Open Journal of Blood Diseases
Vol.3 No.3A(2013), Article ID:37445,6 pages DOI:10.4236/ojbd.2013.33A003
SEPT10 Expression in Chronic Lymphocytic Leukemia. Correlation with Clinical and Biological Prognostic Factors
![]()
1Laboratorio de Genética de Neoplasias Linfoides, Instituto de Medicina Experimental, CONICET-Academia Nacional de Medicina, Buenos Aires, Argentina; 2División Patología Molecular, Instituto de Investigaciones Hematológicas, Academia Nacional de Medicina, Buenos Aires, Argentina; 3Servicio de Hematología, Hospital General de Agudos “Dr Teodoro Álvarez”, Buenos Aires, Argentina.
Email: islavutsky@hematologia.anm.edu.ar
Copyright © 2013 Ana Travella et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received August 10th, 2013; revised September 10th, 2013; accepted September 18th, 2013
Keywords: Chronic Lymphocytic Leukemia; SEPT10 Expression; FISH; Cytogenetics; IGHV Mutational Status
ABSTRACT
Chronic lymphocytic leukemia (CLL) is characterized by a highly variable clinical course. Microarray studies allowed highlight genes differentially expressed in this pathology. In this study, we have evaluated the prognostic significance of SEPT10 expression in CLL patients. Results were correlated with immunoglobulin heavy-chain variable (IGHV) genes mutational status, genomic rearrangements and clinical parameters. SEPT10 mRNA levels were determined by quantitative real-time PCR in 70 newly diagnosed CLL patients consecutively referred to our Institution. A wide heterogeneity for SEPT10 expression was found. Gene upregulation was observed in 18.5% of cases. The univariate analysis showed a positive association between gen expression and platelet count (p < 0.0001) and a negative correlation with hemoglobin levels (p = 0.0094). Although no significant differences were observed, mean treatment free survival was shorter in patients with high expression (31 months) with respect to those with low mRNA levels (72 months). Cases with abnormal karyotypes had increased expression compared to those with normal karyotypes and no association between gene expression and FISH (fluorescence in situ hybridization) risk groups and IGHV mutational status was found. Cases using IGHV3-23 gene rearrangement had low SEPT10 expression. Our results showed an association between SEPT10 expression and features of adverse outcome but without independent prognostic value. The study of SEPT10 expression may be important for a better understanding of disease heterogeneity, adding further information to those provided by established prognostic factors.
1. Introduction
Chronic lymphocytic leukemia (CLL) is the most frequent type of adult leukemia in Western world. The disease is characterized by a highly variable clinical course, with time to progression ranging from months to decades [1]. Clinical staging systems have been very useful in guiding disease management and treatment decisions. However, these staging systems have shown a limited capacity to predict clinical outcome at an early stage of the disease. Based on this situation, a variety of biomarkers have been investigated as potential prognostic factors in CLL. They include genomic aberrations, mutational status of immunoglobulin heavy-chain variable (IGHV) genes and expression profiles of different genes. Particularly, fluorescence in situ hybridization (FISH) analysis allowed the identification of distinct cytogenetic risk groups, in which patients with deletion 13q14 as a single alteration have a better outcome, while patients with deletions 11q22 or 17p13 show the shortest median survival and, cases with trisomy 12 have an intermediate prognosis [2]. In addition, IGHV defines two disease subgroups: one expressing mutated (M) IGHV segments with a more favorable clinical course, and the other expressing unmutated (UM) IGHV segments, associated to a poor outcome [3,4].
