World Journal of Cardiovascular Diseases
Vol.07 No.10(2017), Article ID:79712,18 pages
10.4236/wjcd.2017.710032
MicroRNA and Its Role in Cardiovascular Disease
Suvash Pokhrel1, Yin Guotian2*
1Department of Internal Medicine, Third Affiliated Hospital of Xinxiang Medical University, Xinxiang, China
2Department of Cardiology, Third Affiliated Hospital of Xinxiang Medical University, Xinxiang, China
Copyright © 2017 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: September 4, 2017; Accepted: October 15, 2017; Published: October 19, 2017
ABSTRACT
MicroRNAs play a key role in regulation of gene expression during cardiac development and cardiac remodeling. MicroRNAs that present in bodily fluids may be useful for screening, diagnosis or therapeutic implication as a treatment. MicroRNAs are relatively new approach targets for researchers and clinicians in today world. MicroRNAs are small noncoding RNA (ncRNA) having approximately 21 to 25 nucleotides in length, and they mainly act as a transcriptional regulators of gene expression in diverse biological processes such as cellular proliferation, differentiation, tumorigenesis to death and so on. There is no doubt that lethiferous cardiac disease is one of the most common causes of deaths worldwide. MicroRNAs may regulate in several cardiovascular pathologies, not only limited to hypertrophy, heart failure, arrhythmias, hypertension, myocardial infarction, dyslipidemias and congenital heart diseases, but in circulation and bodily fluids are potential novel biomarkers for above mentioned cardiac pathologies. Knowing abnormalities in genetic level, early and accurate detection, effective treatment and prevention is the ideal management of cardiovascular diseases in today’s world. However, every detail of an individual microRNA and their system is huge and beyond the scope of this article. Therefore, in this review we try to cover the overall major aspects of the microRNAs and its role in cardiovascular system.
Keywords:
MicroRNA, Noncoding RNAs, Cardiovascular Disease

1. Introduction
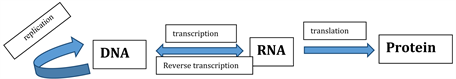
Since the discovery of Deoxyribonucleic acid (DNA) double helix structure by Watson and Crick in 1953 [1] , the regulation of gene expression may occur through the following process as shown in Figure 1.
Transcription is the first stage of gene expression in which information contained in double standard DNA molecule gets transfer into single standard Ribonucleic acid (RNA) molecule. Messenger RNA (mRNA) translates the information in the RNA base sequence to the amino acid sequence of a protein through the process of translation [2] .
With the completion of Human genome project in 2003 till now, it has been concluded that human genome contains approx. 20,000 to 25,000 protein coding genes, more appropriately 22,333 which corresponds to the total current protein coding gene sequences are available on National center for biotechnology information reference sequence (NCBI’s RefSeq) database [3] .
More interestingly it has been found, only less than 2% of RNA transcripts are protein coding RNAs (mRNA) and remaining are noncoding RNAs (ncRNAs) [4] .
The term ncRNA is commonly used for RNA that does not encode a protein, but they have diverse functions from mRNA splicing and RNA modifications to translational regulation. This hidden layer of gene regulation has been uncovered recently which has brought the tremendous research opportunity in the field of human genetics and molecular biology [5] .
Non coding RNAs are broadly divided into two main groups; the short noncoding RNAs (sncRNAs) which have less than 30 nucleotide base pairs and long noncoding RNAs (lncRNAs) which have more than 200 nucleotide base pairs. Examples of short noncoding RNAs are microRNAs (miRNAs), short interfering RNAs (siRNAs), and piwi-interacting RNAs (piRNAs). Long noncoding RNAs are long intergenic ncRNAs (lincRNAs), braveheart, Fendrr, transcribed ultra- conserved regions (T-UCRs), pseudogenes and antisense RNAs [6] [7] .
Micro RNAs are the most widely studied and well known classes of ncRNAs which are only ~21 to 25 nucleotides (nt) in length and they function through expression in diverse biological processes acting as a transcriptional regulators of gene expression including cellular proliferation, differentiation, tumorigenesis to death [8] . Primary transcript of miRNA is cleaved by enzyme nuclear RNase III also called Dorsha which produces 70 - 90 nt stem-loop structured pre-miRNAs and then it transports into cytoplasm by exportin [9] (Figure 2).
Mature miRNAs having ~22 nt are cleaved from 70 - 90 nt hairpin pre miRNA precursors by RNase III Dicer in the cytoplasm and finally incorporated into the RNA-induced silencing complex which are regarded as negative regulators of gene expression by inhibiting translation and/or promoting mRNA degradation by base pairing to complementary sequences within the 3' UTR region of protein-coding mRNA transcripts [10] [11] [12] . Recently it has been found that
Figure 1. Central dogma of molecular biology.
Figure 2. miRNA synthesis.
miRNA-binding sites are also located in 5' UTRs [13] [14] thereby regulating translation process. MicroRNAs can also up-regulate gene expression, likely via the suppression of transcriptional repressors [15] .
The first miRNA, lin-4 which reduces the amount of Lin 14 protein was discovered in 1993 by Lee et al. in Caenorhabditis elegans. Lin-4 binds to its complementary sequences element in 3’-UTR of Lin 14 mRNA, thereby negatively regulating Lin-14 translation [16] . Mutation in Lin14 gene causes elimination of larval stage specific cellular events in C.elegans [17] .
The second miRNA or the first miRNA in Human let-7 was discovered in 2000 with its target protein lin-41 [18] . Unlike first miRNA, let-7 is highly conserved in many species including human which is regulated in transcriptional and post transcriptional level. Let-7 recognizes 3’UTR in lin-41 gene and binds to it and hence down regulates lin-41 translation. Lin-41 is associated with different protein ubiquitin ligase in an autoubiqui-tylation assay [19] . Additionally mouse lin-41 is found to be more pronounced during early development of mouse limb bud, nervous tissue and craniofacial tissue [20] .
Let-7 family of miRNAs also found to modulate immune response against opportunistic human pathogen like Pseudomonas aeruginosa [21] . Since the discovery of lin-4 and let-7, a lot of attention of researchers has been found towards this new RNA revolution. According to the “miRBase” till now 2588 mature miRNA exists in human targeting thousands of genes within the body [22] . Among these different miRNA, they function as cell or tissue specific manner resulting in normal development of cell, tissue and an organ to their disease progression upon alteration of miRNA hemostasis [23] so as heart issue is of no exception.
