid Water DNA Isolation Kit from MO-BIO Laboratories (Carlsbad, CA, USA) was used for the extraction of the DNA. Following, the filter membrane was removed and placed in a tube with beads used for mechanical removal of the microorganisms, after, for the DNA extraction; the steps of the technique were followed as established by the manufacturer. The molecular marker of H. pylori, glm M, was used, and to optimize the qPCR technique, annealing temperature, concentration of primers and probe were standardized.
2.1. Preparation of Genomic DNA of H. pylori for Elaboration of Standard Curves
Genomic DNA of the H. pylori strain ATCC 51932 was used with an initial concentration of 215.1 ng/µL and A260/A280 absorbance relation of 1.9, quantified with the NanoDrop Lite (Thermo Scientific, Wilmington, USA) spectrophotometer. DNA extraction was performed previously with the QIAmp® DNA Mini (QIAGEN, Hilden, Germany) kit, following the specifications of the manufacturer.
Following, the calculations for determining the amount of genome copies of H. pylori equivalent to 215.1 ng/ µL were carried out, considering that one bacterium corresponds to 1.815 fg of DNA; that the H. pylori genome has a size of 1,655,849 pb and, in addition, that there exists one unique copy of the glmM gene per bacterium [36] [37] . The amount of genome copies calculated per reaction is shown in Table 1. Once the theoretical calculation was obtained, serial 1:10 dilutions up to 10−8 were performed. The last six dilutions were chosen for elaboration of the respective standard curves.
2.2. Optimization of Reactions
In order to establish the best temperature for alignment of the primers, temperatures of 52˚C, 54˚C and 56˚C were evaluated.
For optimization of primer concentration, the concentrations of 1 µM, 0.25 µM and 500 nM per reaction were evaluated with a standard concentration of the hydrolysis probe of 0.25 µM in all three cases, as well as the same alignment temperature for all cases.
Regarding optimization for probe concentration, due to the fact that the reaction had been previously evaluated with a concentration of the glmMPb probe of 1 µM and 0.25 µM at a temperature of 52˚C, a unique standard
Table 1. Theoretical concentration of H. pylori per dilution for elaboration of standard curves.
curve was generated with a probe concentration of 0.1 µM in order to assess efficiency of the reaction while using lower probe concentrations.
For each one of the reactions, a master mix of 7.5 µL was employed, containing: 5 µL of LightCycler® 480 Probes Master (2X) (Roche Diagnostics GmbH); 1.5 µL of water for PCR LightCycler® 480 Probes Master (Roche Diagnostics GmbH); and 1 µM of glmMFw and glmMRv primers (Integrated DNA Technologies, Coralville, Iowa, USA). For fluorescence emission, a ZEN™ doubled-quencher (Integrated DNA Technologies) type hydrolysis probe was employed, purified through HPLC and marked with fluoresceine (FAM) and two quenchers: Iowa BlackFQ and ZEN.
To each one of the reactions 2.5 µL of the corresponding DNA dilution were added, for a final reaction volume of 10 µL. All reactions were made by duplication in 96-well white plates, specific for the equipment. Additionally, a negative control with 2.5 µL of DNA from Pseudomonas aeruginosa ATCC 27856 was mounted by duplication in each standard curve.
In order to verify the formation of PCR products, the amplicons were run in an agarose gel at 4% (m/v) during 1.5 h at 80 V; the gel was dyed with GelRed (PHENIX Research Products, USA). Visualization was performed with a EM 0079 (Major Science, Saratoga, USA) transilluminator.
After the reactions concluded, results were analyzed by means of Absolute Quantification by Second Derivative Maximum.
The sampling strategy in Costa Rica included sampling the aqueducts in areas of high prevalence of gastric cancer, by collecting samples of chlorine-treated water with chlorine content lower than the established by the Water Quality Regulation in Costa Rica. As for Panamá, analysis of the bacteria was based on samples from aqueducts supplying untreated water for human consumption in the province of Chiriquí.
3. Results
The software version # LCS480 1.5.0.39 was used in the analysis of the technical parameters of the qPCR optimization technique.Statistical analysis of the data for optimal value, error, efficiency and the slope of the equations was performed using the software of the equipment according to the indications of the manufacturer. The statistical method used was the Second Derivative Maximum and, for calculation of R2, the program Microsoft Excel 2010 was employed, plotting the logarithm of the concentration against the Cp obtained for each of the samples from the optimization. The results are shown in Table 2.
