Open Journal of Soil Science
Vol.04 No.10(2014), Article ID:51041,7 pages
10.4236/ojss.2014.410037
Genetic Differentiation Caused by Chromium Treatment in Leersia hexandra Swartz Revealed by RAPD Analysis
X. W. Cai*, Y. Shao, Z. M. Lin
College of Earth Science, Guilin University of Technology, Guilin City, China
Email: *monkeycxw@tom.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 26 August 2014; revised 18 September 2014; accepted 25 September 2014
ABSTRACT
Randomly amplified polymorphic DNA (RAPD) technique was applied to assess the genetic variations and phylogenetic relationships in genetic differentiation within 4 Chromium-treatment Leersia hexandra. The fresh leaves of Leersia hexandra cultivated on the condition of chrome pollution and exogenous organic acids were used as experimental material. The genomic DNA of Leersia hexandra was extracted by using CTAB method. The results showed that different samples of Leersia hexandra exhibited DNA polymorphism when using the random primer S43, S51and S55 as the primers in the RAPD reaction. One specific DNA band about 1000 bp was found in the sam- ple which treated with 10 mmol/L concentration EDTA when used the S43 primer to RAPD. The obvious differences between different EDTA-treatment levels suggest that EDTA has certain ef- fects on enrichment to heavy metals of Leersia hexandra, it will be more favored to Leersia hexan- dra accumulation of chromium when EDTA concentration increased.
Keywords:
Chromium Treatment, Genetic Differentiation, Randomly Amplified Polymorphic DNA (RAPD), Leersia hexandra Swartz

1. Introduction
Leersia hexandra Swartz, a perennial marshy plant, has been reported to be a Cr-accumulating plant with high tolerance to Cr. Under nutrient solution culture, it did not show any obvious symptoms of Cr toxicity when Cr concentrations in the leaves reached 5608 mg∙kg−1 dry weight [1]
Previous and continuous researches show that environmental factors have a significant impact on the plants [2] . Excessive Cr in soil has negative impact on plant growth. At the same time, it can also accumulate in plants by roots, and enter human and animal bodies and harm them through the food chain [3] .
Random Amplified Polymorphic DNA (RAPD) is a kind of molecular marker based on PCR. It uses l0 bp random primers amplifying the different DNA fragments in genome to show the polymorphism [4] [5] . One RAPD amplification is actually a simple PCR reaction, and it suits for a large number of samples for rapid anal- ysis. The required DNA template is very small amounts, generally an amplification of only 10 ng to 50 ng DNA [6] . RAPD was used to determine genetic variability of ten populations of alfalfa [7] and the relationships be- tween RAPD markers and 22 quantitative traits of caraway (Carum carvi L.) were analyzed [8] . The RAPD- PCR method was used to describe the pattern of DNA band variation in the samples influenced by the environ- mental pollution, to describe the level of pollution in an area contaminated with smoke and waste from an iron- steel factory, and to reveal the level of potential [9] .
At present, there are not molecular biology reports on heavy metal chromium enrichment of L. hexandra, and little molecular ecology study on plants to heavy metal pollutants under the long -term effect. Reports on using RAPD to analyze heavy metal pollution on plant population genetic diversity are increasing recently, Li et al. (2007) used RAPD to analyze genetic diversity and genetic differentiation of Dicranopteris dichotoma popula- tions and lead-zinc mine tailings in population which grow in nature potential [10] . Wen et al. (2001) planted 4 Datura seeds in different regions and do analysis of this 4 Datura habitats potential [11] . Gu et al. (2008) used RAPD to analyze genetic diversity of clethroides populations which grow in lead zinc tailings in the storing time of 10 years and 20 years in natural and contrast soil potential [12] . L. hexandra is a kind of important heavy me- tal chromium enrichment plant, so it is important to study its molecular enrichment mechanism. Therefore, this study used L. hexandra treated with heavy metal pollution in the laboratory as experimental materials, analyzed the genetic diversity of L. hexandra under different EDTA treatment of heavy metal chromium pollution through RAPD, and tried to explore the differentiation and evolution of L. hexandra population under long-term persis- tence of toxic heavy metals pollution from molecular level.
