Open Journal of Medical Microbiology
Vol.4 No.2(2014), Article
ID:45988,9
pages
DOI:10.4236/ojmm.2014.42013
Isolation and Identification of Multi-Drug Resistant Strains of Non-Lactose Fermenting Bacteria from Clinical Isolates
Minhas Akbar1, Muhammad Zahid1*, Pir Asmat Ali1, Aftab Alam Sthanadar1,2, Mudassir Shah1,3, Iram Alam Sthanadar1, Muhammad Kaleem1, Muhammad Aslam1, Khayyam1, Zahirullah4, Syeda Mahreen Ul Hassan5, Noor Jehan1, Muhammad Ismail Khan1
1Department of Zoology, Islamia College University, Peshawar, Pakistan
2Department of Zoology and Animal Sciences, Post Graduate College Dargai, Malakand, Pakistan
3Government Degree College Dara Adam Kheil, FR Kohat, Pakistan
4Lady Reading Hospital, Peshawar, Pakistan
5Department of Microbiology, Shaheed Benazir Bhutto Women University, Peshawar, Pakistan
Email: *mzahidsafi75@yahoo.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 17 April 2014; revised 5 May 2014; accepted 12 May 2014
ABSTRACT
Purpose: We studied the drug resistance of different microbes from clinical isolates. The morphological characteristics of bacteria were observed through culture characteristics and by carrying out gram staining techniques while the biochemical characteristics of bacteria were carried out by biochemical test. Methods: A total of 324 samples were collected from suspected patients visiting different hospitals at district Peshawar. For morphological identification, samples of clinical isolates were analyzed by blood agar, MacConkey agar and Eosine Methylene Blue, identified by gram staining and characterized by different biochemical tests. Antibiotic Sensitivity test by Modified Kirby-Bauer Disc diffusion method was used to test the in-vitro susceptibility of the identified isolates to different antibiotics such as Ceftazidime, Ceftazidime, Ceftriaxone, Cefepime and Imipenem. Results: These resistant non-lactose fermenting gram negative bacteria were isolated from samples of pus/wound (33.30%, n = 108/324), blood (33.30%, n = 108/324), urine (23.30%, n = 75/324) and from ascetic/pleural fluids (10.20%, n = 33/324). The study revealed that the percentage of non-fermenting bacterial infection was higher in females (53%) as compared to males (47%) along with higher infection observed in the age group of 11 - 30 years. Pseudomonas aeroginosa showed high resistance against Cefepime (88.80%), followed by Cefoperazone (55.50%), Ceftazidime (48.10%), Ceftriaxone (33.30%). Imipenem was active with low resistance (7.40%). More resistance was seen in Morganella morganii against Imipenem (66.70%) followed by Cefoperazone (55.50%), Ceftriaxone (55.50%). Cefepime showed low resistance (11%). Multi-drug resistant Proteus mirabillis was highly resistance to Ceftriaxone (74.07%), followed by Cefepime (59.20%), Cefoperazone (44.40%) and low resistance for Imipenem (25.90%). Salmonella typhi demonstrated high resistance against Imipenem (74.07%), followed by Ceftriaxone (40.70%), Ceftazidime (37.03%). Cefepime showed low resistance (3.70%), hence it is more active against S. typhi. Conclusions: The different species of non-lactose fermenting gram negative bacteria have shown a different resistivity pattern in the present study. Therefore identification of non-lactose fermenting gram negative bacteria and looking after their resistivity/susceptibility pattern are important for suitable management of the infections caused by them.
Keywords:Multidrug Resistant, Non-Lactose Fermenting Gram Negative Bacteria, Disc Diffusion Technique

1. Introduction
Developing resistance to antibiotics is natural to microbes, which cannot primarily be ceased of as constantly evolving nature of microbes to chemicals around them. The phenomenon is very important regarding its practical and economic implications. It is because of this resistance people cannot be affectively treated and remain ill for a longer period of time. The development of tolerance of microbes to more than one drug is multi drug resistance (MDR) [1] . In the last few decades, antibiotic resistance is becoming a major problem across the globe [2] .