The number of copies of the gene in each of the dilutions used was calculated using the standard curve by plotting the logarithm of the concentration calculated by the equipment vs Cp value of each standard.To determine the detection limit of the technique, the minimum number of copies/mL quantified by the equipment in Standard 6―which, theoretically, contained three copies of the gen―was analyzed.
As for Costa Rica, 25% of the samples showed an average value of residual chlorine of 0.1 mg/L; the remaining 75% of the samples showed no residual chlorine, despite having gone through a chlorination process in the respective aqueducts; also, slight water turbidity was observed in some samples. Ignoring one sample of extreme
Table 2. Values of the optimization of qPCR technical processes.
value (outlier) in each country, the water samples of Costa Rica presented an average value of 3.6 × 103 copies of H. pylori bacteria/100 mL of water, while samples from Panamá had an average value of 3.3 × 102 copies of the bacteria/100 mL.
The units expressed by the equipment correspond to copies/mL as reported by Roche Diagnostics GmbH in the LC480II Operator’s Manual. The results were then calculated and expressed to a volume of 100 mL from an initial sample volume of 250 mL. In the case of Costa Rica’s samples, it was determined that 79.5% were positive for H. pylori, while 20.5% samples had negative results; removing outliers, high average quantification of bacteria was determined at 3.6 × 103 copies/100 mL. For Panama, 86% of the samples were found positive for the presence of H. pylori; removing outlier, high average quantification of bacteria was determined at 3.3 × 102 copies/100 mL. Quantitative results of the positive samples for both countries are shown in Figure 1; also shown is the relation between the positive and negative (controls) curves of the samples. Note the difference between the averages found for both countries.
4. Discussion
After recognizing that the environment plays a preponderant role in acquisition of the infection and that the association between chronic H. pylori infection and the increased risk to develop gastric cancer is well established, it can be affirmed that the mix of factors among persons with H. pylori infection―strain-specific components, host immune responses, and environmental factors―influences the risk of gastric disease, including adenocarcinoma of the stomach [38] ; this is of particular epidemiological relevance given the high incidence of gastric cancer in Costa Rica.
In countries with high incidence of gastric cancer, it has been observed that the presence of infections among people seems to differ depending on whether they inhabit the mountain range or the littoral. This variation seems to be due to the provenance of the strains of H. pylori [9] . Specifically in Costa Rica, it was found that at average heights around 1500 meters above sea level, gastric cancer incidence rates range from 40 to 50 cases per 100,000 inhabitants. A Pearson correlation of 0.86 was determined for this finding, strongly influenced by who the Aqueduct Operating Entity was, either the Instituto Nacional de Aguas, the ASADAS (Asociaciones Administradoras de Sistemas de Acueductos y Alcantarillados Sanitarios de Costa Rica) or the Municipalities [39] .
Review of updated 2011 information from the Ministry of Health in Panama [40] , shows overall adjusted rates between 9 and 10 cases per 100,000 inhabitants, similar to those of cantons with low gastric cancer incidence in Costa Rica. Regarding Costa Rica, gastric cancer high incidence data correspond to people living in the central part of the country, in the provinces of Cartago and San Jose. The last available statistical data from 2003 for men shows results of 31.87 cases per 100,000 inhabitants, and 17.42 cases per 100,000 inhabitants for women in the same year [41] .
Although water in high gastric cancer incidence zones in Costa Rica is dispensed to the population by the Municipalities or ASADAS, it is considered of good quality according to health parameters regarding presence of fecal coliforms. According to findings from this research, water chlorination treatments applied by the Operating Entities have proved to be in sufficient in some cases, with important concentrations of viable bacteria still remaining in the water. Research should be conducted as to the causes of strong resistance of H. pylori to chlorination. However, the likelihood of occurrence of this situation has already been published [42] . In addition, marked apparent turbidity exceeding 5 NTU (nephelometric turbidity units), which is the maximum permitted by the Water Quality Regulation in Costa Rica, was determined in the samples analyzed. Solids in suspension, such as soil particles, can serve to hide the bacteria.