2. Material and Methods
2.1. Materials
Plant material handling
L. hexandra collected from Guilin city, China, and artificial cultivated in sunlight greenhouse. 400 mg chrome metal Cr3+ (CrCl3) to per kg were added to soil when L. hexandra planted. EDTA solution of 0 mmol/L, 2.5 mmol/L, 5 mmol/L and 10 mmol/L concentration were prepared to handle L. hexandra plant, 3 repeats for every kind of concentration gradient, joined the appropriate EDTA solutions to L. hexandra plant every other week, and added a total of 3 times.
Reagents and solutions
2 × CTAB extracting buffer (2% CTAB, 1.4 mol/L NaCl, and 20 mmol/L EDTA, and 100 mmol/L Tris-Cl, pH 8); phenol/chloroform/isoamyl alcohol (25:24:1), chloroform/isoamyl alcohol (24:1), β-mercaptoethanol; 3 mol/L NaAc, (pH 5.2) isopropyl alcohol; TE solution (10 mmol/L Tris-Cl, 1 mmol/LEDTA, pH 8); 75% alcohol; alcohol; 1 × TAE buffer (40 mmol/L Tris, and 20 mmol/L HAc, 1 mmol/L EDTA, pH 8.0), Taq DNA polyme- rase; agarose; Lambda DNA/EcoR I + Hind III Marker.
Instruments and equipments
General refrigerators; HVE-50 high pressure sterilization pot (Japan Hirayama), LEGEND MICRO17 high speed centrifuge (United States saimofei Fisher); trace moving liquid (Japan Nichipet EX); T1Thermocyler am- plification apparatus (Germany BIOMETRA); DYY-12 Sanheng Multiple use Electrophoresis apparatus (Bei- jing Liuyi); DYCP
The primer sequences
The primer sequences used in this study are shown in Table 1.
2.2. Experimental Methods
Extraction and purification for L. hexandra genomic DNA
Table 1. RAPD random primer of Leersia hexandra Swartz
Applied an improved ctab method potential [13] [14] , to extract total DNA from leaves of L. hexandra dried by silica gel, and used phenol/chloroform/isoamyl alcohol (25:24:1) to extract total DNA, anhydrous ethanol to purified total DNA, 0.8% agarose gel electrophoresis to detect DNA extraction and effects.
Primers screening
Used L. hexandra genomic DNA extracted as DNA templates for RAPD random primers screening. Primers are used in this research process all synthesized by Shanghai bio-engineering technology services company li- mited. Random primers with clear RAPD amplification and stability response were screened for final primers of L. hexandra RAPD molecular markers.
RAPD amplification reaction of L. hexandra
We used L. hexandra genomic DNA extracted as DNA templates for RAPD amplification reaction. Opti- mized reaction system: 25 μl total volume, 18.5 μl double distilled water, 2.5 μl 10 uffer liquid, 2 μl Mg2+ con- centration for 20 mmol/L, 0.5 μl dNTP concentration for 200 μmol/L, 0.5 μl primer concentration for 0.4 μmol/L, 1 μl template DNA originated from DNA extraction diluted 4 times with TE buffer liquid, 2U/25 μl Taq enzyme. Circulation system after optimized: for 5 min 94˚C predegeneration, then for 40 cycles, followed by 1 min 94˚C predegeneration, 1 min 36˚C annealing, 1 min 72˚C stretch. Last, 7 min 72˚C complete the extension potential [15] . All RAPD reactions were in T1 Thermocyler amplification a paratus (Germany BIOMETRA). Results of PCR amplified products detected by 0.8% agarose gel electrophoresis, and used Lambda DNA/Hind III + EcoR I Markers as molecular standard, and electrophoresis in 120 V constant-voltage for 40 min then do EB dyeing for 10 min, electrophoresis buffer of 1 × TAE, observed by Furi FR-980 gel imaging device, and then taken pic- tures by the gel image analysis system.
