Open Journal of Immunology
Vol.06 No.01(2016), Article ID:64699,20 pages
10.4236/oji.2016.61003
Proteomics and Its Applications in Diagnosis of Auto Immune Diseases
Gebrehiwot Gebretsadik1*, M. K. C. Menon2
1Department of Medical Biochemistry, College of Health and Medical Sciences, Haramaya University, Haramaya, Ethiopia
2Department of Medical Biochemistry, College of Health Sciences, Addis Ababa University, Addis Ababa, Ethiopia

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 11 September 2015; accepted 15 March 2016; published 18 March 2016
ABSTRACT
Although the proteomics and its applications in detecting autoimmune diseases are a prominently discussed issue, this review will focus particularly some prominent aspects regarding clinical utility of these techniques in prognosis, diagnosis, and treatment of these diseases. The impact of immunofluorescent techniques, enzyme immunoassays and use of proteomics biomarkers in the characterization of the auto immune diseases is briefly discussed. The necessity of adopting existing technologies of protein chemistry, predisposition testing, targeted monitoring and prevention of diseases through nutrition coupled with lifestyle changes will be focused as modern diagnostic tools in realizing the changeover from isolated medicine to personalized medicine. Use of biological fluids, in order to identify low abundance proteins as biomarkers in detecting autoimmune diseases is attempted in the study of serum/plasma proteomics.
Keywords:
Proteomics, Autoimmune Diseases, Biomarkers, Genomics

1. Introduction
1.1. Proteomics
Proteomics is the study of proteins, particularly their structures and functions [1] [2] . Proteins are vital parts of living organisms, as they are the main components of the physiological metabolic pathways of cells. The term proteomics was first coined in 1997 [3] to make an analogy with genomics, the study of the genome.
Proteomics is much more complicated than genomics. Most importantly, while the genome is a rather constant entity, the proteome differs from cell to cell and is constantly changing through its biochemical interactions with the genome and the environment. One organism has radically different protein expression in different parts of its body, in different stages of its life cycle and in different environmental conditions [4] .
The entirety of proteins in existence in an organism throughout its life cycle, or on a smaller scale the entirety of proteins found in a particular cell type under a particular type of stimulation, are referred to as the proteome of the organism or cell type respectively. Since proteins play a central role in the life of an organism, proteomics is instrumental in discovery of biomarkers, such as markers that indicate a particular disease.
Proteomics is the comprehensive study of expression, function and interactions of proteins in a cell, tissue and biological fluid. In recent years, proteomics has being vastly used in research of autoimmune diseases for its powerful capability of parallel detection of thousands of different proteins or antigens using biological fluids, tissues and cells from the patients [5] .
1.1.1. Current Proteomic Technologies
Proteomics has steadily gained momentum over the past decade with the evolution of several approaches. Few of these are new and others build on traditional methods. Mass spectrometry-based methods and micro arrays are the most common technologies for large-scale study of proteins.
There are two mass spectrometry-based methods currently used for protein profiling. The more established and widespread method uses high resolution, two-dimensional electrophoresis to separate proteins from different samples in parallel, followed by selection and staining of differentially expressed proteins to be identified by mass spectrometry. Despite the advances in 2DE and its maturity, it has its limits as well. The central concern is the inability to resolve all the proteins within a sample, given their dramatic range in expression level and differing properties [6] .
The second quantitative approach uses stable isotope tags to differentially label proteins from two different complex mixtures. Here, the proteins within a complex mixture are labeled first isotopically, and then digested to yield labeled peptides. The labeled mixtures are then combined, the peptides separated by multidimensional liquid chromatography and analyzed by tandem mass spectrometry. Isotope coded affinity tag (ICAT) reagents are the widely used isotope tags. In this method, the cysteine residues of proteins get covalently attached to the ICAT reagent, thereby reducing the complexity of the mixtures omitting the non-cysteine residues.
Quantitative proteomics using stable isotopic tagging is an increasingly useful tool in modern development. Firstly, chemical reactions have been used to introduce tags into specific sites or proteins for the purpose of probing specific protein functionalities. The isolation of phosphorylated peptides has been achieved using isotopic labeling and selective chemistries to capture the fraction of protein among the complex mixture. Secondly, the ICAT technology was used to differentiate between partially purified or purified macromolecular complexes such as large RNA polymerase II pre-initiation complex and the proteins complexed with yeast transcription factor. Thirdly, ICAT labeling was recently combined with chromatin isolation to identify and quantify chromatin- associated proteins. Finally ICAT reagents are useful for proteomic profiling of cellular organelles and specific cellular fractions [6] .