This review mainly focuses on recent development on microRNAs related to various aspect of cardiac system from development, remodeling and diagnostic to disease progression.
2. MicroRNA in Cardiac Development and Physiology
From the embryonic period to fully developed heart, miRNAs are constantly found in cardiac tissues. miR-21, miR-29a, miR-129, miR-210, miR-211, miR- 320, miR-423 and let-7c are highly expressed in normal fetal heart tissue which plays a major role in embryonic and post-natal cardiac development [24] .
Decreased level of above mentioned miRNAs are due to depleted level of Dicer, an enzyme whose role is essential for miRNA processing lead to pericardial edema, poorly developed ventricular myocardium and rapid lethality in mouse models. Similarly Tamoxifen inducible Cre recombinase deletion of dicer causes heart failure and death in adult heart [25] . In other word inhibition of miRNA expression is due to dicer deletion. Deletion of dicer during embryonic phase resulted in hypo plastic ventricular muscle, pericardial edema [26] . Post-natal deletion of dicer in heart muscle cell may result in cardiomyocyte contractility dysfunction and disarray which ultimately result in dilated cardiomyopathy, heart failure and related consequences [27] .
miRNA 1 and miRNA 133 plays a crucial role in cardiac development which has been proved experimentally in mice model. miRNA-1 (miR 1-1, miR 1-2) and miR-133 (miR 133a-1, miR 133a-2 and miR 133b) and miR-206 are muscle specific microRNA which are regulated by different enzymes in different tissue. miR-1 is regulated by serum response factor (SRF) in heart whereas by Myogenic differentiation 1 (MyoD 1) and myocyte enhancer factor 2 (Mef 2) in skeletal muscles [28] . Main functions of miRNA-1 on heart development are favoring of apoptotic mechanisms [29] and inhibition of cardiac development [30] .
miRNA-1 targets the Hand2, a transcription factor which favors the proliferation of cardiomyocytes in zebra fish embryo heart [31] . This suggest during cardiac development rise in miRNA-1 level causes decrease in Hand2 transcription factor level and for a healthy heart this need to balance in a miRNA homeostasis. If there is excessive increase in miRNA-1 in early embryogenesis, it may lead to dysregulation of cardiac myocyte proliferation, myoblast differentiation, ventricular septal defect (VDS) by interfering cardiac morphogenesis [32] [33] .
Histone deacetylates 4 (HDAC4) is another transcriptional repressor protein of muscle gene expression and miR-1 targets this protein and promotes myogenesis [15] . The role of miR-133 in cardiac development is as a regulator of cardiogenesis. It acts on Nelf-A/WHSC2 which is a nuclear factor involved in cardiogenesis [34] . It has been found that loss of miR-1 and miR-133 lead to ventricular septal defect (VSD) and cardiac chamber dilatation which resulted in embryonic and neonatal lethality [35] . miR-196a which is also found in human heart is found to be associated with HOXB8-Sonic hedgehog signaling pathway and this pathway is important for cardiac septation, valve formation and outflow tract morphogenesis [29] .
3. MicroRNA in Cardiac Remodeling
The term “remodeling” is adaption in response to any chronic stress in heart, for example chronic hypertension can lead to hypertrophy in cardiac myocytes which is required to maintain adequate cardiac output. However if cardiac remodeling process remains for long duration it may lead to heart failure, myocardial infarction and other cardiac disease [24] .
miR-1 and miR-133 are found to be downregulated in a heart where cardiac remodeling processing is active especially in pressure overload-induced hypertrophy, Akt overexpression-induced hypertrophy and adaptive cardiac hypertrophy [36] .
Calcium dependent gene expressions are crucial for the myocyte growth and differentiation. Calmodulin, Mef2a and Gata4 are the main calcium dependent transcription factors and miR-1 inhibits these transcription factors as shown by a study in a mice model [37] . miR-133 regulates transcription factors like RhoA, a GDP-GTP exchange protein, Cdc42 which are associated with cardiac hypertrophy and Nelf-A/WHSC2 related to cardiogenesis [34] . Other classes of miRNA like miR-208, miR-195, miR-214, and miR-21 are over expressed in hypertrophied heart or simply upregulated [38] . During pressure induced stress, cardiac specific intron of alpha myosin heavy chain (αMHC6) gene 6 encodes a cardio specific miR-208 and then miR-208 further upregulates beta myosin heavy chain thereby promoting cardiac hypertrophy [39] [40] .
Recent works reported by Bang et al. [41] showed that in response to stress oncadio fibroblast secrete miRNA enriched exosomes, miR-21 also called “star” microRNA which are a potent paracrineacting RNA molecule that responsible for cardiomyocyte hypertrophy. If miR-195 is over expressed, it may leads to cardiac hypertrophy. Upregulation of microRNA-195 in failing heart can result in dilated cardiomyopathy as suggested by Rooij et al. in transgenic mice model [42] .
4. MicroRNA in Cardiac Disease
4.1. Arrhythmia
miRNA levels are found to be altered from its normal hemostasis in both atrial or ventricular arrhythmia. Both upregulation and down regulation of miR-1 may cause arrhythmic condition especially in ventricle and atria respectively.
In Ventricular arrhythmia, miR-1 represses KCNJ2 and GJA1. KCNJ2 encodes Kir2.1 which is a potassium channel subunit and GJA1 encodes connexin 43 which is a cardiac gap junction. This leads to slowed conduction and depolarization of cytoplasmic membrane which has arrhythmogenic potential and elimination of miR-1 has shown to improvement in arrhythmic potential in infarcted rat hearts. This has opened a window for future anti arrhythmic therapy targeting miR-1 [43] .
Another mechanism by which overexpression of miR-1 causes arrhythmia is by CaM-KII-dependent phosphorylation of the L-type Ca2+ channels and ryanodine receptors 2 (RyR2) leading to burst of calcium from sarcoplasmic reticulum responsible for arrhythmia [44] .
A study by Zhao et al. suggested that homozygous mutant for miR-1-2 in mice have high incidence of electro physio abnormalities resulting sudden cardiac death due to repression of potassium channel subunit which is involved in transient K+ current out flux [26] . miR-328 is associated with atrial fibrillation. Study by Lu et al. [45] found that miR-328 have found to be elevated by 3.5 fold in atrial fibrillation patient in comparison to those without and it opened the door for potential therapeutic for atrial fibrillation targeting miR-328. miR-1 levels are found to be reduced in atrial fibrillation. Reduced level of miR-1 leads to increased inward potassium current and this is responsible for the generation of arrhythmia in atrial muscle.(46).