The current water quality Regulation proposes treating the water with residual chlorine from 0.1 mg/L to 0.3

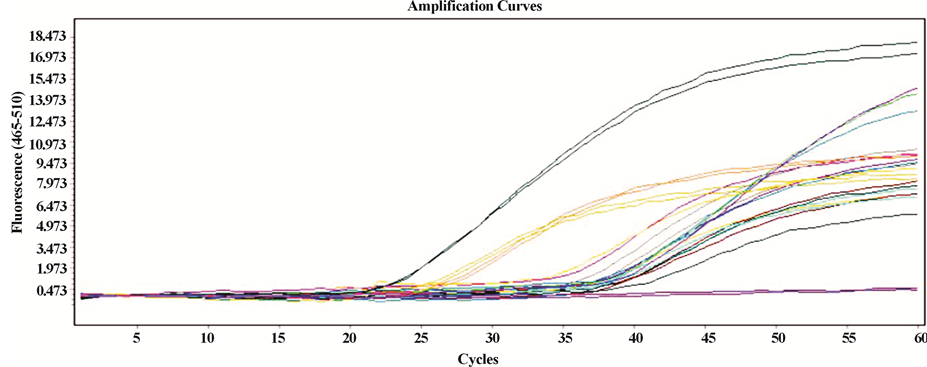

Figure 1. (a) Amplification curves generated by the LC480II equipment for positive samples in Costa Rica; upper curves: positive control; lower lines: negative controls; (b) amplification curves generated by LC480II for positive samples in Panamá; upper curves: positive control; lower lines: negative controls; (c) and (d) frequency distribution for positive samples in Panama and Costa Rica respectively.
mg/L up to a maximum of 0.6 mg/L concentration. However, consistent with our results, this concentrationis totally insufficient to eradicate the bacteria.
As for the results obtained from the quantification of the bacteria in drinking water, significant differences were observed in water management in both countries that may result in differences in quality. While non-chlo- rinated water from Chiriquí Panamá presents 3.3 × 102 copies of the bacteria per 100 mL, in Costa Rica chlorinated water shows 3.6 × 103 copies/100 mL on average.
Continuous and efficient water chlorination determines non-formation of biofilms in piping conducting water from the sites of chlorination to consumers. According to specific research, the bacterium is capable of resisting in biofilm-like environments, as in the stomach, where the resistance of this bacterium to conventional therapies has suggested the role of biofilm-growing bacteria, which are recalcitrant to many antimicrobial agents. This has led to use of substances degrading components of the biofilm [43] .
With respect to formation of biofilms in piping and water storage tanks, it is known that when H. pylori adheres to the surface, activation of genes related to “quorum sensing” starts, thus activating the mechanisms of formation of biofilms. In particular, Azevedo and colleagues [28] [44] [45] have worked with biofilms in pipes and water deposits, and they consider this to be the way bacteria can remain viable in the water. Additionally, biofilms developed by multiple H. pylori strains are, in general terms, more complex than those associated with single strains; such condition might promote the genetic exchange favoring the generation of more virulent strains [46] .
With the recent determination and quantification of H. pylori in the soil by Pérez and colleagues, water treatment aiming at achieving elimination of apparent turbidity gains special relevance, as turbidity influences survival of the bacteria in the water and its resistance to chlorination.
Consequently, for the sake of public health, special relevance is given to treating and dispensing drinking water containing no less than 0.6 mg/L to 0.8 mg/L residual chlorine, with apparent turbidity less than 5 NTU, particularly in the case of chlorine-treated superficial waters in areas of high gastric cancer incidence.
5. Conclusions
Further studies to determine augmented resistance to strains of H. pylori are deemed to be of immediate importance, given the amounts found in treated waters in Costa Rica.
The possibility of modifying the Regulations for Drinking Water Quality must also be explored, allowing for greater residual chlorine if necessary, especially in treated water coming from superficial water systems, as it may possess higher contents of bacteria as well as higher contents of soluble organic matter.
Quantitative analyses of the bacterium in water systems intended for the population can be implemented, as well as avoiding dispensation of water with turbidity over 5 NTU.
Acknowledgements and Disclosures
We would like to thank the Vicerrectoría de Investigación y Extensióndel Instituto Tecnológico de Costa Rica and the University of Chiriquí, Panamá for their financial support.
We are also grateful to student José Pablo Cerdas Velasquez for his assistance.
Conflict of Interest Statement
The authors declare that there is no conflict of interest.