More recently, microarray studies allowed the establishment of a gene expression profile characteristic of CLL which highlights differentially expressed genes in patients with stable or progressive disease. Among them, SEPT10 (septin 10), located at 2q13 band, has been proposed as a probable prognostic marker in CLL. This gene belongs to septins family, an evolutionarily conserved group of at least 14 distinct genes encoding GTP (guanosine triphosphate)-binding proteins and filament-forming proteins, involved in multiple cellular processes like vesicle trafficking, apoptosis, remodeling of the cytoskeleton, infection, neurodegeneration and neoplasia [5]. Many evidences have indicated that some septins were abnormally expressed in certain kinds of tumors and the altered expressions were related to the process of carcinogenesis [6]. In addition, mammalian septins modulate microtubule dynamics through the interaction with a microtubule associated protein, MAP4 [7], and particularly SEPT10 is an important regulator of micro-tubule stability [8]. In CLL, the literature showed that SEPT10 mRNA expression correlates with both high lipoprotein lipase (LPL) expression and UM IGHV gene status [9,10]. However, its association with other characteristics of the disease has been scarcely explored. In this study we have evaluated the prognostic significance of SEPT10 expression in CLL patients in relation to IGHV mutational status, genomic rearrangements and clinical parameters.
2. Material and Methods
2.1. Patients
Our cohort included 70 newly diagnosed CLL patients (43 males; median age: 65 years, range: 38 - 83 years) consecutively referred to our Institution. Patients were diagnosed according to the International Workshop on Chronic Lymphocytic Leukemia criteria [11]. Stage was assessed according to the Rai classification [12]. Clinical stages showed the following distribution: 0: 25 patients (36%), I: 10 (14%), II: 22 (31%), III: 7 (10%) and, IV: 6 (9%). All individuals provided their informed consent. The study was approved by the local Ethics Committee.
2.2. Cytogenetic and FISH Analysis
Chromosome analysis was performed on peripheral blood lymphocytes, cultured for 96 hours at 37˚C in F-12 medium supplemented with 15% of fetal calf serum, stimulated with Pokeweed mitogen and Lipopolysaccharide, or in presence of the immunostimulatory CpG-oligonucleotide (2 μM) and interleukin 2 (200 U/ml) for 72 hs. Slides were prepared by conventional method. G-banding technique was used.
For FISH analysis, slides were hybridized with LSI TP53/ATM/13q14/13q34/CEP12 DNA probes (VysisAbbott), according to manufacturer’s protocol. Two hundred interphase nuclei were analyzed for each probe. The cut-off for positive values (mean of normal control + 3 standard deviations), determined from samples of ten cytogenetically normal donors, were: 3.02%, 10.2%, 7.7% and 5.1% for trisomy 12, monosomies of D13S319 (13q14), ATM (11q22) and TP53 (17p13), respectively.
2.3. mRNA Extraction and SEPT10 Expression Analysis
Total RNA was extracted with Trizol reagent (Invitrogen, Buenos Aires, Argentina) from mononuclear cells isolated on a Ficoll-Paque Plus (GE Healthcare Bio-Sciences, Uppsala, Sweden) density gradient from peripheral blood samples of patients and healthy controls. The cDNA synthesis was carried out using Moloney Murine Leukemia Virus Reverse Transcriptase and random primer (Promega, Madison, WI, USA). SEPT10 mRNA levels were determined by quantitative real-time PCR (qRT-PCR) in a LightCyclerÒ 2.0 system (Roche Diagnostics, Mannheim, Germany), based on SYBR Green I methodology, and utilizing the Lightcycler software. Briefly, 1 μg of total RNA was amplified in the presence of 0.5 μM of specific primers for SEPT10 [10] or the housekeeping gene GAPDH (Glyceraldehyde 3-phosphate dehydrogenase) [13] with the following protocol: 10 min at 95˚C for initial denaturation, 50 cycles of 15 s at 95˚C, 10 s at 62˚C and 15 s at 72˚C, followed by the respective melting curve. All measurements included a determination of the standards and the use of a no-template as a negative control.