4.2. Hypertension
Role of miRNA in blood pressure regulation has been investigated through several studies in recent years.
miR-155 negatively regulates angiotensin II type 1 receptor expression at the post transcriptional level. Increased level of miR-155 is found to be associated with decreased blood pressure by binding to Angiotension II type 1 receptor at 3’-UTR [47] .
An hypothesis proposed by Bähring et al. [48] suggest that autosomal-domi- nant hypertension and brachydactyly, a multiphenotype Mendelian syndrome, are caused by a chromosomal rearrangement on 24p which features multiple splicing and noncoding RNA resembling miRNA sequence. This gives rays to scientists for future study opportunity on hypertension related to miRNA. The role of miRNA in hypertension has further supported by Marques et al. [49] in which they establish the relationship between renin mRNA over expression and miRNA in human hypertensive kidney. Their discovery found that downregulation of 2 miRNAs that bind to and affect reninmRNA levels. Out of 850 miRNAs they screened two miRNAs; miR-181a and miR-663 are responsible for downregulation of renin expression in human kidney.
Chronic Ang II-mediated hypertension in rats increases miR-132/-212 in aorta, heart and kidney. Eskidsen et al. [50] infused Angiotensin II in rat for 10 days and they have reported increased in blood pressure, cardiac hypertrophy and fibrosis in rat model. Similarly level of miR-132, miR-212 were found to be down regulated in the internal mammary artery of patients treated with angiotensin II receptor type 1 blocker.
Relationship between regulation of 11-β hydroxysteroid dehydrogenase type 2 (11β-HSD2) and miRNA has recently been done by Rezaei et al. [51] . 11β-HSD2 is responsible for conversion of 11β-hydroxy glucocorticoids to inactive 11-ke- tosteroid and also inactivation of mineralocorticoid receptor in human by cortisol. In this study they found that overexpression of miR-20a reduces 11β HSD2 activity thereby facilitating the mineralocorticoid effect especially in salt sensitive hypertension.
4.3. Heart Failure
No doubt that heart failure remains the significant burden to modern society and treating physician with leading cause of hospital admission in adults over the age of 65. According to Health Management Organization, Kaiser Permanente Coronary artery disease, hypertension, diabetes mellitus, atrial fibrillation and valvular heart disease are the five major factors contributing heart failure [52] .
With the advancement in miRNA study, it has raised the possibility of its appearances in heart failure cases using circulating miRNAs are biomarkers. Study by Goren et al. [53] found that level of specific miRNAs like miR-423-5p, miR- 320a, miR-22 and miR-92b are found to be elevated in patient with systolic heart failure. Further study by Devaux et al. [54] revealed that panels of 4 miRNAs namely miR-16, miR-27a, miR-101, and miR-150 are associated with improved prediction of left ventricular contractility six months after acute myocardial infarction (AMI).
Among various miRNAs, expressions of miR-122/423-5p/210/499/622 are found to be increased in patient with heart failure and are liked to more than one study [24] . The significance of increased level of miR-423-5p has been further supported by Tijsen et al. [55] where 6 other miRNAs were found to be elevated but miR-423-5p was strongly correlated with clinical diagnosis of heart failure. miR-1 plasma levels were found to be elevated in acute myocardial induced heart failure (AMI-HF) [56] .
A recent study done in myocardial biopsy of 34 Chinese patients with recent heart failure and then compared it with a group of patient without heart failure undergoing routine cardiac surgery. It has been found that miR-1, -21, -23, -29, -130, -195 and -199 were high in the heart failure group [57] . It has to be noted that other miRNAs who are responsible for cardiac hypertrophy and remodeling are also contributing factor in heart failure. More consistent RNA profiles in heart failure cases needed to be developed and it demands more studies in large populations which shall be overcome soon in near future and microRNA profiling in case of heart failure shall be mature in coming days.
4.4. Myocardial Infarction
Myocardial infarction, being a medical emergency is leading cause of deaths and long term complication worldwide. Early diagnosis and therapeutic intervention can significantly reduce its mortality and future risk and complications.
Post infarct patient can go into significant cardiac remodeling depending upon infarct size. As suggested by da costa et al. [25] , disruption of miRNA genesis due to deletion of dicer can lead to significant maladaptive cardiac remodeling leading to heart failure. Myocardial infarction itself is a long term hypoxic environment to cardiomyocyte. Wu et al. [58] described the role of hypoxia in miRNA mediated gene silencing potentiation via post translational modification of Argonaute-2. Hypoxia causes change in gene expression of miR-210 and miR-181b through hypoxia-inducible factor-1 (HIF-1) whereas Ago-2 causes dicer independent cleavage of precursor miR-451 to mature miR- 451. The cardio protective Ischemic preconditioning (IPC) in murine heart is associated with increased level of miR-144/451 which modulates reactive oxygen species (ROS) production by targeting Rac-1 which is a key component of Nicotinamide adenine dinucleotide phosphate oxidase (NADPH oxidase) [59] .
Cardiac specific miRNAs, miR-208b and miR-499 were found to be elevated in circulation with 1600 and 100 fold respectively in patient with acute myocardial infarction when comparing it with the control. However, leukocyte specific miRNAs were not increased in significant amount [60] . Further to this, the role of miR-499 in AMI diagnosis has been supported by Adachi et al. in which they measured the plasma concentration of miR-499 in 14 patients with acute coronary syndrome, 15 patients with congestive heart failure (CHF) and 10 patients without any cardiovascular disease. They concluded that miR-499 was exclusive to cardiac tissue with significant increase in plasma level following acute myocardial infarction (AMI) than below the limit of detectable range in plasma with other groups mentioned above [61] .