3. Results and Discussion
3.1. L. hexandra DNA Extraction Results
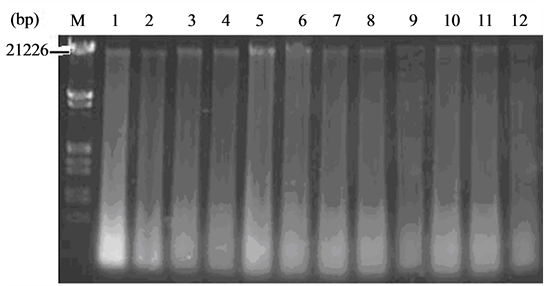
Agarose gel electrophoresis results of L. hexandra leaves genomic DNA is shown in Figure 1. Test combined with common CTAB method for genomic DNA extraction, and extracted genomic DNA respectively for 12 samples, high quality DNA have great influence on stability and reliability experiment results. Seen from Figure 1, the primary belt of L. hexandra genomic DNA is at about 21,226 bp. After agarose gel electrophoresis for DNA extracted from L. hexandra leaves, its band isn’t very bright and clear but with much tail, which showed that process of DNA extraction degradated in severe, proteins and RNA degradation was not very thorough. To illustrate, extraction of DNA purity in this experiment was not high, but in little amount of DNA in RAPD experiment, generally, expansion just need 10 ng to 50 ng DNA, purity requirements were not very high. The RAPD results showed that the extraction genomic DNA from L. hexandra can be applied to RAPD reaction.
3.2. L. hexandra RAPD Primers Screening Results
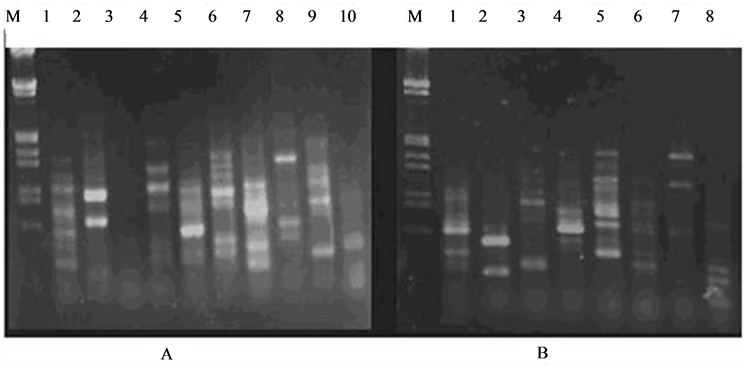
Used L. hexandra genomic DNA as a template, for screening of the 46 UBC random primers, results showed that 29 UBC hadn’t amplified products or obscured and couldn’t tell, other 17 UBC random primers amplified a strip of clear and stable, that account for 36.9% of total primers. They varied approximately 200 bp or 3000 bp size, each primer amplification of fragments between changes in 1 - 7. Part L. hexandra leaves DNA RAPD primers screened results are as shown in Figure 2.
3.3. L. hexandra RAPD Amplification Results
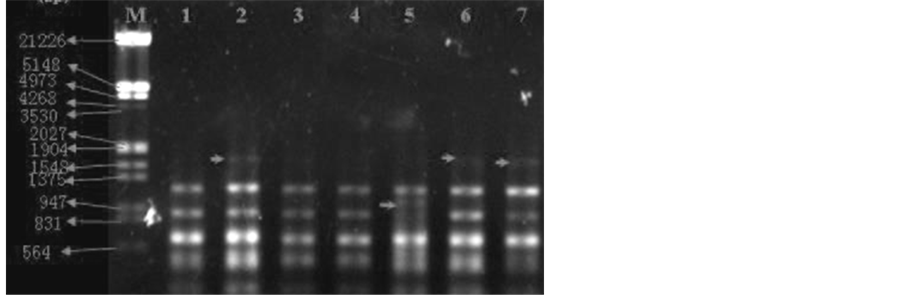
Screened 46 random primers (see Table 1) preliminary, and used primers that can amplify stability and clear DNA bans as DNA amplification primers of L. hexandra polluted by chromium, and screened random primers S43, S51, S55 from tests results for RAPD reaction in this study. RAPD analysis was used for different samples of L. hexandra which under heavy metals chromium of 400 mg/kg concentration pollution and then added dif- ferent concentration exogenous EDTA, sample RAPD results detected by agarose gel electrophoresis as shown in Figure 3, Figure 4 and Figure 5.