2.4. IGHV Mutational Status
The IGHV gene sequences were determined as previously described [14]. Briefly, amplification of IGHV regions by PCR was performed on cDNA using VH framework region 1 consensus family specific primers (VH1-VH6) and JH primers [15]. When amplifications of these primers were unsuccessful, an alternative set of primers that anneal to sequences in the leader region (LH1-LH6) and one antisense Cµ-primer [16] were used. Thermal cycling conditions were 3 min at 93˚C, followed by 33 cycles at 94˚C for 30 s, 62˚C for 30 s, 72˚C for 30 s, elongation at 72˚C for 7 min, and a final step at 4˚C for 10 min. PCR products were purified in 1% agarose gels, sequenced bi-directionally and analyzed on an automated DNA sequence analyzer (377 ABI Prism, PE Biosystem, Foster City, CA). Sequence data were analyzed using IgBLAST (http://www.ncbi.nlm.nih.gov/igblast) and the ImMunoGeneTics database (IMGT; http://imgt.cines.fr). IGHV sequences with less than 98% homology with respect to the germline counterpart were considered as M while those with homology of 98% or higher were classified as UM [3,4].
2.5. Statistical Analysis
Statistical evaluation of mRNA expression data was performed using the Mann-Whitney test. Correlations between gene expression and clinical variables were assessed by using the Kendall’s coefficient. The cut-off point for SEPT10 expression was selected according to receiver operating characteristic (ROC) analysis. Group wise comparison of the distribution of clinical and laboratory variables was performed with the Student t test (for quantitative variables) and the χ2 or Fisher’s exact test (for categorical variables). Overall survival and treatment free survival (TFS) were estimated by the Kaplan-Meier method and compared with the log-rank test.
3. Results
Analysis of data showed a wide heterogeneity for SEPT10 expression in CLL patients, with cases that overexpressed this gene and others with low levels of transcripts (Figure 1). For a better analysis, patients were divided into two groups: high expression (SEPT10high) and low expression (SEPT10low), according to the cut-off value (221.05) obtained by ROC curves. It was quite similar to the baseline value of SEPT10 expression observed in control samples with our approach (220.3 ± 107.5). Upregulation of SEPT10 was found in 13 cases (18.5%) (mean ± SE: 720.9 ± 190.5) while the remaining patients showed low levels of mRNA transcript (mean ± SE: 44.26 ± 6.99).
Furthermore, we would like to determine if SEPT10 expression profile had clinical significance. The analysis according to established prognostic factors in CLL did not show differences between patients with high and low gene expression, with the exception of age, that was reduced in SEPT10low patients compared to those with high mRNA levels (p = 0.006) (Table 1). The univariate analysis showed a positive association between gen expression and platelet count (p < 0.0001) and a negative correlation with hemoglobin levels (p = 0.0094). In reference to outcome, although no significant differences were observed, mean TFS was shorter in SEPT10high patients (31 months) with respect to those with low gene expression (72 months).
We also analyzed SEPT10 expression in relation to cytogenetic alterations, FISH risk groups and IGHV mu-

Figure 1. Scatter plot showing the wide distribution of SEPT10 mRNA levels in chronic lymphocytic leukemia (CLL) patients.
tational status, and a similar distribution of patients in the different categories was observed (Table 2). However, when SEPT10 mRNA levels were evaluated, patients with abnormal karyotypes had increased expression compared to those with normal karyotypes (Figure 2) and no association between gene expression and FISH risk groups and IGHV mutational status were found. Expression levels were also evaluated in different IGHV families and a similar distribution among VH1, VH3 and VH4 was observed. More detailed analysis of IGHV segments usage showed that IGHV3-23 gene rearrangement was the most represented in our cohort (7 cases); six of them (85.7%) showing low SEPT10 expression, as well as the only one case of our series using IGHV3-21 segment.