Elevated level of miR-1 and miR-133a were found to be present in acute MI, unstable angina pectoris and takotsubo cardiomyopathy without elevation of serum cardiac troponin or serum creatinine phosphokinase (CPK). So it can be used as a marker of cardiomyocyte injury or death [62] . Future risk of heart failure (HF) in patient who suffered acute myocardial infarction can be measured using miRNAs as an indicator. For this Matsumoto et al. [63] found that patient who suffered heart failure after acute myocardial infarction had elevated p53 responsive microRNAs particularly miR-34a, miR-192, miR-194 in their early convalescent stage of AMI.
miR-15 family, containing 6 related miRNAs, are upregulated in the infarcted region of the heart in response to ischemia-reperfusion injury in mice and pigs and upon successful therapy with locked nucleic acid (LNA)-modified anti-miR-15, cardiac infarct size and cardiac remodeling has found to be reduced. This has opened the door for potential therapeutic implementation of anti-miR therapy in myocardial infarction (MI) [64] . Post myocardial infarction induced heart failure is a vulnerable environment for atrial fibrillation and atrial remodeling. On one recent study by Cardin et al. [65] establish the relationship between atrial remodeling and level of miR-21 in a post infarct in experimental rat model. Knockdown of miR-21 expression in atrial muscle leads to prevention of atrial fibrillation and remodeling. This further opens a door for potential future therapy/intervention designed to prevent atrial fibrillation (AF).
The therapeutic role of miR-24 in cardiac fibrosis and remodeling after myocardial infarction has been recently described by Wang et al. [66] . miR-24 expression is down regulated after post myocardial infarction. They described that in vivo delivery of lentivirus miR-24 can interact with tissue growth factor β1 (TGF-β1) to decrease the cardiac fibrosis which resulting from various heart conditions like MI, cardiomyopathies and heart failure.
4.5. Atherosclerosis
Atherosclerosis is a complex and chronic inflammatory interaction of blood vessel wall with altered lipoprotein monocyte-derived macrophages, T cells, and the normal arterial elements. It is one of the most common causes of deaths in western society [67] .
Patient with coronary artery disease have reduced expression of different miRNAs like miR-126, miR-145, miR-155 in their blood [68] . The detail role of microRNAs 126/145/ and 155 in vivo have been described by Wei et al. [69] . They found that downregulation of miR145 in smooth muscle cell disrupts smooth muscle cell normal hemostasis and favors atheroma formation. Similarly upregulation of miR-155 in macrophage improves advanced atherosclerosis and miR-126 reduces atheroma lesion formation by increasing lesional CXCL12 expression which has atheroprotective effect.
miR-33 which is located in SREBF2 gene is responsible for reduce expression of cholesterol transporter ABC transporter A1 (ABCA1) which leads to decreased level of High density lipoprotein (HDL), a good cholesterol. Similarly anti miR-33 therapy has found to be atheroprotective by increasing circulating HDL in mice model [70] . A study done by Gao et al. [71] in 355 Chinese patients with hyperlipidemia found that level of miR-122 and miR-370 were positively correlated with total cholesterol, Triglycerides and Low density lipoprotein cholesterol C (LDL-C) and coronary artery disease (CAD) progression.
4.6. Congenital Heart Disease
miRNA plays important role from embryogenesis to fully developed adult heart. Several congenital cardiac defects have been found to be associated with altered expression of miRNA. Alteration of miR-1and miR-181c levels is found to be associated with Ventricular septal defects (VSD). In human presenting with VSD, their cardiac cells show decreased expression of miR-1 and increased expression of miR-181c [72] [73] . Congenital heart disease in Down syndrome patients have been found to have over expressed 5 major miRNAs namely miR-99a, let-7c, miR-125b-2, miR-155 and miR-802 in chromosome 21. [74] .
Several cadio-facial neural crest defects may cause due to abnormalities in neural crest cell migration and dicer deletion, which is mainly responsible for the miRNA maturation. Noonans Syndrome, DiGeorge Syndrome, LEOPARD syndrome, cardio-facio-cutaneous syndrome and Costello syndrome are some of the examples for such diseases [72] .
Tetralogy of Fallot (TOF) is the most common congenital cyanotic heart disease. The association of TOF with miRNAs has been recently reported by O’Brien et al. [75] in 33 infant with non-syndromic TOF without a 22q11.2 deletion. They found that 33 miRNAs were down regulated in patients who have non syndromic TOF. Again in a separate study in 2015 by this group found that over expression of miR-421 in ventricular cell is associated with TOF [76] .
In a study, by Liang et al. [77] concluded that miRNA-940 is most significantly down regulated in patients with TOF. Bicuspid aortic valve being a common congenital valvular anomaly, miR-26a, miR-30b and miR-195 levels were found to be low in a study by Ni-gam et al. [78] . Two years later, study by Yanagawa et al. [79] reveled that miR-141 is a key regulator of bone morphogenetic protein 2 (BMP-2 dependent) dependent aortic valve calcification. miR-141 was found to be downregulated in patient with bicuspid aortic valve.
4.7. Circulating miRNA as a Diagnostic Indicator of Cardiovascular Disease
The detection of miRNAs in body fluids is done by various methods namely real time PCR or micro array assay. Circulating miRNAs are stable nucleic acids whose sequences can be amplified and are easily detectable with appropriate methods in wide range of bodily fluids from blood, urine, saliva, cerebrospinal fluid (CSF) and breast milk. Despite the advancement in medical science, early and effective diagnosis or treatment for myocardial infarction still remains a clinical challenge [80] .
Role of cardiac specific miRNAs for the detection of acute myocardial infarction (AMI) have been described by various researchers. Cardiac specific miRNAs-1/133/208b/499 may have the future for upcoming diagnostic markers for acute myocardial infarction [62] [60] . miR-1/21/22/23/29/122/130/195/199/320a/423-5p/499/622 levels were found to be overexpressed in heart failure patients. Particularly increased level of miR-423-5p was well correlated with heart failure in different studies [53] [56] [24] [55] [56] [57] . Similarly level of miR-122 and miR-370 were positively correlated with total cholesterol, Triglycerides and LDL cholesterol (LDL C) and coronary artery disease (CAD) progression [71] .
4.8. Circulating miRNAs as Therapeutic Role
Since single miRNA can target multiple mRNA making protein on gene regulation, so now it is feasible to use miRNA as therapeutic target. Elimination of miR-1 has shown to improvement in arrhythmic potential in infarcted rat hearts and this has opened a new window for future anti arrhythmic therapy targeting miR-1 [43] . Knockdown of miR-21 expression in atrial muscle leads to prevention of atrial fibrillation and remodeling. This further opens a door for potential future therapy/intervention designed to prevent atrial fibrillation (AF) [65] .
In vivo delivery of lentivirus miR-24 can interact with tissue growth factor β1 (TGF-β1) to decrease the cardiac fibrosis resulting from various heart conditions like MI, cardiomyopathies and heart failure [66] . miRNAs based therapeutics have been more studied in atherosclerosis. Atheroprotective effect of miR-126 has been explained by [69] . Similarly anti miR-33 therapy has found to be atheroprotective by increasing circulating HDL in mice model [70] (Table 1).