3.4. RAPD Amplification Results of Primer S43
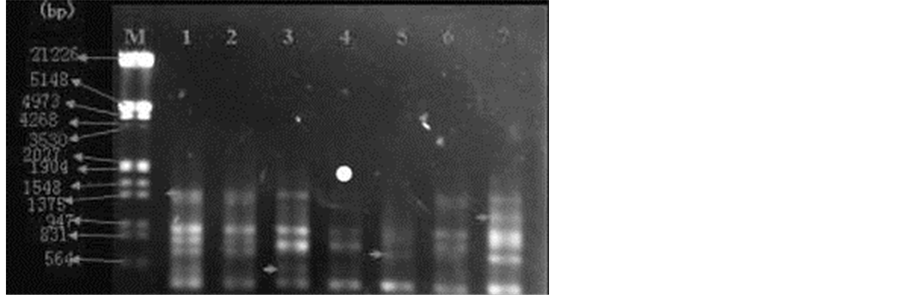
Figure 3 shows the amplification results of S43 RAPD random primers of L. hexandra samples. EDTA concen- tration of the first sample was 0 mmol/L (contrast). EDTA concentration of the second and third sample was 2.5
Figure 1. Leaves general DNA electrophoresis spectrums of L. hexandra, 1 - 3: EDTA 0 mmol/L; 4 - 6: EDTA 2.5 mmol/L; 7 - 9: EDTA 5.0 mmol/L; 10 - 12: EDTA 10 mmol/L. M is Lambda DNA/Hind III + EcoR I Markers.
Figure 2. Filtered results of RAPD part primers in L. hexandra leaves DNA, A: Lane 1 - 10 Respectively Express Primer S51, S52, S57, S59, S60, S93, S94, S95, S96 and S97 amplification results; B: Lane 1 - 8 Respectively Express Primer S4, S9, S98, S99, S43, S44, S55 and S56 amplification results; M is Lambda DNA/Hind III + EcoR I Markers.
Figure 3. Amplification results in primer 43(GTCGCCGTCA) RAPD, 1: EDTA 0 mmol/L (Contrast); 2,3: EDTA 2.5 mmol/L; 4,5: EDTA 5.0 mmol/L; 6,7: EDTA 10 mmol/L; M is Lambda DNA/Hind III + EcoR I Markers.
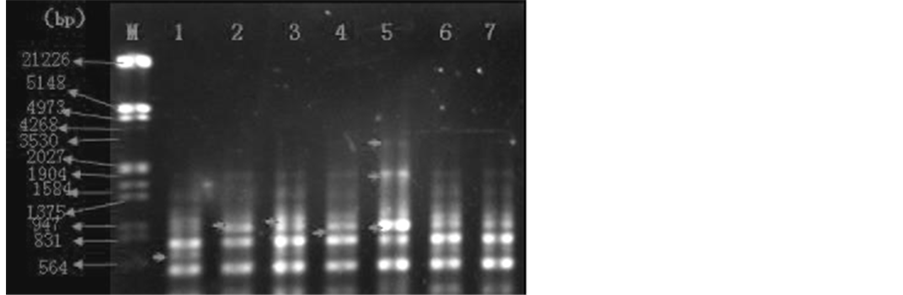
Figure 4. Amplification results in primer 51(GTCGCCGTCA) RAPD. The treatment of samples 1 - 7 is the same as Figure 3.