Factors leading to antibiotic resistance include a widespread and aggressive use of broad-spectrum antibiotics, wrong investigated and diagnosis, using drugs without proper prescription by physicians and doctors, further including misuse of drugs by patients. With the resistant strains of bacteria the treatments of common infections become difficult or impossible. Non-fermenters are gram-negative bacteria that cannot ferment sugars to produced energy for cell physiology. Gram negative non-fermenting bacteria (NFGNB) were isolated from different clinical samples. Because of extreme multidrug resistance problems, species of this group offer a serious challenge for healthcare management [3] . As mostly, non-fermenting (gram-negative) bacteria are niche pathogens that cause infections in critically ill or immune-compromised patients. As they are primarily healthcare associated pathogens, they rarely cause infection in healthy individuals [4] .
There are different mechanisms for resistance in non-fermenting gram-negative bacteria, including (i) production of enzymes (ii) enzymatic inactivation of antimicrobial agents (iii) specific targeted enzyme that is inhibited by antimicrobial agents (iv) alterations in target sites, (v) production of efflux pumps (vi) loss of outer membrane proteins or porins (vii) reduced uptake of the antimicrobial agent. That is because of these different resistance mechanisms that the therapeutic options are severely limited to treat infections caused by them [3] [4] . Non-fermenters include many species belonging to several genera. Previous studies suggest four species rarely found in hospitals, significant problems in hospital practice, including Pseudomonas aeroginosa, Morganella morganii, Proteus mirabilis and Salmonella typhi [5] . The emergence of multidrug resistance P. aeruginosa has emerged as a severe health problem [6], which is of low permeability of the cell wall, mutation in chromosomal genes regulating resistance genes and also acquiring resistance genes from other organisms via plasmids, transposons and bacteriophages [7] [8] . As the infection caused by multiple antibiotics resistant P. aeroginosa may result in worse clinical outcomes, this bacterium has gained particular importance [9] . Multi-antibiotic resistant of M. morganii strain is due to change in outer membrane permeability and mutation of the major porin or by a change in the number of porins in the outer membrane [10] . Increasing resistance to β-Lactam antibiotics in P. mirabillis, is mediated by the production of acquired β-lactamases. Plasmid-mediated ESBLs, including TEMtype derivatives active against expanded spectrum cephalosporin are also spreading in P. mirabilis [11] .
In Pakistan the first case of multidrug resistance of S. typhi was reported in 1987 [12] . Since 1991, the cases of infections caused by Salmonella were increasingly resistant to extended-spectrum cephalosporins and fluoroquinolones [13] . Salmonella typhi resistance to Fluoroquinolone is associated with point mutations occuring mostly within a domain of gyrA. Cephalosporins resistance is usually mediated by extended spectrum β-lactamases derived from TEMand SHV-type enzymes [14] . The present study aimed to assess the multi-drug resistance of non-lactose fermenting bacteria in a low economic country like Pakistan, where people are not well aware of health issues. Our investigation will be a real contribution to the society and human health for further proper medication and human ease.
2. Materials and Methods
2.1. Sample Collection
A total 350 clinical samples were collected from suspected patients in different hospitals of Peshawar, KP Pakistan. Samples were collected from urine, pus/wound, blood, ascetic/plural fluids and were analyzed for colonial morphology and routine biochemical identification. The isolation of clinical samples was carried out according to standard protocol [15] . The collected samples from urine, pus/wound, blood and ascetic/plural fluids were spread on blood, MacConkey and Eosine Methyline Blue (EMB) agar plates and incubated at 37˚C for 24 - 48 hours. Gram staining was carried out as early described [16] to identify the NFGNB bacteria.
2.2. Biochemical Characterizations
Biochemical characterizations were performed through biochemical tests of clinical isolates. The protocol for clinical sample’s identification was according to Cheesbrough et al. [15] . Indole, Methyl red, Citrate utilization, Triple sugar iron, Oxidase, Urease and Nitrate tests were also carried out.
2.3. Antibiotic Sensitivity Test
The Kirby-Bauer Disc Diffusion Method was used to test the in vitro susceptibility of the identified isolates to Ceftazidime (30 µg), Cefoperazone (75 µg), Ceftriaxone (30 µg), Cefepime (30 µg), Imipenem (10 µg). Pseudomonas aeroginosa colonies were picked up from the culture plate with the help of a sterile platinum wire loop and emulsified in 4 ml of sterile peptone water to match with 0.5 McFarland turbidity standards (1.5 ´ 108 cfu/ml). The surface of Mueller Hinton Agar (Oxoid, Basingstoke, UK) in a Petri dish was inoculated evenly through a sterile swab and for 10 minutes was allowed the agar to dry. A multichannel disc dispenser (Oxoid, Basingstoke, UK) was used to deposit the antibiotics discs onto the surface of the inoculated medium. The plate was then incubated at 37˚C for 24 hours. With measuring scale the diameters of zone of inhibition were measured in millimeters after 24 hours of period of incubation [17] [18] . The above procedure was repeated thrice for each P. mirabillis, M. morganii and for S. typhi isolates.