4. Discussion
CLL is characterized by a heterogeneous clinical course, with some patients experiencing rapid disease progression and others living for decades without requiring treatment. However, classical prognostic parameters have a limited capacity to predict clinical outcome at an early stage of the disease. In recent years, numerous genetic approaches have provided new markers for prognosis and response prediction [17]. Among them, a considerable number of genes differentially expressed in CLL were identified, including SEPT10 [18,19], which role as prognostic factor in this pathology was scarcely evaluated [9,10]. In this study, we have analyzed SEPT10 mRNA expression in a cohort of newly diagnosed CLL patients. Our series showed a low percentage of patients with SEPT10 overexpression (18% of cases), value close to the 27% observed by Van’t Veer et al. [10], indicating that deregulation of this gene is limited to a specific subgroup of patients. Besides, expression analysis showed a wide heterogeneity of mRNA levels, which could be related to variations in expression control at the transcripttional, post-transcriptional and translational levels or
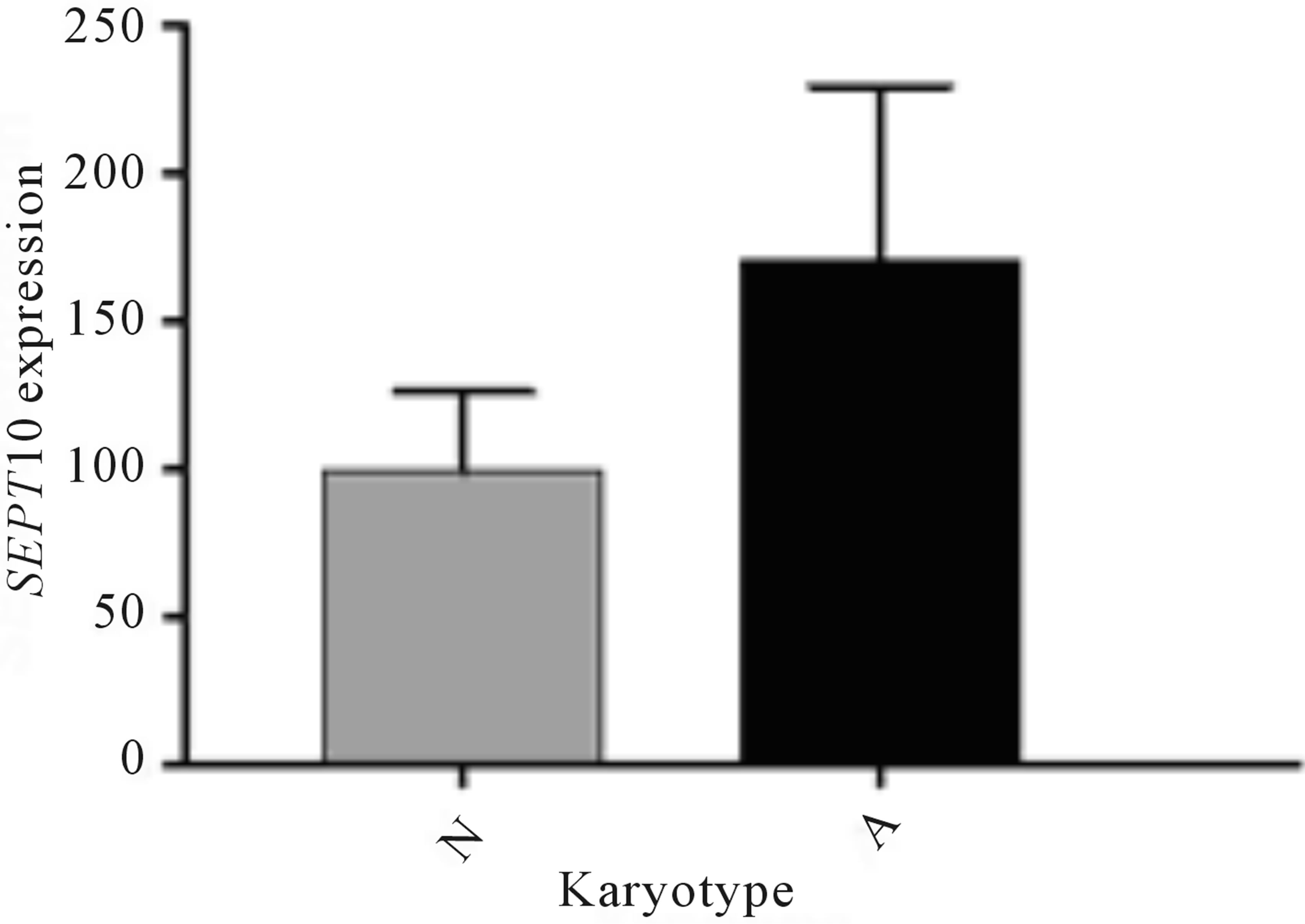
Figure 2. SEPT10 mRNA expression profile in chronic lymphocytic leukemia patients with normal (N) and abnormal (A) karyotypes.