Figure 5. Amplification results in primer 55(GTCGCCGTCA)RAPD. The treatment of samples 1 - 7 is the same as Figure 3.
mmol/L and the third sample appeared DNA polymorphism at about 564 bp (as shown by the arrow in the 3rd sample). EDTA concentration of the fourth and fifth sample was 5 mmol/L, and the fifth sample appeared DNA polymorphism at about 700 bp (as shown by the arrow in the 5th sample). EDTA concentration of the sixth and seventh sample was 10 mmol/L, and the 7th appeared DNA polymorphism at about 1000 bp (as shown by the arrow in the 7th sample).
EDTA concentration of the first was 0 mmol/L (contrast). Comparison between the second and third and the first samples, the third appeared DNA polymorphism at about 564 bp (as shown by the arrow in the 3rd sample). Comparison between the fourth and fifth sample and the first sample, both appeared strip absence at about 1375 bp (absence strip as shown by the arrow in the first sample), in addition, the fourth sample appeared strip ab- sence at about 700 bp (absence strip as shown by the arrow in the first sample). Comparison between the sixth and seventh and the first sample, the sixth sample appeared strip absence at about 700 bp (as shown by the arrow in the first sample), and the seventh sample appeared DNA polymorphism at about 1000 bp (as shown by the arrow in the 7th sample).
RAPD amplification results of primer S51
Figure 4 shows the amplification results of S51 RAPD random primers of L. hexandra samples. EDTA con- centration of the first was 0 mmol/L (contrast). EDTA concentration of the second and third sample was 2.5 mmol/L and compared with each other, the third sample appeared DNA polymorphism at about 1000 bp (as shown by the arrow in the second sample). EDTA concentration of the fourth and fifth sample was 5 mmol/L, and compared with each other, the fifth sample appeared DNA polymorphism at about 3530bp. EDTA concen- tration of the sixth and seventh sample was 10 mmol/L, and compare result shown that both were almost no dif- ference.
EDTA concentration of the first was 0 mmol/L (contrast). Comparison between the second and third and the first sample, both appeared DNA polymorphism at about 947 bp (as shown by the arrow in the 2nd and 3rd sample). Comparison between the fourth and fifth sample and the first sample, both appeared DNA polymor- phism at about 947 bp (as shown by the arrow in the 4th and 5th sample), in addition, the 5th sample appeared DNA polymorphism at about 1904 bp and 3530 bp ( as shown by the arrow in the 5th sample), and both the samples appeared strip absence at about 700 bp (strip absence as shown by the arrow in the first sample). Compared to the sixth and seventh sample, the contrast showed no difference.
RAPD amplification results of primer S55
Figure 5 shows the amplification results of S55 RAPD random primers of L. hexandra samples. EDTA con- centration of the first was 0 mmol/L (contrast). EDTA concentration of the second and third sample was 2.5 mmol/L and compared with each other, the 2nd sample appeared DNA polymorphism at about 1548 bp (as shown by the arrow in the 2nd sample). EDTA concentration of the fourth and fifth sample was 5 mmol/L, and the 5th sample appeared DNA polymorphism at about 947 bp (as shown by the arrow in the 5th sample). EDTA concentration of the sixth and seventh sample was 10 mmol/L, and compared with each other, result shown that both were almost no difference.
EDTA concentration of the first was 0 mmol/L (contrast). Comparison between the second, the third and the first sample, the second sample appeared DNA polymorphism at about 1548 bp (as shown by the arrow in the 2nd sample). Comparison between the fourth, the fifth sample and the first sample, the fifth sample appeared DNA polymorphism at about 947 bp (as shown by the arrow in the 5th sample). Comparison between the sixth and seventh and the first sample, both samples appeared DNA polymorphism at about 1548 bp (as shown by the arrow in the 6th and 7th sample).
Study on the evolution of pollution can help to understand how plants adapt to the environment and evolve, to understand biological adaptation mechanism under pollution. Knowledge on the genetic diversity can be used in future breeding programs potential [16] [17] . Students concluded that DNA polymorphism detected by RAPD analysis in conjunction with other biochemical parameters could be a powerful eco-toxicological tool in bio- monitoring arsenic pollution potential [18] .