3. Results
A total of 324 multiple-drug resistant gram negative non-fermenters were isolated from 350 clinical samples processed at Microbiology Laboratory of Services Hospital Lahore. These bacterial strains were then identified on the basis of their cultural, morphological and biochemical characteristics (Table 1).
The occurrence of isolates was higher in females (53%, n = 171 in 324) as compared to males (47%, n = 315 in 324), while highest frequency of MDR-NFGNB was observed among young patients of age 11 - 30 (n = 171). The distribution of isolates among different age group showed that overall infection rate was higher in young individuals (11 - 30 years) but P. aeroginosa infection was higher in old age (61 - 74) (Table 1).
The Distribution of MDR-NFGNB P. aeroginosa, M. morganii, P. mirabillis and S. typhi in different clinical specimens (urine, pus/wound, blood, ascetic/pleural fluids) are shown in (Table 2). The results were also presented on simple cylindrical bar graph in Figure 1.
It was observed that Pseudomonas aeriginosa was found to be 88.80% resistant against Cefepime. The resistance found in MDR—P. aeroginosa against other antibiotics included Cefoperazone (55.50%); Ceftazidime (48.10%); Ceftriaxone (33.30%); Imipenem (7.40%). Imipenem is most active against P. aeroginosa (Table 3).
MDR-Patteren in M. morganii has observed 66.60% resistance against Imipenem. The resistance found against other antibiotics included Cefoperazone (55.50%); Ceftriaxone (55.50%); ceftazidime (48%); Cefepime (11%). M. morganii show lowest resistance to Cefepime, thus it is active against M. morganii (Table 3).
MDR-Patteren in P. mirabilis, a higher degree of resistance to Ceftriaxone (74.07%) was detected in P. mirabillis isolates. The frequency of resistance against was Cefepime (59.20%); Cefoperazone (44.4%); ceftazidime (37.03%); Imipenem (25.90%); (Table 3). As Imipenem showed lower resistance to P. mirabilis.
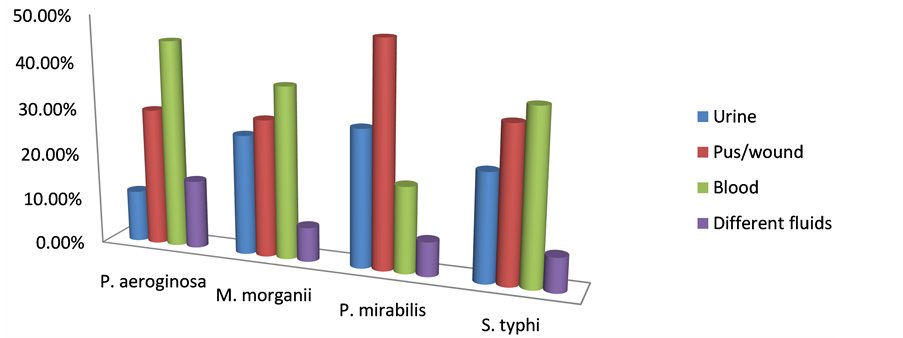
Figure 1. MRD-Bacteria percent isolated samples from different parts of the body of Patients.
Table 1. Distribution of MDR-bacteria in relation to gender and age wise in present study.
*n for numbers: % for percentage.
MDR-Patteren in S. typhi Salmonella typhi isolates exhibited high resistance against Imipenem (74.07%). Resistance to other antibiotics included Ceftriaxone (40.70%); Ceftazidime (37.03%); Cefoperazone (25.90%); Cefepime (3.70%) and thus a lower resistance to Cefepime was observed (Table 3). For the sake of convenience, results in Table 3 were presented on simple horizontal bar graph (Figure 2).