Cite this paper
VirginiaMontero-Campos,ShirleyArias-Cordero,BenedictoValdés-Rodríguez,MonserratJarquín-Cordero, (2015) Quantitative Detection of Helicobacter pylori by Real Time PCR in Drinking Water—Environmental and Public Health Risk Significance. Open Journal of Medical Microbiology,05,118-127. doi: 10.4236/ojmm.2015.53015
References
- 1. Azevedo, N., Guimares, N., Figuereido, C. and Kevil, C. (2007) A New Model for the Transmission of Helicobacter pylori: Role of Environmental Reservoirs as Gene Pools to Increase Strain Diversity. Critical Reviews in Microbiology, 33, 157-169.
http://dx.doi.org/10.1080/10408410701451922 - 2. Perez, G., Rothenbacher, D. and Brenner, H. (2004) Epidemiology of Helicobacter pylori Infection. Helicobacter, 9, 1-6.
http://dx.doi.org/10.1111/j.1083-4389.2004.00248.x - 3. Megraud, F. (2003) When and How Does Helicobacter pylori Infection Occur? Gastroenterologie Clinique et Biologique, 27, 374-379.
- 4. Pounder, R. and Ng, D. (1995) The Prevalence of Helicobacter pylori Infection in Different Countries. Alimentary Pharmacology & Therapeutics, 9, 33-39.
- 5. Goodman, K., O’Rourke, K., Day, R., Wang, C., Nurgalieva, Z., Phillips, C.V., Aragaki, C., Campos, A. and De la Rosa, J.M. (2005) Dynamics of Helicobacter pylori Infection in a US-Mexico Cohort during the First Two Years of Life. International Journal of Epidemiology, 34, 1348-1355.
http://dx.doi.org/10.1093/ije/dyi152 - 6. Haggerty, T., Perry, S., Sanchez, L., Perez-Perez, G. and Parsonnet, J. (2005) Significance of Transiently Positive Enzyme-Linked Immunosorbent Assay Results in Detection of Helicobacter pylori in Stool Samples from Children. Journal of Clinical Microbiology, 43, 2220-2223.
http://dx.doi.org/10.1128/JCM.43.5.2220-2223.2005 - 7. Graham, D. (2007) The Year in Helicobacter. Helicobacter, 12, 14-28.
- 8. Kanbay, M. (2005) Helicobacter pylori Seroprevalence in Patients with Coronary Artery Disease. Digestive Diseases and Sciences, 50, 2071-2074.
http://dx.doi.org/10.1007/s10620-005-3009-7 - 9. Peter, A. and Montenero, A. (2006) Is Helicobacter pylori a Cause of Atrial Fibrillation? Future Cardiology, 2 429-439.
http://dx.doi.org/10.2217/14796678.2.4.429 - 10. Migneco, A., Ojetti, V., Specchia, L., Franceschi, F., Candelli, M., Mettimano, M., Montebelli, R., Savi, L. and Gasbarrini, G. (2003) Eradication of Helicobacter pylori Infection Improves Blood Pressure Values in Patients Affected by Hypertension. Helicobacter, 8, 585-589.
http://dx.doi.org/10.1111/j.1523-5378.2003.00180.x - 11. Kaptan, K. (2006) Helicobacter pylori and Cobalamin Deficiency. Clinical & Laboratory Haematology, 28, 360-360.
http://dx.doi.org/10.1111/j.1365-2257.2006.00799_1.x - 12. Aydemir, S. (2005) The Effect of Helicobacter pylori on Insulin Resistance. Digestive Diseases and Sciences, 50, 2090-2093.
http://dx.doi.org/10.1007/s10620-005-3012-z - 13. Ojetti, V. (2005) The Role of H. pylori Infection in Diabetes. Current Diabetes Reviews, 1, 343-347.
http://dx.doi.org/10.2174/157339905774574275 - 14. Ojetti, X. and Chen, D. (2006) Helicobacter pylori and Hepatocellular Carcinoma: Correlated or Uncorrelated? Journal of Gastroenterology and Hepatology, 21, 345-347.
http://dx.doi.org/10.1111/j.1440-1746.2006.04245.x - 15. Queiroz, D.M., Rocha, A.M., Rocha, G.A., Cinque, S.M., Oliveira, A.G., Godoy, A. and Tanno, H. (2006) Association between Helicobacter pylori Infection and Cirrhosis in Patients with Chronic Hepatitis C Virus. Digestive Diseases and Sciences, 51, 370-373.
http://dx.doi.org/10.1007/s10620-006-3150-y - 16. Brawner, K., Morrow, C. and Smith, P. (2014) Gastric Microbiome and Gastric Cancer. Cancer Journal, 20, 211-216.
http://dx.doi.org/10.1097/PPO.0000000000000043 - 17. Cao, L. and Yu, J. (2015) Effect of Helicobacter pylori Infection on the Composition of Gastric Microbiota in the Development of Gastric Cancer. Gastrointestinal Tumors, 2, 14-25.