Balancing the use of mass spectrometers in proteomics and in medicine is the use of protein micro arrays. The aim behind protein micro arrays is to print thousands of protein detecting features for the interrogation of biological samples. Antibody arrays are an example in which a host of different antibodies are arrayed to detect their respective antigens from a sample of human blood. Another approach is the arraying of multiple protein types for the study of properties like protein-DNA, protein-protein and protein-ligand interactions. Ideally, the functional proteomic arrays would contain the entire complement of the proteins of a given organism. The first version of such arrays consisted of 5000 purified proteins from yeast deposited onto glass microscopic slides. Despite the success of first chip, it was a greater challenge for protein arrays to be implemented. Proteins are inherently much more difficult to work with than DNA. They have a broad dynamic range, are less stable than DNA and their structure is difficult to preserve on glass slides, though they are essential for most assays. The global ICAT technology has striking advantages over protein chip technologies [6] .
This is a promising and newer microarray application for the diagnosis, study and treatment of complex diseases such as cancer. The technology merges laser capture microdissection (LCM) with micro array technology, to produce reverse phase protein microarrays. In this type of microarrays, the whole collection of protein themselves are immobilized with the intent of capturing various stages of disease within an individual patient. When used with LCM, reverse phase arrays can monitor the fluctuating state of proteome among different cell population within a small area of human tissue. This is useful for profiling the status of cellular signaling molecules, among a cross section of tissue that includes both normal and cancerous cells. This approach is useful in monitoring the status of key factors in normal prostate epithelium and invasive prostate cancer tissues. LCM then dissects these tissue and protein lysates were arrayed onto nitrocellulose slides, which were probed with specific antibodies. This method can track all kinds of molecular events and can compare diseased and healthy tissues within the same patient enabling the development of treatment strategies and diagnosis. The ability to acquire proteomics snapshots of neighboring cell populations, using reverse phase microarrays in conjunction with LCM has a number of applications beyond the study of tumors. The approach can provide insights into normal physiology and pathology of all the tissues and is invaluable for characterizing developmental processes and anomalies [6] .
1.1.2. The Proteomic Analysis and Identification of Biomarkers in Detecting Diseases
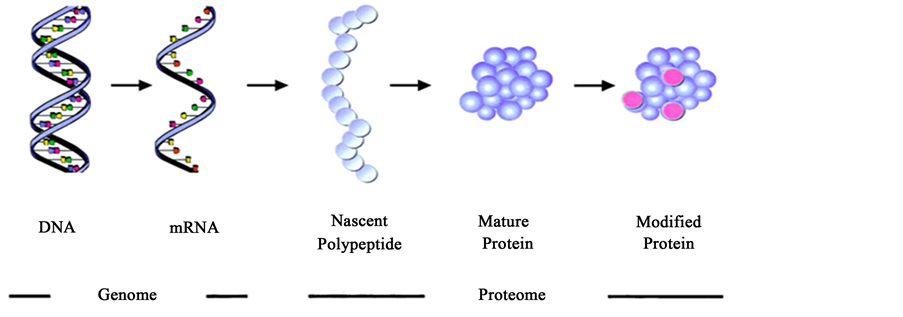
“Genomics” and “Genome” are “-omics” terms which focused on individual genomes in various organisms, which resulted in genome-sequencing projects and characterization of individual genes and proteins derived from them (Figure 1). Northern Blots and Western blots were the techniques then available, and only a handful of genes or proteins could be analyzed for their status as producers of diseases. Subsequent developments changed the situation, and several excellent reviews appeared during the 1990’s which focused on the growth of the gene, expressed sequence tag (EST), and protein-sequence data bases. The genome-sequencing projects yielded complete sequences of numerous organisms including human genome, and produced browser-based bioinformatics, catalogs containing databases which can predict the functions of protein products. Subsequent development of oligonucleotide microarray, including gene specific oligonucleotides can probe various genomic sites for abnormalities [7] - [9] .
Table 1 provides a short summary of the key features that distinguishes protein biochemistry with that of the proteomics. Physical Biochemists and enzymologists typically study the relationship between protein structure and function using one protein, or a multisubunit protein complex by amino acid sequence analysis, structure determination and modeling studies.