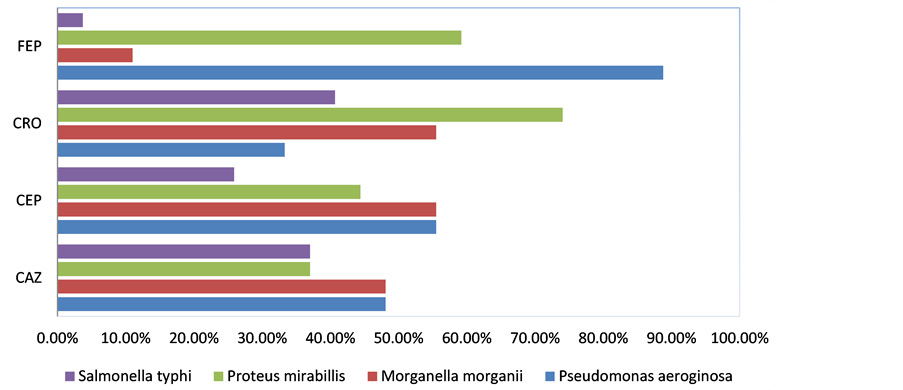
Figure 2. Antibiotics percent resistance in multi drug resistant bacteria from isolated samples.
Table 2. Clinical isolates of gram negative non-fermenters bacteria from different sites in hospitals.
*n for numbers: % for percentage.
Table 3. Antibiotic resistance percentage of gram negative non fermented bacteria (n = 81) isolated from different clinical samples.
4. Discussion
Multi-drug resistance in bacteria is of very much concern to clinicians. Not only these organisms are very resistant but they are also rapidly spreading [19] . In clinical practices the antibiotic resistant pathogens causes financial burden, treatment failure and can spread to other patients [20] . In the last few decades, due to the widespread use of antibiotics, non-fermentative gram negative bacilli have emerged important health care-associated pathogens. Recent studies conducted on important areas like identification of non-fermentative gram negative bacilli, and monitoring their susceptibility patterns, which is important for the appropriate management of the infections caused by them, and to make clear the fact that it is important to establish the clinical relevance of the isolated non fermentative gram negative bacilli, before they are considered as pathogens. This would prevent unnecessary usage of antibiotics and the rise of drug-resistant strains [21] . There is limited data on the prevalence and resistance pattern of NFGNB. The current study was intended to address these issues.
In this study MDR-NFGNB was isolated from patients of different age groups (11 - 74). The highest number of MDR-NFGNB was isolated from age group of 11 - 30 years, followed by age group of 31 - 60 and 61 - 74 years. People were prone to infection at the age of 11 - 30 years because at this age the people are more active, have more social contacts, more journey from one place to another so have more chances of getting infection. Chances of getting infection do not depend upon time but depend on number of exposure to the injurious bacteria, viruses and toxins. At old age the people are restricted from social contact so have less chances of developing infection.
In the present study, Gender wise distribution of MDR-NFGNB was isolated both from males and females and highest numbers are from females. This could be due to the social activity of females in their life in developing countries like Pakistan. Where females are much ignored as compared to male so their food cleanliness is not good as males, as a result their immune system is weak. Furthermore, females give birth child’s so admitted frequently to hospitals and thus has more chances of infection.
In present study P. aeroginosa, M. morganii, P. mirabillis and S. typhi were isolated and identified from blood, pus/wound, urine and ascetic fluids. These findings were in line with the results reported by other investigators [6] [22] and Javeed et al., [2] for P. aeroginosa, Singla et al. [23] , Falagas et al. [24] and Lee et al. [25] , for M. morganii. The present investigations showed higher frequency of P. mirabillis in pus (48.2%). Different researchers targeted S. typhi in blood, stool and bone marrow [26] [27] . The present study is unique in the sense as it identified S. typhi from urine (29.60%), blood (37.03%), pus/wound (25.90%), ascetic and pleural fluids (7.40%).
In-vitro MDR-Patteren in P. aeroginosa in present study to Imipenem was 7.40% which is consistence with the results of Romao et al. [28] who found 11% resistance against Imipenem, Tripathi et al. [29] found 10.80% resistance and Manian et al. [30] found 8% resistance against Imipenem in P. aeroginosa. Pseudomonas aeriginosa showed 88.80% resistance against Cefepime in the present study which is not consistent with the results of Zehra et al. [31] Romao et al. [28] and Tripathi et al. [29] who all detected 77.70%, 66%, 71%, 36.27% resistance against Cefepime respectively. The reason in the difference could be because of geographical differences. The fact is antibiotic resistance vary in different regions, environmental conditions and time to time.