Table 1. SEPT10 gene expression according to the known prognostic factors in CLL.

Table 2. SEPT10 gene expression according to cytogenetic, FISH and IGHV groups.
associated to the presence of alternative splice variants [5]. Citoquines or mitogens that induce proliferation and differentiation through different signaling pathways may also influence the activity of this gene.
As previously referred, clinical significance of SEPT10 mRNA expression was scarcely evaluated in the literature [9,10,20]. In this aspect, our results add new information about it. Thus, we found association between SEPT10 expression and age, platelet count and hemoglobin levels. In addition, although no significant differences were found, longer TFS was observed in our SEPT10low subgroup, supporting a previous report showing that patients with high SEPT10 expression required earlier treatment [9]. Simultaneously, we have evaluated for the first time the association between cytogenetic alterations and SEPT10 expression and cases with abnormal karyotypes had a non-significant SEPT10 upregulation compared to those with normal karyotypes. These findings are of interest taking into account the adverse clinical impact of structural abnormalities and complex karyotypes in CLL [21-23], suggesting a relationship between SEPT10 expression and accumulation of chromosome alterations in leukemic cells. In addition, we did not observe correlations between SEPT10 levels and FISH risk groups, coincident with the sole report in the literature that analyzed this aspect [10].
Unlike to those observed in the literature [9,10,20], we did not find association between SEPT10 expression and IGHV mutational status. A study in a larger series may be necessary to clarify this point. In reference to IGHV segment usage, association between low SEPT10 mRNA levels and IGHV4-34 and high expression with IGHV1- 69 gene rearrangement, were reported [10]. Although the number of our cases is not large, an association between IGHV3-23 and low expression was observed. Interestingly, different authors [10,24] have detected lower SEPT10 expression in IGHV3-21 expressing CLL, suggesting the inability of SEPT10 expression to recognize the poor prognosis of this CLL subset. The only one case of our series expressing IGHV3-21 segment supports these findings.
In conclusion, our results showed a wide heterogeneity in the expression profile of SEPT10 with overexpression in a subgroup of CLL patients. The correlation with clinical characteristics showed association with features of adverse outcome but without independent prognostic value. Thus, the study of this gene may be important for a better understanding of the disease variability, and could be used in combination with other markers, adding further information to those provided by established prognostic factors. In this line, an association between SEPT10 and LPL [9], which overexpression was related to adverse clinical outcome in CLL [25], have been previously described, suggesting that SEPT10 could direct LPL to the cell surface by establishing a link between proteoglycans (the cell surface LPL receptor) and the actin cytoskeleton [5,9]. The understanding of biological mechanism of the disease will contribute to future risk stratification strategies with potential value in treatment decisions.
5. Acknowledgements
This work was supported by grants from the National Research Council (CONICET) and the National Agency of Scientific and Technical Promotion (ANPCyT), Argentina.
REFERENCES
- N. Chiorazzi, K. R. Rai and M. Ferrarini, “Chronic Lymphocytic Leukemia,” New England Journal of Medicine, Vol. 352, No. 8, 2005, pp. 804-815. http://dx.doi.org/10.1056/NEJMra041720
- H. Döhner, S. Stilgenbauer, A. Benner, E. Leupolt, A. Kröber, L. Bullinger, et al., “Genomic Aberrations and Survival in Chronic Lymphocytic Leukemia,” New England Journal of Medicine, Vol. 343, No. 26, 2000, pp. 1910-1916. http://dx.doi.org/10.1056/NEJM200012283432602
- R. N. Damle, T. Wasil, F. Fais, F. Ghiotto, A. Valetto, S. L. Allen, et al., “Ig V Gene Mutation Status and CD38 Expression as Novel Prognostic Indicators in Chronic Lymphocytic Leukaemia,” Blood, Vol. 94, No. 6, 1999, pp. 1840-1847.