http://dx.doi.org/10.1159/000380893 - 18. Ferlay, J., Hai-Rim, S., Bray, F., Forman, D., Mathers, C. and Maxwell, D. (2010) Estimates of Worldwide Burden of Cancer in 2008: GLOBOCAN 2008. International Journal of Cancer, 127, 2893-2917.
http://dx.doi.org/10.1002/ijc.25516 - 19. Engstrand, L. and Lindberg, M. (2013) Helicobacter pylori and the Gastric Microbiota. Best Practice & Research Clinical Gastroenterology, 27, 39-45.
http://dx.doi.org/10.1016/j.bpg.2013.03.016 - 20. Dicksved, J., Lindberg, M., Rosenquist, M., Enroth, H., Jansson, J. and Engstrand, L. (2008) Molecular Characterization of the Stomach Microbiota in Patients with Gastric Cancer and in Controls. Journal of Medical Microbiology, 58, 509-516.
http://dx.doi.org/10.1099/jmm.0.007302-0 - 21. Fernández-Delgado, M., Contreras, M., García-Amado, M., Michelangeli, F. and Suárez, P. (2007) Evidencias de la transmisión acuática de Helicobacter pylori. Interciencia, 33, 412-417.
- 22. Israel, D. (2001) Genetic Exchange. In: Mobley, H.L.T., Menz, G.L. and Hazell, S.L., Eds., Helicobacter pylori: Physiology and Genetics, American Society for Microbiology Press, Washington DC, Chapter 28.
http://www.ncbi.nlm.nih.gov/books/NBK2430/ - 23. Dewhirst, F., Shen, Z., Scimeca, M., Stokes, L., Boumenna, T., Chen, T., Paster, B. and Fox, J. (2005) Discordant 16S and 23S rRNA Gene Phylogenies for the Genus Helicobacter: Implications for Phylogenetics Interference and Systematics. Journal of Bacteriology, 187, 6106-6118.
http://dx.doi.org/10.1128/JB.187.17.6106-6118.2005 - 24. Shahamat, M., Alavi, M., Watts, J.E., González, J.M., Sowers, K.R., Maeder, D.W. and Robb, F.T. (2004) Development of Two PCR-Based Techniques for Detecting Helical and Coccoid Forms of Helicobacter pylori. Journal of Clinical Microbiology, 42, 3613-3619.
http://dx.doi.org/10.1128/jcm.42.8.3613-3619.2004 - 25. Ogura, M., Perez, J.C., Mittl, P.R.E., Lee, H.-K. and Dailide, G. (2007) Helicobacter pylori Evolution: Lineage-Specific Adaptations in Homologs of Eukaryotic Sel1-Like Genes. PLoS Computational Biology, 3, e151.
http://dx.doi.org/10.1371/journal.pcbi.0030151 - 26. UNIPROT CONSORTIUM (2011) P55980 (CAGA_HELPY) Version 60. (en línea). Consultado 11 marzo de 2015.
http://www.uniprot.org/uniprot/P55980 - 27. Fux, C., Costerton, J.W., Stewart, P. and Stoodley, P. (2005) Survival Strategies of Infectious Biofilms. Trends in Microbiology, 13, 34-40.
http://dx.doi.org/10.1016/j.tim.2004.11.010 - 28. Azevedo, N., Pacheco, A., Keevil, C. and Vieira, M. (2006) Adhesion of Water Stressed Helicobacter pylori to Abiotic Surfaces. Journal of Applied Microbiology, 101, 718-724.
http://dx.doi.org/10.1111/j.1365-2672.2006.03029.x - 29. Touati, E., Michel, V., Thiberge, J. and Wuscher, N. (2003) Chronic Helicobacter pylori Infections Induce Gastric Mutations in Mice. Gastroenterology, 124, 1408-1419.
http://dx.doi.org/10.1016/S0016-5085(03)00266-X - 30. She, F.F., Lin, J.Y., Liu, J.Y., Huang, C. and Su, D.H. (2003) Virulence of Water-Induced Coccoid Helicobacter pylori and Its Experimental Infection in Mice. World Journal of Gastroenterology, 9, 516-520.
- 31. Yañez, M., Barberá, V., Soria, E. and Catalán, V. (2009) Quantitative Detection of Helicobacter pylori in Water Samples by Real Time PCR Amplification of the Cag Pathogenicity Island Gene, cagE. Journal of Applied Microbiology, 107, 416-424.
http://dx.doi.org/10.1111/j.1365-2672.2009.04219.x - 32. Santiago, P., Moreno, Y. and Ferrus, M.A. (2015) Identification of Viable Helicobacter pylori in Drinking Water Supplies by Cultural and Molecular Techniques.