Morganella morganii in-vitro MDR-Patteren showed 11% resistance against Cefepime. The finding of the current study is in agreement with the results of Falagas et al. [24] , who found 8% resistance against Cefepime. Similarly Xiao-Bo et al. [32] found 14.30% resistance to Cefepime. In this study, the resistance of MDR-M. morganii against Imipenem 66.60% followed by Cefoperazone and Ceftriaxone 55.50% equals while Ceftazidime have 48.10%. These results were quite different from the results of Falagas et al. [24] who showed resistance 80% against Ceftriaxone and 90% against Ceftazidime. Xiao et al. [32] found 16.50% resistance against Imipenem and 5.50% against Amikacin. Similarly, Lee et al. [25] found 19.40% resistance against Imipenem. The difference is due to geographical locations, specimens types and extraction protocols.
In this study in-vitro MDR-P. mirabillis showed 74.07% resistance against Ceftriaxone followed by 59.20% against Cefepime, 44.40% against Ceftazidime, 37.30% against Ceftazidime and 25.90% against Imipenem. These results are in contradiction with the some of the earlier studies conducted by Falagas et al. [24] observed 100% resistance against Cefepime , Ceftazidime and 14.30% against Imipenem in P. mirabillis. Sharma et al. [33] examined 9.60% resistance against Cefoperazone and 4.80% resistance against Cefepime in P. mirabillis. Xiao et al. [32] detected 7.60% resistance against Ceftriaxone, 6.40% against Ceftazidime, 2.40% against Cefepime, 0.60% against Imipenem and 0.30% against Cefoperazone in P. mirabillis. Here also the difference in our results and others are because of sampling size and difference in specimen types.
5. Conclusion
The present study in-vitro MDR-Patteren S. typhi showed 40.70% resistance against Ceftriaxone, 37.03% Ceftazidime, 25.90% Cefoperazon, 74.07% Imipenem and 3.70% Cefepime. These results are quite in agreement with the findings of Mustaq [34] , while different from the results of Nagshetty et al. [26] who showed 6.31% resistance against Ceftriaxone. Kumar et al. [35] found 12.10% resistance against Ceftriaxone. The difference may be due to the overuse of Ceftriaxone in our region. The study of multi-drug resistance is still an unsolved issue and needs further future endures [36] [37] . If this scenario is not properly tackled, there is a time coming, when there will be no more medication effect in terms of antibiotic use in order to cure and save human health and life.
Acknowledgements
The authors acknowledge with gratitude the invaluable support and assistance provided by Professor Dr. Asad Ullah Khan for valuable guidance and proof reading of the manuscript. We are also thankful to Microbiology laboratories in Services hospital Lahore for providing us lab facilities.
References
- White, D.G., Hudson, C., Maurer, J.J. and Ayers, S. (2000) Characterization of Chloramphinicol and Florfenical Resistance in Escherichia coli Associated with Bovine Diarrhea. Journal of Clinical Microbiology, 38, 4593-4598.
- Javeed, I., Hafeez, R. and Anwar, M.S. (2011) Antibiotic Susceptibility Pattern of Bacterial Isolates from Patients Admitted to a Tertiary Care Hospital in Lahore. Biomedica, 27, 19-23.
- John, E. and McGrowan, J.R. (2006) Resistance in Non-Fermenting Gram-Negative Bacteria: Multidrug Resistance to the Maximum. American Journal of Infection Control, 34, 29-37.http://dx.doi.org/10.1016/j.ajic.2006.05.226
- Fluit, A.C., Visser, M.R. and Schmitz, F.J. (2002) Molecular Detection of Antimicrobial Resistance. Journal of Clinical Microbiology, 14, 836-871. http://dx.doi.org/10.1128/CMR.14.4.836-871.2001
- Enoch, D.A., Birkett, C.I. and Ludlam, H.A. (2007) Non Fermentative Gram-Negative Bacteria. International Journal of Antimicrobial Agents, 3, 33-41.
- Meenakumari, S., Verma, S., Absar, A. and Chaudhary, A. (2011) Antimicrobial Susceptibility Pattern of Clinical Isolates of Pseudomonas aeruginosa in an Indian Cardiac Hospital. International Journal of Engineering Science and Technology, 3, 7117-7124.