- T. J. Hamblin, Z. Davis, A. Gardiner, D. G. Oscier and F. K. Stevenson, “Unmutated Ig VH Genes Are Associated with a More Aggressive Form of Chronic Lymphocytic Leukaemia,” Blood, Vol. 94, No. 6, 1999, pp. 1848-1854.
- P. A. Hall, K. Jung, K. J. Hillan and S. E. Russell, “Expression Profiling the Human Septin Gene Family,” Journal of Pathology, Vol. 206, No. 3, 2005, pp. 269-278. http://dx.doi.org/10.1002/path.1789
- M. Liu, S. Shen, F. Chen, W. Yu and L. Yu, “Linking the Septin Expression with Carcinogenesis,” Molecular Biology Reports, Vol. 37, No. 7, 2010, pp. 3601-3608. http://dx.doi.org/10.1007/s11033-010-0009-2
- B. E. Kremer, T. Haysted and I. G. Macara, “Mammalian Septins Regulate Microtubule Stability through Interaction with the Microtubule-Binding Protein MAP4”, Molecular Biology of the Cell, Vol. 16, No. 10, 2005, pp. 4648-4659. http://dx.doi.org/10.1091/mbc.E05-03-0267
- M. Xu, M. Takanashi, K. Oikawa, H. Nishi, K. Isaka, T. Yoshimoto, et al., “Identification of a Novel Role of Septin 10 in Paclitaxel-Resistance in Cancers through a Functional Genomic Screen,” Cancer Science, Vol. 103, No. 4, 2012, pp. 821-827. http://dx.doi.org/10.1111/j.1349-7006.2012.02221.x
- M. Bilban, D. Heintel, T. Scharl, T. Woelfel, M. M. Auer, E. Porpaczy, et al., “Deregulated Expression of Fat and Muscle Genes in B-Cell Chronic Lymphocytic Leukemia with High Lipoprotein Lipase Expression,” Leukemia, Vol. 20, No. 6, 2006, pp. 1080-1088. http://dx.doi.org/10.1038/sj.leu.2404220
- M. B. Van’t Veer, A. M. Brooijmans, A. W. Langerak, B. Verhaaf, C. S. Goudswaard, W. J. Graveland, et al., “The Predictive Value of Lipoprotein Lipase for Survival in Chronic Lymphocytic Leukemia,” Haematologica, Vol. 91, No. 1, 2006, pp. 56-63.
- M. Hallek, B. D. Cheson, D. Catovsky, F. Caligaris-Cappio, G. Dighiero, H. Döhner, et al., “Guidelines for the Diagnosis and Treatment of Chronic Lymphocytic Leukemia: A Report from the International Workshop on Chronic Lymphocytic Leukemia Updating the National Cancer Institute-Working Group 1996 Guidelines,” Blood, Vol. 111, No. 12, 2008, pp. 5446-5456.
- K. R. Rai, A. Sawitsky, E. P. Cronkite, A. D. Chanana, R. N. Levy and B. S. Pasternack, “Clinical Staging of Chronic Lymphocytic Leukemia,” Blood, Vol. 46, No. 2, 1975, pp. 219-234.
- L. H. Hu, F. H. Chen, Y. R. Li and L. Wang, “Real-Time Determination of Human Telomerase Reverse Transcriptase mRNA in Gastric Cancer,” World Journal of Gastroenterology, Vol. 10, No. 23, 2004, pp. 3514-3517.
- C. Stanganelli, A. Travella, R. Bezares and I. Slavutsky, “Immunoglobulin Gene Rearrangements and Mutational Status in Argentinean Patients with Chronic Lymphocytic Leukemia,” Clinical Lymphoma Myeloma and Leukemia, Vol. 13, No. 4, 2013, pp. 447-457. http://dx.doi.org/10.1016/j.clml.2013.02.019
- O. Pritsch, C. Magnac, G. Dumas, C. Egile and G. Dighiero, “V Gene Usage By Seven Hybrids Derived From CD5+ B-Cell Chronic Lymphocytic Leukemia and DisPlaying Autoantibody Activity,” Blood, Vol. 82, No. 10, 1993, pp. 3103-3112.