Heavy metal pollution is an important form of soil pollution, the past researches focused on heavy metal toxic effect and mechanism of plants, and study on migration, accumulation, distribution and other aspects on heavy metals in the plant’s tissues, organs and ecological systems potential [19] . The accumulating ability of Leersia hexandra Swartz may be due to they live in electroplating waste water pollution in streams for a long time, in the process of adapting to this environment produced a variation, some genetic characteristics have changed, been the emergence of new forms of Chromium patience and Enrichment capacity also increased. And there are few studies on molecular ecology about plant under long-term effects of heavy metals. Through RAPD analysis of genetic diversity of L. hexandra under the heavy metals pollution, we can understand fundamental changes that is DNA level changes in the cells under tolerance in heavy metals pollution, so as to reform L. hexandra gene by using modern molecular techniques, to make greatly ability on enrichment and accumulation to heavy metals, so as to develop ability of restoring soil .
This study first screen primers of L. hexandra RAPD polymorphicing, and screen random primers S43 (GTCGCCGTCA), S51 (AGCGCCATTG), S55 (CATCCGTGCT) from the 46 random primers to be random primers of L. hexandra RAPD molecular markers preliminary. And then used S43, S51, S55 as primers on the polymorphism of different samples for research. The combined use of both RAPD and ISSR markers for the genetic analysis of the cultivar varieties have been performed earlier potential [20] [21] (Gupta, et al., 2008, Lu, et al., 2009). Our study utilizes a total of 55 primers to disclose the genetic similarity between the selected varie- ties.
Compared EDTA concentration of
4. Conclusion
Different L. hexandra samples exhibited DNA polymorphism when using the random primer S43, S51 and S55 as the primers in the RAPD reaction. One specific DNA band about 1000 bp was found in the sample which was treated with 10 mmol/L concentration EDTA when used the S43 primer to RAPD. The obvious differences be- tween different EDTA-treatment levels suggest that EDTA has not much effects on enrichment to heavy metals of L. hexandra, so we should do more work to research the L. hexandra accumulation of chromium mechanism when EDTA concentration increased.
Acknowledgements
This research was funded by Guangxi Nature Science of Foundation (2011GXNSFA018012), Key Discipline Physical Geography Established in Guilin University of Technology and Guangxi Key Laboratory of Hidden Metallic Ore Deposits Exploration Guilin University of Technology, Guilin, China.
References
- Zhang, X.H., Liu, J., Huang, H.T., Chen, J., Zhu, Y.N. and Wang, D.Q. (2007) Chromium Accumulation by the Hyper- accumulator Plant Leersia hexandra Swartz. Chemosphere, 67, 1138-1143. http://dx.doi.org/10.1016/j.chemosphere.2006.11.014
- Wang, T., Zhang, Q.B. and Ma, K.P. (2006) Treeline Dynamics in Relation to Climatic Variability in the Central Tianshan Mountains, Northwestern China. Global Ecology and Biogeography, 15, 406-415. http://dx.doi.org/10.1111/j.1466-822X.2006.00233.x
- Zhang, X.W. and Liu, B. (2010) Effect of Chromium on Crops Growth. Environmental Science, 23, 48-51.
- Hu, Y.Q. and Zhao, S.J. (2010) RAPD Technology and Its Application in Plant Research. Biotechnology Bulletin, 5, 74-77.
- Williams, J.G.K., Kubelik, A.R., Licak, J.A., et al. (1990) DNA Polymorphisms Amplified by Arbitrary Primers Are Useful as Genetic Marker. Nucleic Acids Research, 18, 6531-6535. http://dx.doi.org/10.1093/nar/18.22.6531
- Wen, C.H., Wang, Z.H. and Duan, C.Q. (1998) Pollution Resistant Differentiation and Evolution in Plants and the Application of Molecular Biology Technique. Ecological Science, 17, 19-24.