5. Conclusions
It seems yesterday since the discovery of miRNAs, but within a decade it has
Table 1. Summary of various miRNAs with its possible roles in cardiovascular disease.
shown very detail rays of insight into the biology of cardiovascular system. Current increase in interest of miRNAs in cardiovascular diseases among researchers signifies its bright future in coming days. Although much has happened within this short period of time but still need much more research on miRNAs and findings especially in larger studies. Contrary to its name “microRNA”, it has wider macro function in heart from cardiac embryogenesis, differentiation, hypertrophy to disease. Days are not far for cure of heart failure due to various conditions; prevent cardiac remodeling, prediction of future heart failure in post MI patients. miRNA might outweigh cardiac sensitivity troponins in coming days for the early and accurate diagnosis of acute myocardial infarction (AMI) with predictive future complications risk and their treatment. Therefore, it seems there is a possibility to treat cardiomyopathies in the near future. This will definitely be an exciting gift to needy patients and availability of more durable and permanent treatment options for treating cardiovascular diseases.
Acknowledgement
We would like to thank Dr. Pranita Bhatta for proofreading of the manuscript.
Cite this paper
Pokhrel, S. and Guotian, Y. (2017) MicroRNA and Its Role in Cardiovascular Disease. World Journal of Cardiovascular Diseases, 7, 340-357. https://doi.org/10.4236/wjcd.2017.710032
References
- 1. Watson, J.D. and Crick, F.H.C. (1953) A Structure for Deoxyribose Nucleic Acid. Nature, 171, 737-738. https://doi.org/10.1038/171737a0
- 2. Watson, J.D., Baker, T.A., Bell, S.P., Gann, A., Levine, M., Losik, R., et al. (2014) Molecular Biology of the Gene. 7th Edition, Benjamin-Cummings Publishing Company, Boston.
- 3. Pertea, M. and Salzberg, S.L. (2010) Between a Chicken and a Grape: Estimating the Number of Human Genes. Genome Biology, 11, 206.
- 4. Wilusz, J.E., Sunwoo, H. and Spector, D. (2009) Long Noncoding RNAs: Functional Surprises from the RNA World. Genes & Development, 23, 1494-1504. https://doi.org/10.1101/gad.1800909
- 5. Mattick, J.S. and Makunin, I.V. (2006) Non-Coding RNA. Human Molecular Genetics, 15, R17-R29.
- 6. Esteller, M. (2011) Non-Coding RNAs in Human Disease. Nature Reviews Genetics, 12, 861-874. https://doi.org/10.1038/nrg3074
- 7. Huang, Z.P., Ding, Y., Chen, J., Wu, G., Katoaka, M., Hu, Y., et al. (2016) Long Non-Coding RNAs Link Extracellular Matrix Gene Expression to Ischemic Cardiomyopathy. Cardiovascular Research, 112, 543-554. https://doi.org/10.1093/cvr/cvw201
- 8. Wahid, F., Shehzad, A., Khan, T. and Kim, Y.Y. (2010) MicroRNAs: Synthesis, Mechanism, Function, and Recent Clinical Trials. Biochimica et Biophysica Acta, 1803, 1231-1243. https://doi.org/10.1016/j.bbamcr.2010.06.013
- 9. Zorio, E., Medina, P., Rueda, J., Millán, J.M., Arnau, M.A., Beneyto, M., et al. (2009) Insights into the Role of microRNAs in Cardiac Diseases: From Biological Signalling to Therapeutic Targets. Cardiovascular & Hematological Agents in Medicinal Chemistry, 7, 82-90. https://doi.org/10.2174/187152509787047676
- 10. Kiriakidou, M., Tan, S., Lamprinaki, S., Planell-Saguer, M., Nelson, T. and Mourelatos, Z. (2006) An mRNA m7G Cap Binding-like Motif within Human Ago2 Represses Translation. Cell, 129, 1141-1151. https://doi.org/10.1016/j.cell.2007.05.016
- 11. Bagga, S., Bracht, H., Massirer, K., Holtz, J., Eachus, R., et al. (2005) Regulation by let-7 and lin-4 miRNAs Results in Target mRNA Degradation. Cell, 122, 553-563.
- 12. Humphreys, D.T., Westman, B.J., Martin, I.K. and Preiss, T. (2005) MicroRNAs Control Translation Initiation by Inhibiting Eukaryotic Initiation Factor 4E/cap and poly (A) Tail Function. PNAS, 102, 16961-16966. https://doi.org/10.1073/pnas.0506482102
- 13. Grey, T.R., Meyers, H., Wu, M.S., Hook, L., et al. (2010) A Viral microRNA Down-Regulates Multiple Cell Cycle Genes through mRNA 5’UTRs. PLOS Pathogens, 6, e1000967.
- 14. Forman, J., Legesse-Miller, A. and Coller, H. (2008) A Search for Conserved Sequences in Coding Regions Reveals That the let-7 microRNA Targets Dicer within Its Coding Sequence. PNAS, 105, 14879-14884. https://doi.org/10.1073/pnas.0803230105
- 15. Thum, C. and Bauersachs, J. (2008) MicroRNAs: Novel Regulators in Cardiac Development and Disease. Cardiovascular Research, 79, 562-570. https://doi.org/10.1093/cvr/cvn137
- 16. Lee, C., Feinbaum, R.L. and Ambros, V. (1993) The C. Elegans Heterochronic Gene lin-4 Encodes Small RNAs with Antisense Complementarity to lin-14. Cell, 75, 843-854.