- M. Campbell, A. Zelenetz, A. Levy and R. Levy, “Use of Family Specific Leader Region Primers for PCR Amplification of the Human Heavy Chain Variable Region Gene Repertoire,” Molecular Immunology, Vol. 29, No. 2, 1992, pp. 193-203. http://dx.doi.org/10.1016/0161-5890(92)90100-C
- C. Moreno and E. Montserrat, “New Prognostic Markers in Chronic Lymphocytic Leukemia,” Blood Reviews, Vol. 22, No. 4, 2008, pp. 211-219. http://dx.doi.org/10.1016/j.blre.2008.03.003
- A. Rosenwald, A. A. Alizadeh, D. Widhofp, R. Simon, R. E. Davis, X. Yu, et al., “Relation of Gene Expression Phenotype to Immunoglobulin Mutation Genotype in B Cell Chronic Lymphocytic Leukemia,” Journal of Experimental Medicine, Vol. 194, No. 11, 2001, pp. 1639- 1647. http://dx.doi.org/10.1084/jem.194.11.1639
- U. Klein, Y. Tu, G. A. Stolovitzky, M. Mattioli, G. Cattoretti, H. Husson, et al., “Gene Expression Profiling of B Cell Chronic Lymphocytic Leukemia Reveals a Homogeneous Phenotype Related to Memory B Cells,” Journal of Experimental Medicine, Vol. 194, No. 11, 2001, pp. 1625-1638. http://dx.doi.org/10.1084/jem.194.11.1625
- D. Kienle, A. Benner, C. Läufle, D. Winkler, C. Schneider, A. Bühler, et al., “Gene Expression Factors as Predictors of Genetic Risk and Survival in Chronic Lymphocytic Leukemia,” Haematologica, Vol. 95, No. 1, 2010, pp. 102-109. http://dx.doi.org/10.3324/haematol.2009.010298
- C. Mayr, M. R. Speicher, D. M. Kofler, R. Buhmann, J. Strehl, R. Busch, et al., “Chromosomal Translocations Are Associated with Poor Prognosis in Chronic Lymphocytic Leukemia,” Blood, Vol. 107, No. 2, 2006, pp. 742- 751. http://dx.doi.org/10.1182/blood-2005-05-2093
- E. Van Den Neste, V. Robin, J. Francart, A. Hagemeijer, M. Stul, P. Vandenberghe, et al., “Chromosomal Translocations Independently Predict Treatment Failure, Treatment-Free Survival and Overall Survival in B-Cell Chronic Lymphocytic Leukemia Patients Treated with Cladribine,” Leukemia, Vol. 21, No. 8, 2007, pp. 1715-1722. http://dx.doi.org/10.1038/sj.leu.2404764
- A. Travella, L. Ripollés, A. Aventin, A. Rodríguez, R. F. Bezares, M. R. Caballín and I. Slavutsky, “Structural Alterations in Chronic Lymphocytic Leukaemia. Cytogenetic and FISH Analysis,” Hematological Oncology, Vol. 31, No. 2, 2013, pp. 339-347. http://dx.doi.org/10.1002/hon.2025
- D. Benedetti, R. Bomben, M. Dal-Bo, D. Marconi, A. Zucchetto, M. Degan, et al., “Are Surrogates of IGHV Gene Mutational Status Useful in B-Cell Chronic Lymphocytic Leukemia? The Example of Septin-10,” Leukemia, Vol. 22, No.1, 2008, pp. 224-226. http://dx.doi.org/10.1038/sj.leu.2404867
- M. A. Kaderi, M. Kanduri, A. M. Buhl, M. Sevov, N. Cahill, R. Gunnarsson, et al., “LPL Is the Strongest Prognostic Factor in a Comparative Analysis of RNA-Based Markers in Early Chronic Lymphocytic Leukemia,” Haematologica, Vol. 96, No. 8, 2011, pp. 1153-1160. http://dx.doi.org/10.3324/haematol.2010.039396