- Živković, B., Radović, J., Sokolović, D., Šiler, B., Banjanac, T. and Štrbanović, R. (2012) Assessment of Genetic Diversity among Alfalfa (Medicago sativa L.) Genotypes by Morphometry, Seed Storage Proteins and RAPD Analysis. Industrial Crops and Products, 40, 285-291. http://dx.doi.org/10.1016/j.indcrop.2012.03.027
- Bocianowski, J. and Seidler-Łożykowska, K. (2012) The Relationship between RAPD Markers and Quantitative Traits of Caraway (Carum carvi L.). Industrial Crops and Products, 36, 135-139. http://dx.doi.org/10.1016/j.indcrop.2011.08.019
- Cansaran-Duman, D., Atakol, O. and Aras, S. (2011) Assessment of Air Pollution Genotoxicity by RAPD in Evernia prunastri L. Ach. from around Iron-Steel Factory in Karabük, Turkey. Journal of Environmental Sciences, 23, 1171- 1178. http://dx.doi.org/10.1016/S1001-0742(10)60505-0
- Li, J.M., Jin, Z.X. and Zhu, H.C. (2007) Heavy Metal Pollution Fern Population Genetic Differentiation Analysis of RAPD. Journal of Ecology, 26, 171-176.
- Wen, C.H., Duan, C.Q., Xiu, C.X., et al. (2001) Heavy Metal Pollution of Differentiation of RAPD Analysis of Datura Stramonium L. Journal of Ecology, 21, 1238-1245.
- Gu, Q.P., Jin, Z.X. and Li, J.M. (2008) Under Heavy Metal Stress Clethroides Populations Genetic Diversity Analysis of RAPD. Jiangsu Agricultural Sciences, 2008, 54-58.
- Yu, Z.X., Ou, G.Z., Chen, Q.X., et al. (2010) Pitaya Total DNA Extraction Method Comparison Research. Agricultural Science Bulletin, 26, 300-303.
- Cheng, S.P., Shi, J., Shi, G.A., et al. (2010) Pistacia chinensis Leaf Genomic DNA Extraction Methods. Journal of Henan University of Science and Technology, 31, 71-73.
- Wang, J.J., Wu, S.L., Zhang, F.C., et al. (2010) Grape Genome DNA Extraction and RAPD Reaction System Optimization. Agricultural Sciences of Xinjiang, 47, 1066-1070.
- Lin, G.M., Sui, F.F., Lin, N., et al. (2010) Wilfordii Leaf DNA Extraction and RAPD Reaction System. Chinese Agricultural Science Bulletin, 26, 40-43.
- Singh, S., Panda, M.K. and Nayak, S. (2012) Evaluation of Genetic Diversity in Turmeric (Curcuma longa L.) Using RAPD and ISSR Markers. Industrial Crops and Products, 37, 284-291.
- Ahmad, M.A., Gaur, R. and Gupta, M. (2012) Comparative Biochemical and RAPD Analysis in Two Varieties of Rice (Oryza sativa) under Arsenic Stress by Using Various Biomarkers. Journal of Hazardous Materials, 217-218, 141-148. http://dx.doi.org/10.1016/j.jhazmat.2012.03.005
- Tang, Y., Wang, P.P. and Zhang, N. (2006) Research in Heavy Metal Toxicity Mechanism in Plant. Journal of Shengyang Agricutural University, 37, 551-555.
- Tao, J.X., Zhang, X.H., Luo, H., et al. (2010) Leersia hexandra Swartz for Electroplating Sludge Contaminated Soil Chromium Copper Nickel Uptake and Accumulation. Journal of Guilin University of Technology, 30, 144-147.
- Gupta, S., Srivastava, M., Mishra, G.P., Naik, P.K., Chauhan, R.S., Tiwari, S.K., Kumar, M. and Singh, R. (2008) Analogy of ISSR and RAPD Markers for Comparative Analysis of Genetic Diversity among Different Jatropha curcas Genotypes. African Journal of Biotechnology, 7, 4230-4243.
NOTES

*Corresponding author.