- 17. Johnson, R.W., Liu, L.Y., Hanna-Rose, W. and Chamberlin, H.M. (2009) The Caenorhabditis Elegans Heterochronic Gene lin-14 Coordinates Temporal Progression and Maturation in the Egg-Laying System. Developmental Dynamics, 238, 394-404. https://doi.org/10.1002/dvdy.21837
- 18. Lagos-Quintana, R.R. and Lendeckel, T. (2001) Identification of Novel Genes Coding for Small Expressed RNAs. Science, 294, 853-858. https://doi.org/10.1126/science.1064921
- 19. Rybak, A., Fuchs, H., Hadian, K., Smirnova, L., Wulczyn, A.M., et al. (2009) The let-7 Target Gene Mouse lin-41 Is a Stem Cell Specific E3 Ubiquitin Ligase for the miRNA Pathway Protein Ago2. Nature Cell Biology, 11, 1411-1420. https://doi.org/10.1038/ncb1987
- 20. Schulman, B.R.M., Esquela-Kerscher, A. and Slack, F.J. (2005) Reciprocal Expression of lin-41 and the microRNAs let-7 and mir-125 during Mouse Embryogenesis. Developmental Dynamics, 234, 1046-1054. https://doi.org/10.1002/dvdy.20599
- 21. Ren, Z. and Ambros, V.R. (2015) Caenorhabditis Elegans microRNAs of the let-7 Family Act in Innate Immune Response Circuits and Confer Robust Developmental Timing against Pathogen Stress. PNAS, 112, E2366-E2375. https://doi.org/10.1073/pnas.1422858112
- 22. Kozomara, A. and Griffiths-Jones, S. (2014) miRBase: Annotating High Confidence microRNAs using Deep Sequencing Data. Nucleic Acids Research, 42, D68-D73. https://doi.org/10.1093/nar/gkt1181
- 23. Hausser, B., Rodak, J. and Wirth, Z.M. (2009) MirZ: An Integrated microRNA Expression Atlas and Target Prediction Resource. Nucleic Acid Research, 37, W266-W272.
- 24. Romain, S.P.R., Tomaszewski, C.G. and Samani, N. (2015) MicroRNAs in Cardiovascular Disease: An Introduction for Clinicians. Heart, 101, 921-928. https://doi.org/10.1136/heartjnl-2013-305402
- 25. Da Costa Martins, P.A., Bourajjaj, M., Gladka, M., Kortland, M., Oort, R.J., Pinto, Y.M., et al. (2008) Conditional Dicer Gene Deletion in the Postnatal Myocardium Provokes Spontaneous Cardiac Remodeling. Circulation, 118, 1567-1576. https://doi.org/10.1161/CIRCULATIONAHA.108.769984
- 26. Zhao, R.J.F., Li, V., Drehle, M., Muth, A.N., et al. (2007) Dysregulation of Cardiogenesis, Cardiac Conduction, and Cell Cycle in Mice Lacking miRNA-1-2. Cell, 129, 303-317.
- 27. Chen, J.F., Murchison, E.P., Tang, R., Callis, E., Tatsuguchi, D., et al. (2008) Targeted Deletion of Dicer in the Heart Leads to Dilated Cardiomyopathy and Heart Failure. PNAS, 105, 2111-2116. https://doi.org/10.1073/pnas.0710228105
- 28. Rao, P.K., Kumar, R.M., Farkhondeh, B.S. and Lodish, F. (2006) Myogenic Factors That Regulate Expression of Muscle-Specific microRNAs. PNAS, 103, 8721-8726. https://doi.org/10.1073/pnas.0602831103
- 29. Xu, C., Lu, Y., Pan, Z., Chu, L., Lin, X., et al. (2007) The Muscle-Specific microRNAs miR-1 and miR-133 Produce Opposing Effects on Apoptosis by Targeting HSP60, HSP70 and Caspase-9 in Cardiomyocytes. Journal of Cell Science, 120, 3045-3052. https://doi.org/10.1242/jcs.010728
- 30. Zhao, S. and Srivastava, E. (2004) Serum Response Factor Regulates a Muscle-Specific microRNA That Targets Hand2 during Cardiogenesis. Nature, 436, 214-220. https://doi.org/10.1038/nature03817
- 31. Schindler, G.M., Wang, J., Firulli, A., Firulli, B., Poss, K.D., et al. (2014) Hand2 Elevates Cardiomyocyte Production during Zebrafish Heart. Development, 141, 1-11. https://doi.org/10.1242/dev.106336
- 32. Mishima, S.C. and Giraldez, J. (2007) miR-1-2 Gets to the Heart of the Matter. Cell, 129, 247-249.
- 33. Sayed, D., Hong, C., Chen, I.Y. and Lypowy, A. (2007) MicroRNAs Play an Essential Role in the Development of Cardiac Hypertrophy. Circulation Research, 100, 416-424. https://doi.org/10.1161/01.RES.0000257913.42552.23
- 34. Carè, C., Felicetti, F., Bonci, A.A., Gallo, P., et al. (2007) MicroRNA-133 Controls Cardiac Hypertrophy. Nature Medicine, 13, 613-618. https://doi.org/10.1038/nm1582
- 35. Liu, B., Williams, H., Qi, R.A., Bassel-Duby, et al. (2008) microRNA-133a Regulates Cardiomyocyte Proliferation and Suppresses Smooth Muscle Gene Expression in the Heart. Genes and Development, 22, 3242-3254. https://doi.org/10.1101/gad.1738708
- 36. Xiao, C.Y.H. (2010) MicroRNAs: Novel Regulators of the Heart. Journal of Thoracic Disease, 2, 43-47.
- 37. Ikeda, H., Kong, L., Bejar, B., et al. (2009) MicroRNA-1 Negatively Regulates Expression of the Hypertrophy-Associated Calmodulin and Mef2a Genes. Molecular and Cellular Biology, 29, 2193-2204. https://doi.org/10.1128/MCB.01222-08
- 38. Tian, J. and An, N.L. (2017) Role of microRNAs in Cardiac Development and Disease. Experimental and Therapeutic Medicine, 13, 3-8. https://doi.org/10.3892/etm.2016.3932
- 39. Rooij, V., Sutherland, B., Qi, R.A. and Hill, O. (2007) Control of Stress-Dependent Cardiac Growth and Gene Expression by a MicroRNA. Science, 316, 575-579. https://doi.org/10.1126/science.1139089
- 40. Callis, E., Pandya, K., Seok, H.Y., Tang, R.H., Tatsuguchi, M., Huang, Z.P., et al. (2009) MicroRNA-208a Is a Regulator of Cardiac Hypertrophy and Conduction in Mice. The Journal of Clinical Investigation, 119, 2772-2786. https://doi.org/10.1172/JCI36154
- 41. Bang, C., Batkai, D., Gupta, F., Holzmann, A., et al. (2014) Cardiac Fibroblast-Derived microRNA Passenger Strand-Enriched Exosomes Mediate Cardiomyocyte Hypertrophy. The Journal of Clinical Investigation, 124, 2136-2146. https://doi.org/10.1172/JCI70577
- 42. Rooij, E., Sutherland, B., Liu, W.H., McAnally, G.R.D., et al. (2006) A Signature Pattern of Stress-Responsive microRNAs That Can Evoke Cardiac Hypertrophy and Heart Failure. PNAS, 103, 18255-18260. https://doi.org/10.1073/pnas.0608791103
- 43. Yang, L., Xiao, L., Luo, L.B., et al. (2007) The Muscle-Specific microRNA miR-1 Regulates Cardiac Arrhythmogenic Potential by Targeting GJA1 and KCNJ2. Nature Medicine, 13, 486-491. https://doi.org/10.1038/nm1569
- 44. Terentyev, B.E., Terentyeva, M., Malana, E., Kuhn, E., et al. (2009) miR-1 Overexpression Enhances Ca2+ Release and Promotes Cardiac Arrhythmogenesis by Targeting PP2A Regulatory Subunit B56α and Causing CaMKII-Dependent Hyperphosphorylation of RyR2. Cellular Biology, 104, 514-521.