- Lambert, P.A. (2002) Mechanisms of Antibiotic Resistance in Pseudomonas aeruginosa. Journal of the Royal Society of Medicine, 95, 22-26.
- Poole, K. (2004) Efflux-Mediated Multiresistance in Gram-Negative Bacteria. Clinical Microbiology and Infection, 10, 12-26.
- Merlo, C.A., Boyle, M.P., Diener, W.M., Marshall, B.C., Goss, C.H. and Lechtzin, N. (2007) Incidence and Risk Factors for Multiple Antibiotic-Resistant Pseudomonas aeruginosa in Cystic Fibrosis. Chest, 132, 562-568.http://dx.doi.org/10.1378/chest.06-2888
- Rojas, L., Vinuesa, T., Tubau, F., Truchero, C., Benzc, R. and Vinas, M. (2006) Integron Presence in a Multiresistant Morganella Morganii Isolate. International Journal of Antimicrobial Agents, 27, 505-512. http://dx.doi.org/10.1016/j.ijantimicag.2006.01.006
- Biendo, M., Thomas, D., Laurans, G., Daoudi, F.H., Canarelli, B., Rousseau, F. and Castelain, S. (2005) Molecular Diversity of Proteus mirabilis Isolates Producingextended-Spectrum Beta-Lactamases in a French University Hospital. Clinical Microbiology and Infection, 11, 395-401. http://dx.doi.org/10.1111/j.1469-0691.2005.01147.x
- Butt, T., Ahmad, R.N., Salman, M. and Kazmi, S.Y. (2005) Changing Trends in Drug Resistance among Typhoid Salmonellae in Rawalpindi, Pakistan. Eastern Mediterranean Health Journal, 11, 1038-1044.
- Su, L.H., Wu, T.L., Chia, J.H., Chu, C., Kuo, A.J. and Chiu, C.H. (2005) Increasing Ceftriaxone Resistance in Salmonella Isolates from a University Hospital in Taiwan. Journal of Antimicrobial Chemotherapy, 55, 846-852.http://dx.doi.org/10.1093/jac/dki116
- Wain, J. and Kidgell, C. (2004) The Emergence of Multidrug Resistance to Antimicrobial Agents for the Treatment of Typhoid Fever. Transactions of the Royal Society of Tropical Medicine and Hygiene, 98, 423-430.http://dx.doi.org/10.1016/j.trstmh.2003.10.015
- Cheesbrough, M. (2000) District Laboratory Practice Manual in Tropical Countries Part 2. Cambridge University Press, Cambridge, 178-179.
- Harley, J. (1990) Prescott L. Laboratory Exercises in Microbiology. Wm. C. Brown Publishers, 49-53.
- Gloria, A., Cheryl, B., John, E., Richard, F., Joan, S.K., Tanja, P., Joy, W. and Scott, F.D. (2003) Manual for the Laboratory Identification and Antimicrobial Susceptibility Testing of Bacterial Pathogens of Public Health Importance in the Developing World. Centers for Disease Control and Prevention, Atlanta, Georgia, USA and World Health Organization: Department of Communicable Disease Surveillance and Response, 103-118.
- Struve, C., Forestier, C. and Krogfelt, K.A. (2003) Application of a Novel Multi-Screening Signature-Tagged Mutagenesis Assay for Identification of Klebsiella pneumoniae Genes Essential in Colonization and Infection. Micro, 149, 167-176. http://dx.doi.org/10.1099/mic.0.25833-0
- Roshan, M., Ikram, A., Mirza, I.A., Malik, N., Abbasi, S.A. and Alizai, S.A. (2011) Susceptibility Pattern of Extended Spectrum B-Lactamase Producing Isolates in Various Clinical Specimens. Journal of the College of Physicians and Surgeons Pakistan, 21, 342-346.
- Mahmod, A. (2000) Bacteriology of Surgical Site Infections and Antibiotic Susceptibility Pattern of the Isolates at a Tertiary Care Hospital in Karachi. Journal of Pakistan Medical Association, 50, 256-259.
- Upgade, A., Prabhu, N., Gopi, V. and Soundararajan, N. (2012) Current Status of Antibiotic Resistant Non-Fermentative Gram Negative Bacilli among Nosocomial Infections. Advances in Science and Research, 3, 738-742.