Figure 1. Biochemical context of genomics and proteomics (Source: Loscalzo J, Circulation, 2003; 108: 380-383).
Table 1. Differences between protein chemistry and proteomics.
(Source: Daniel C. Liebler, introduction to proteomics, Humana press, 2002).
Undoubtedly, Proteomics on the other hand study multiprotein systems, instead of single components or study partial sequence analysis using database matching tools. Therefore, Proteomics is systems biology rather than structural biology and directs the characterization and behavior of systems [10] .
Gene microarrays offer a snapshot of the expression of many or all genes in a cell. Unfortunately, the levels of mRNAs do not necessarily predict the levels of the corresponding proteins in a cell. Differing stability of mRNAs and different efficiencies in translation can affect the generation of new proteins. Once formed, proteins differ significantly in stability and turnover rates. Many proteins involved in signal transduction, transcription-factor regulation, and cell-cycle control are rapidly turned over as a means of regulating their activities. Finally, mRNA levels tell us nothing about the regulatory status of the corresponding proteins, whose activities and functions are subject to many endogenous posttranslational modifications and other modifications by environmental agents.
The sequencing of the human genome is undoubtedly one of the major accomplishments of biomedical science [7] [8] . Knowledge of the precise sequence of all human genes provides unparalleled access to a complete understanding of human biology in all of its complexity. Or does it? The completion of the human genome sequence showed far fewer genes than expected, ≈30,000, which is not that different from that of the lowly earthworm, with at least 17,300 [9] . This number of genes was viewed with surprise by some investigators as too few to account for human biological diversity in form and function. The genome-centric view of the biological universe, however, is rather myopic and fails to take into consideration what protein chemists have known for almost a century. Proteins, the ultimate products of the human genome, define biology. Proteins are directly responsible for all biological form and function. It is estimated that there are ≈6 to 7 times as many distinct proteins (i.e. ≈200,000) as genes in humans, in part owing to splicing and exchange of various structural cassettes among genes during transcription. Proteins are not only more abundant than the genes that encode them, but they are also much more structurally complex with primary, secondary, tertiary, and quaternary structural elements; additionally, they have greatly varied biochemical functions that critically depend on structure. Furthermore, mature proteins are also subject to a host of post-translational modifications, including proteolysis, sulfhydryl oxidation and disulfide bond formation, phosphorylation, glycosylation, S-nitrosation, fatty acylation, and oxidation. These biochemical modifications often yield products with functions different from those of the unmodified parent protein, and many of these modifications, such as oxidation, reflect the consequences of environmental modulation of genetic determinants. Taken together, the many post-translational changes in protein structure and function add incredible complexity to that of the basic genome and constitute what has become the protein equivalent of the genome, namely, the proteome. In parallel to genomics, proteomics is, thus, defined as the sequence, modification, and function of all proteins in a biological system.
1.1.3. Shortcomings of Transcriptional Profiling
Several studies culled genes encoding proteins with pathophysiologic and therapeutic relevance from DNA microarray analysis of tissue derived from autoimmune lesions. Nevertheless, mRNA transcipt profiling has important limitations and will likely prove insufficient to unravel the etiology of, and to develop next-generation therapeutics for, autoimmune disease. A growing number of studies suggest that relatively frequent discordance exists between mRNA and protein expression. As outlined in Table 2, in the study of autoimmune disease, RNA
Table 2. Immunology questions not adequately addressed by transcriptional profiling.
(Source; William H. Robinson, Lawrence Steinman, et al. Protein and Peptide Array Analysis of Autoimmune Disease. BioTechniques 2002).
transcript profiling of certain genes is not informative with regard to net protein expression, protein function, and/or the specificity of antigen receptors. Proteomics, which is the direct study of the expression and function of proteins encoded by these RNA transcripts, circumvents many of the limitations of RNA transcript profiling and will likely provide critical insights into the mechanisms of autoimmune disease in the post genomics era [11] .
1.2. Autoimmune Diseases
Autoimmune diseases are a family of chronic, and often disabling, illnesses that develop when underlying defects in the immune system lead the body to attack its own organs, tissues, and cells [12] . While many of these diseases are rare, collectively they affect 14.7 to 23.5 million people in United States of America, and―for reasons unknown―their prevalence is rising. Since cures are not yet available for most autoimmune diseases, patients face a lifetime of illness and treatment.