- 45. Lu, Y., Zhang, W., Pan, G., Zhang, F., et al. (2010) MicroRNA-328 Contributes to Adverse Electrical Remodeling in Atrial Fibrillation. Circulation, 122, 2378-2387.
- 46. Girmatsion, B., Bonauer, A., Greinecker, G., Scherer, M.A., et al. (2009) Changes in microRNA-1 Expression and IK1 Up-Regulation in Human Atrial Fibrillation. Heart Rhythm, 6, 1802-1809.
- 47. Zhenga, X.C.C., Chena, W.D., Shena, W.L., Ruanb, C.C., Zhua, L.M., et al. (2010) MicroRNA-155 Regulates Angiotensin II Type 1 Receptor Expression and Phenotypic Differentiation in Vascular Adventitial Fibroblasts. Biochemical and Biophysical Research Communications, 400, 483-488.
- 48. Bahring, S., Kann, N., Gong, M., Chitayat, T.R., et al. (2008) Inversion Region for Hypertension and Brachydactyly on Chromosome 12p Features Multiple Splicing and Noncoding RNA. Hypertension, 51, 426-431. https://doi.org/10.1161/HYPERTENSIONAHA.107.101774
- 49. Marques, Z., Campain, E., Tomaszewski, Z.-S., Yang, H.J., Charchar, J., et al. (2011) Gene Expression Profiling Reveals Renin mRNA Overexpression in Human Hypertensive Kidneys and a Role for MicroRNAs. Hypertension, 58, 1093-1098. https://doi.org/10.1161/HYPERTENSIONAHA.111.180729
- 50. Eskildsen, V., Jeppesen, L., Schneider, N.Y., Sandberg, B., Hansen, P.B.L., et al. (2013) Angiotensin II Regulates microRNA-132/-212 in Hypertensive Rats and Humans. International Journal of Molecular Science, 14, 11190-11207.
- 51. Rezaei, A., Neuenschwander, B., Mordasini, D., Frey, F.J., et al. (2014) Regulation of 11β-Hydroxysteroid Dehydrogenase Type 2 by MicroRNA. Hypertension, 64, 860-866. https://doi.org/10.1161/HYPERTENSIONAHA.114.00002
- 52. Goyal, A., Thomas, N., Davis, R.L., Butler, A., et al. (2010) Predictors of Incident Heart Failure in a Large Insured Population: A One Million Person-Year Follow-Up Study. Circulation: Heart Failure, 3, 698-705. https://doi.org/10.1161/CIRCHEARTFAILURE.110.938175
- 53. Goren, K., Zafrir, T., Lewis, S. and Amir, O. (2012) Serum Levels of microRNAs in Patients with Heart Failure. European Journal of Heart Failure, 14, 147-154. https://doi.org/10.1093/eurjhf/hfr155
- 54. Devaux, V., McCann, P., Kelly, C., Ng, L., et al. (2013) A Panel of 4 microRNAs Facilitates the Prediction of Left Ventricular Contractility after Acute Myocardial Infarction. PLoS ONE, 8, e70644. https://doi.org/10.1371/annotation/458a1f6a-6327-429a-81cb-992c97f04bd6
- 55. Tijsen, J., Creemers, E., Moerland, D., Windt, L.J., Wal, C., Kok, E., et al. (2010) MiR423-5p as a Circulating Biomarker for Heart Failure. Circulation Research, 106, 1035-1039. https://doi.org/10.1161/CIRCRESAHA.110.218297
- 56. Zhang, N., Ban, T., Xu, L., Wang, N., et al. (2013) Elevated Plasma microRNA-1 Predicts Heart Failure after Acute Myocardial Infarction. International Journal of Cardiology, 166, 259-260.
- 57. Lai, K.B., Sanderson, E. and Izzat, Y.C.M. (2015) Micro-RNA and mRNA Myocardial Tissue Expression in Biopsy Specimen from Patients with Heart Failure. International Journal of Cardiology, 199, 70-83.
- 58. Wu, C., So, D.-D.N., Qi, H.H., Bloch, B., Shi, et al. (2011) Hypoxia Potentiates MicroRNA-Mediated Gene Silencing through Posttranslational Modification of Argonaute2. Molecular and Cellular Biology, 11, 4760-4774. https://doi.org/10.1128/MCB.05776-11
- 59. Wang, Z., Zhang, L., Chen, M.M., et al. (2012) Loss of the miR-144/451 Cluster Impairs Ischaemic Preconditioning-Mediated Cardioprotection by Targeting Rac-1. Cardiovascular Research, 94, 379-390. https://doi.org/10.1093/cvr/cvs096
- 60. Corsten, F., Dennert, J.S., Kuznetsova, T., Devaux, Y., Hofstra, L., et al. (2010) Circulating MicroRNA-208b and MicroRNA-499 Reflect Myocardial Damage in Cardiovascular Disease. Circulation: Cardiovascular Genetics, 3, 499-506. https://doi.org/10.1161/CIRCGENETICS.110.957415
- 61. Adachi, N., Otsuka, N.K., Hirokawa, G., et al. (2010) Plasma MicroRNA 499 as a Biomarker of Acute Myocardial Infarction. Clinical Chemistry, 1183-1185.
- 62. Kuwabara, O., Horie, T., Nishi, N.K., Kinoshita, et al. (2011) Increased microRNA-1 and microRNA-133a Levels in Serum of Patients with Cardiovascular Disease Indicate the Existence of Myocardial Damage. Circulation: Cardiovascular Genetics, 110, 958-975.