- Moniri, R., Mosayebi, Z., Movahedian, A.H. and Mousavi, G.A. (2005) Emergence of Multi-Drug-Resistant Pseudomonas aeruginosa Isolates in Neonatal Septicemia. Journal of Infectious Disease and Antimicrobial Agents, 22, 39-44.
- Singla, N., Kaistha, N., Gulati, N. and Chander, J. (2010) Morganella morganii Could Be an Important Intensive Care Unit Pathogen. Indian Journal of Critical Care Medicine, 14, 154-155. http://dx.doi.org/10.4103/0972-5229.74176
- Falagas, M.E., Kavvadia, P.K., Mantadakis, E., Kofteridis, D.P., Bliziotis, I.A., Saloustros, E., Marak, S. and Samonis, G. (2006) Morganellamorganii Infections in a General Tertiary. Journal of Hospital Infection, 34, 315-321.
- Lee, I.K. and Liu, J.W. (2006) Clinical Characteristics and Risk Factors for Mortality in Morganella morganii Bacteremia. Microbiology and Infectious Diseases Journals, 39, 328-334.
- Nagshetty, K., Shivannavar, T.C. and Gaddad, S.M. (2010) Antimicrobial Susceptibility of Salmonella Typhi in India. The Journal of Infection in Developing Countries, 4, 70-73.
- Muthu, G., Suresh, A., Sumathy, G. and Srivani, R. (2011) Studies on Antimicrobial Susceptibility Pattern of Salmonella Isolates from Chennai, India. Internships of Pharmaceuticals and Biological Science, 2, 435-442.
- Romao, C.M.C.P.A, Faria, Y.N.D., Pereira, L.R. and Asensi, M.D. (2005) Susceptibility of Clinical Isolates of Multiresistant Pseudomonas aeruginosa to a Hospital Disinfectant and Molecular Typing. Mem Inst Oswaldo Cruz Rio de Janeiro, 100, 541-548.
- Tripathi, P., Banerjee, G., Saxena, S., Gupta, M.K. and Ramteke, P.W. (2011) Antibiotic Resistance Pattern of Pseudomonas aeruginosa Isolated from Patients of Lower Respiratory Tract Infection. African Journal of Microbiology Research, 5, 2955-2959.
- Manian, F.A., Meyer, L., Jenne, J., Owen, A. and Taff, T. (1996) Loss of Antimicrobial Susceptibility in Aerobic Gram-Negative Bacilli Repeatedly Isolated from Patients in Intensive Care Units. Infection Control and Hospital Epidemiology, 17, 222-226. http://dx.doi.org/10.2307/30141024
- Zehra, A., Naqvi, B.S., Bushra, R. and Ali, S.Q. (2010) Comparative Study on Resistance Pattern of Different Pathogens against Cefixime and Cefepime. Journal of Pharmaceutical Sciences, 3, 145-156.
- Xiao, Y.H., Wang, J., Zhu, Y., Qi, H.M., Li, X.Y., Zhao, C.Y. and Xue, F. (2010) Mohnarin of 2008: Surveillance Results of National Bacterial Drug Resistance. Chinical Journal of Nosocom, 16, 6.
- Sharma, S., Gupta, A. and Arora, A. (2012) Cefepime Tazobactam: A New β Lactam/β Lactamase Inhibitor Combination against ESBL Producing Gram Negative Bacilli. International Journal of Pharmaceutical and Biomedical Research, 3, 35-38.
- Mushtaq, M.A. (2006) What after Ciprofloxacin and Ceftriaxone in Treatment of Salmonella typhi. Pakistan Journal of Medical Sciences, 22, 51-54.
- Kumar, S., Rizv, M. and Berry, N. (2008) Rising Prevalence of Enteric Fever Due to Multidrug Resistant Salmonella: An Epidemiological Study. Journal of Medical Microbiology, 57, 1247-1250. http://dx.doi.org/10.1099/jmm.0.2008/001719-0
- Shankar, N., Baghdayan, A.S. and Gilmore, M.S. (2002) Modulation of Virulence within a Pathogenicity Island Vancomycin-Resistant Enterococcus feacalis. Nature, 417, 746-750. http://dx.doi.org/10.1038/nature00802
- Sava, G., Heikens, E. and Huebner, J. (2010) Pathogenesis and Immunity in Enterococcal Infections. Clinical Microbiology and Infection, 16, 533-540.http://dx.doi.org/10.1111/j.1469-0691.2010.03213.x
NOTES

*Corresponding author.