There are over 80 autoimmune diseases known. Multiple mechanisms of autoimmunity induction have been proposed including, among the others, molecular mimicry [13] , viral epitope delivery [14] , generation of neoan- tigenic epitopes after posttranslational modification [15] , unusual TCR-binding properties that permit autoreactive T cells to escape deletion [16] , presence of long regions of extreme structural disorder in the autoantigens [17] , cellular injury and release of self antigens, which generate immune responses [13] .
1.2.1. B-Cell Lymphoma-Extra Large (bcl-xS) B-Cell Lymphoma-Extra Large (bcl-xL)
Autoimmune diseases comprise a wide variety of systemic or organ-specific inflammatory diseases, characterized by aberrant activation of immune cells that target self tissues due to misrecognizing tissue-derived proteins as foreign antigens [18] [19] . The prevalence of autoimmune diseases is approximately 2000 - 3000 per 100,000, although the prevalence varies depending on the diseases, ethnic groups and regions [20] . The etiology and exact pathogenesis of autoimmune diseases remain poorly understood. However, both genetic factors and environmental triggers are profoundly involved in the pathogenesis of autoimmune diseases. Autoimmune diseases are a heterogeneous group of diseases characterized by immune reactions against one or more components of the body itself, causing inflammation and damage to tissues and organs [21] . Autoantibodies are a hallmark of many autoimmune diseases and the presence of autoantibodies is a distinctive and key characteristic of autoimmune diseases. For some systemic autoimmune diseases, moreover, the presence in serum of certain autoantibodies represents one of the classification criteria. It should be underlined that autoantibodies in autoimmune diseases do not appear individually, associating a single, specific autoantibody with a certain well-defined disease; in general, autoimmune diseases are characterized by the presence of several autoantibodies. As an example, Sherer et al. described more than 100 different autoantibodies in systemic lupus erythematosus (SLE) patients [22] .
Although autoimmune diseases affect 3% of the US population, and likely a similar percentage of the population of the industrialized world [23] , it has been posed a challenge to clinicians and research scientists ever since the 19th Century. Despite recent acquisition of extensive information, understanding of the mechanisms leading to pathogenic autoimmunity is fragmentary and incomplete. Recent advances in immuno-diagnostics have begun to assemble the missing pieces of the puzzle. Currently, a large number of autoimmune diseases have been diagnosed clinically with support from laboratory investigations. Increasingly different immunological tests have been evolved with higher specificities and sensitivity to diagnose these diseases. These tests are helpful in defining particular subsets of patients with better or poorer prognosis, different patterns of organ involvement, variety of responses to a given treatment or progression of the disease. The evolution of immunoassays has brought considerable benefits in terms of analytical, operational and clinical outcomes. In this respect, immunodiagnostics has proven to be a critical step in the development of many aspects of modern clinical practice.
Proteomics enables correlations to be established between the range of proteins produced by a cell or tissue and the initiation or progression of a disease state. Proteome research helps in the discovery of new protein markers for diagnostic purposes and of novel molecular targets for drug discovery. The abundant information provided by proteome research is complementary to the genetic information generated by genomic research, which at a broad level attempts to catalogue and characterize these proteins, compare variations and their expression levels in health and disease, study their interactions, and identify their functional role. Field of proteomics and bioinformatics will explore an area of autoantibody profiling of various autoimmune diseases in the coming decades. This will prove to be a beneficial tool for classifying patients with varying degrees of severity, subsets of patients based on autoantibody fingerprint, to characterize autoreactive B cell epitope spreading, isotype usage, identification and characterization of biomarker for candidate autoantigen as well as in designing antigen specific immuno-modulating therapies. In near future, proteomics will broaden our understanding of immunopathogenic mechanisms of autoimmune disorders which will help in early diagnosis; prognosis and management of patients suffering from various autoimmune disorders [24] .
Recent studies to identify antigen specificities of autoantigens, and to identify newer target autoantigens to anti-neutrophil cytoplasmic antibodies (ANCA), myositis specific antibodies, anti-endothelial cell antibodies (AECA), anti-red cell antibodies and anti-endomysial antibodies are based on proteomics. Protein microarrays to identify peptides representing candidate autoantigens is now a powerful technique for immunodiagnosis of Autoimmune diseases like systemiclupus erythematosus, rheumatoid arthritis (RA), polymyositis and systemic sclerosis. Advances in the field of proteomics will open a new wing of autoantibody research and will lead to identification of new auto antigens and corresponding autoantibodies. Studies on new autoantigens will be based on combining traditional techniques like 2-DE and western blotting with latest mass spectrometric (MS) analysis and protein database retrieval using bioinformatics tool. About 1500 different proteins have recently been identified, and a number of potential new markers of diseases have been characterized [25] .