- 63. Matsumoto, S., Suna, S., Nakatani, D., Usami, H.M., et al. (2013) Circulating p53-Responsive MicroRNAs Are Predictive Indicators of Heart Failure after Acute Myocardial Infarction. Circulation Research, 113, 322-326. https://doi.org/10.1161/CIRCRESAHA.113.301209
- 64. Hullinger, G., Montgomery, L., Seto, A.G., Dickinson, A., Semus, M., Lynch, M., et al. (2012) Inhibition of miR-15 Protects against Cardiac Ischemic Injury. Circulation Research, 110, 71-81. https://doi.org/10.1161/CIRCRESAHA.111.244442
- 65. Cardin, S., Guasch, L., Naud, P., Quang, L., Shi, et al. (2012) A Role for MicroRNA-21 in Atrial Profibrillatory Fibrotic Remodeling Associated with Experimental Post-Infarction Heart Failure. Circulation: Arrhythmia and Electrophysiology, 5, 1027-1035. https://doi.org/10.1161/CIRCEP.112.973214
- 66. Wang, J., Huang, W., Xu, R., Nie, Y., Cao, X., Meng, J., et al. (2012) MicroRNA-24 Regulates Cardiac Fibrosis after Myocardial Infarction. Journal of Cellular and Molecular Medicine, 16, 2150-2160. https://doi.org/10.1111/j.1582-4934.2012.01523.x
- 67. Glass, K. and Witztum, L. (2001) Atherosclerosis: The Road Ahead. Cell, 104, 503-516.
- 68. Fichtlscherer, S., De Rosa, S., Fox, H., Schwietz, T., et al. (2010) Circulating MicroRNAs in Patients with Coronary Artery Disease. Circulation Research, 107, 677-684. https://doi.org/10.1161/CIRCRESAHA.109.215566
- 69. Wei, Y., Nazari-Jahantigh, N. and Weber, C.S. (2013) MicroRNA-126, -145, and -155. Arteriosclerosis, Thrombosis, and Vascular Biology, 33, 449-454. https://doi.org/10.1161/ATVBAHA.112.300279
- 70. Rayner, J., Sheedy, J., Esau, C., Hussain, N., Temel, E., Parathath, et al. (2011) Antagonism of miR-33 in Mice Promotes Reverse Cholesterol Transport and Regression of Atherosclerosis. The Journal of Clinical Investigation, 121, 2921-2931. https://doi.org/10.1172/JCI57275
- 71. Gao, W., He, H.W., Wang, Z.M., Zhao, H., Lian, X.Q., Wang, Y.S., et al. (2012) Plasma Levels of Lipometabolism-Related miR-122 and miR-370 Are Increased in Patients with Hyperlipidemia and Associated with Coronary Artery Disease. Lipids in Health and Disease, 11, 55.
- 72. Smith, T., Rajakaruna, C., Caputo, M. and Emanueli, C. (2015) MicroRNAs in Congenital Heart Disease. Annals of Translational Medicine, 3, 333.
- 73. Li, J., Cao, Y., Ma, X.J., Wang, H.J., Zhang, J., Luo, X., et al. (2013) Roles of miR-1-1 and miR-181c in Ventricular Septal Defects. International Journal of Cardiology, 168, 1441-1446.
- 74. Latronico, M.V.G., Catalucci and Condorelli (2008) MicroRNA and Cardiac Pathologies. Physiological Genomics, 34, 239-242. https://doi.org/10.1152/physiolgenomics.90254.2008
- 75. O’Brien, E., Kibiryeva, Zhou, X.G., Marshall, J.A., Lofland, K., Artman, et al. (2012) Noncoding RNA Expression in Myocardium from Infants with Tetralogy of Fallot. Circulation: Cardiovascular Genetics, 5, 279-286. https://doi.org/10.1161/CIRCGENETICS.111.961474
- 76. Bittel, C., Kibiryeva, N., Marshall, A. and O’Brien, J.E. (2014) MicroRNA-421 Dysregulation Is Associated with Tetralogy of Fallot. Cells, 3, 713-723. https://doi.org/10.3390/cells3030713
- 77. Liang, D., Xu, X., Deng, F., Feng, J., Zhang, H., Liu, Y., et al. (2014) miRNA-940 Reduction Contributes to Human Tetralogy of Fallot Development. Journal of Cellular and Molecular Medicine, 18, 1830-1839. https://doi.org/10.1111/jcmm.12309
- 78. Nigam, V., Sievers, H., Jensen, C., Sier, A., Simpson, C., Srivastava, D., et al. (2010) Altered Micrornas in Bicuspid Aortic Valve: A Comparison between Stenotic and Insufficient Valves. Journal of Heart Valve Disease, 19, 459-465.
- 79. Yanagawa, B., Lovren, F., Pan, Y., Garg, V., Quan, A., Tang, G., et al. (2012) MiRNA-141 Is a Novel Regulator of BMP-2-Mediated Calcification in Aortic Stenosis. The Journal of Thoracic and Cardiovascular Surgery, 144, 256-262.
- 80. Boon, A. and Dimmeler, S. (2015) MicroRNAs in Myocardial Infarction. Nature Reviews Cardiology, 12, 135-142. https://doi.org/10.1038/nrcardio.2014.207
Abbreviations and Acronyms
αMHC6: alpha myosin heavy
11βHSD 2: 11-β hydroxysteroid dehydrogenase 2
AF: Atrial fibrillation
AMI: Acute myocardial infarction
AMI-HF: Acute myocardial induced heart failure
BMP 2: Bone morphogenetic protein 2
CAD: Coronary artery disease
CHF: Congestive heart failure
CPK: Creatinine phosphokinase
CSF: Cerebrospinal fluid
DNA: Deoxyribonucleic acid
HF: Heart failure
HDAC4: Histone deacetylates 4
IPC: Ischemic preconditioning
LNA: Locked nucleic acid
lnc RNA: Long noncoding RNA
linc RNA: Long intergenic noncoding RNA
Mef-2: Myocyte enhancer factor 2
mRNA: messenger RNA
MI: Myocardial infarction
miRNA : MicroRNA
miR: MicroRNA
NADPH: Nicotinamide adenine dinucleotide phosphate
ncRNA: Noncoding RNA
nt: nucleotide
NCBIRefSeq: National Center for Biotechnology Information Reference Sequence
RyR2: Ryanodine receptors 2
RNA: Ribonucleic acid
ROS: Reactive oxygen species
SiRNA: Short noncoding RNA
TGFβ1: Tissue growth factor β1
TOF: Tetralogy of fallot
VSD: Ventricular septal defect