- 33. Pérez, L.M., Codony, F., López, D., Fittipaldi, M., Adrados, B. and Morató, J. (2010) Quantification of Helicobacter pylori Levels in Soil Samples from Public Playgrounds in Spain. Journal of Zhejiang University SCIENCE B, 11, 27-29.
http://dx.doi.org/10.1631/jzus.B0900238 - 34. Green, M. and Sambrook, J. (2012) Molecular Cloning: A Laboratory Manual. 4th Edition, Vol. II, Cold Spring Harbor Laboratory Press, New York.
- 35. Bio-Rad Laboratories, Inc. (s.f.). Real-Time PCR Application Guide. En línea. el 5 de febrero de 2015.
http://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_5279.pdf - 36. Lascols, C., Lamarque, D., Costa, J., Copie-Bergman, C., Le Glaunec, J., Deforges, L., Soussy, C., Petit, J., Delchier, J. and Tankovic, J. (2003) Fast and Accurate Quantitative Detection of Helicobacter pylori and Identificaction of Clarithromycin Resistance Mutations in H. pylori Isolates from Gastric Biopsy Specimens by Real-Time PCR. Journal of Clinical Microbiology, 41, 4573-4577.
http://dx.doi.org/10.1128/JCM.41.10.4573-4577.2003 - 37. Olaya, S. (2008) Comparación entre una técnica estandarizada de PCR en tiempo real y PCR convencional para la detección del gen cagA de Helicobacter pylori. Tesis de Maestría, Pontificia Universidad Javeriana, Colombia.
- 38. Azevedo, N., Guimares, N., Figuereido, C. and Kevil, C. (2007) A New Model for the Transmission of Helicobacter pylori: Role of Environmental Reservoirs as Gene Pools to Increase Strain Diversity. Critical Reviews in Microbiology, 33, 157-169.
http://dx.doi.org/10.1080/10408410701451922 - 39. Montero, V., Hernández, A. and Camacho, J. (2014) Culture and Molecular Identification of Helicobacter pylori in Drinking Water from Areas of High and Low Incidence of Gastric Cancer in Costa Rica. OJMM, 4, 261-269.
http://dx.doi.org/10.4236/ojmm.2014.44030 - 40. Boletín del Registro Nacional de Cáncer de Panamá, del Ministerio de Salud de Panamá, 2011.
- 41. Vargas, R., Ortiz, A. and Muñoz, G. (2007) Incidencia y Mortalidad del cáncer en Costa Rica 1995-2005. Ministerio de Salud. Dirección Vigilancia de la Salud Unidad de Estadística-Registro Nacional de Tumores.
- 42. Montero, V., Moya, L. and Porras, M. (2004) Aislamiento de bacterias patógenas resistentes al cloro en agua potable. Cámara Costarricense de la Industria Alimentaria, 71, 20-26.
- 43. Cammarota, G., Sanguinetti, M., Gallo, A. and Posteraro, B. (2012) Review Article: Biofilm Formation by Helicobacter pylori as a Target for Eradication of Resistant Infection. Alimentary Pharmacology & Therapeutics, 36, 222-230.
http://dx.doi.org/10.1111/j.1365-2036.2012.05165.x - 44. Azevedo, N., Vieira, M. and Keevil, C. (2003) Establishment of a Continuous Model System to Study Helicobacter pylori Survival in Potable Water Biofilms. Water Science & Technology, 47, 155-160.
- 45. Azevedo, N., Pinto, A., Reis, N. and Vieira, M. (2006) Shear Stress, Temperature, and Inoculation Concentration Influence the Adhesion of Water-Stressed Helicobacter pylori to Stainless Steel 304 and Polypropylene. Applied and Environmental Microbiology, 72, 2936-2941.
http://dx.doi.org/10.1128/AEM.72.4.2936-2941.2006 - 46. Grande, R., Di Campli, E., Di Bartolomeo, S., Verginelli, F., Di Giulio, M., Baffoni, M., Bessa, L. and Cellini, L. (2012) Helicobacter pylori Biofilm: A Protective Environment for Bacterial Recombination. Journal of Applied Microbiology, 113, 669-676.
http://dx.doi.org/10.1111/j.1365-2672.2012.05351.x