Early immunoassays capable of multiplex analysis include: ELISAs, fluorescence-based immunoassays, and radio-immunoassays performed in microtiter plates; arrays of peptides synthesized on plastic pins [19] , western blot analysis; and genetic plaque-based and colony-based assays. All of these technologies are limited by requirements for relatively large quantities of reagents and of clinical samples. Genetic plaque-based and colony- based assays are further limited by incomplete addressability; DNA sequence analysis is required to determine the identity of the antigens at each location on the array.
Nowadays in modern clinical laboratories chemical, hematologic and immune-chemical parameters can be efficiently measured using automated equipment. Advances in technology with improved understanding of molecular pathology, allow for a fast expansion of diagnostic methods and multiplication of the parameters measured in the laboratory. The development of miniature laboratory chips will reduce the clinical sample and reagent volumes and allow more data to be obtained in a shorter period of time [26] . Currently laboratory tests are used for diagnosis and monitoring of disease activity or therapy. The future tests will focus on predisposition testing, targeted monitoring, and prevention of diseases through nutrition, lifestyle and drug therapy [27] . Modern diagnostics is changing from isolated medicine to personalized medicine [27] . This approach may create a greater opportunity to prevent diseases. The rapid expansion of the diagnostic tools can be attributed to developments in proteomic and genomic technologies.
1.2.2. Expansion to New Autoimmune Disease Areas
Autoimmune diseases not previously investigated on a genomic or proteomic scale are now the subject of high throughput screening studies and include scleroderma, systemic sclerosis, dermatomyositis, juvenile idiopathic arthritis, and Behçet’s disease. Studies on other less common diseases have been conducted, results of which have provided the first piece of evidence of distinct proteomic patterns in autoimmune diseases that otherwise exhibit quite similar phenotypic presentation. It is hoped that other less common but equally malignant autoimmune inflammatory conditions, including orphan diseases such as retroperitoneal fibrosis and relapsing polychondritis, will soon be subjected to genomic and proteomic investigations. New molecular insights gleaned from genomic and proteomic profiling of rare diseases will provide pharmaceutical companies with new incentives to test their pathway-specific therapies in this neglected field, ultimately leading to more and better therapeutic choices for patients with these serious conditions [28] .
2. Current Trends in Proteomics
The analysis of complex protein mixtures such as serum, other body fluids or tissues by profiling hundreds of proteins in the same time, creates pattern of response characteristic for various cellular states or disease conditions. Detailed proteome analysis has become more realistic today with the high?resolution mass spectrometers capable of faster sequencing in a high?throughput fashion and with the emergence of new techniques such as microarrays. A promising area is the application of advanced mass spectrometric and other quantitative proteomic methodologies to laboratory diagnostics [29] [30] . The major proteomic projects of the last decade have shaped proteome-wide sequencing, mapping, and analysis [31] . For example, the creation of the Human Proteome Organization’s Human Brain Proteome Project fosters the effective international exchange of brain related proteomic data [31] . Complex diseases are now rapidly investigated by novel high?throughput biochemical tech- nologies to uncover disease activity, clinical markers, and drug targets. Such diagnostic technologies will lead to personalized medicine.
Opposite to the genome, the proteome is composed of an active array of molecules constantly being modified and with special localization. Proteomic approaches are able to characterize also post?translational modifications, by which the cell quickly modifies protein functions. Protein profiling and identification techniques using advanced mass spectrometry and bioinformatics can lead to the discovery, identification, and characterization of protein biomarkers [32] . Comprehensive proteomic profiling is able to identify thousands of proteins from various clinical samples. Comparison of the proteomes of patient’s and control sample may result in the identification of diseases specific proteins. Similar to gene expression profiling, several protein profiling techniques do not require a priori knowledge of candidate proteins. New tools for highly specific, sensitive, parallel protein analyses both in body fluids and tissue extracts will make a profound impact on clinical diagnostics in the near future. Profiling the proteomes of diseased and healthy tissues allows for the discovery of peptide or protein molecular change, which potentially reveals information on pathogenesis or diagnosis, or both [31] .
Current proteomic research follows at least two pathways. In the first, the identification of proteins in patient’s samples can be used as diagnostic or prognostic disease markers. The second goal is to discover cellular proteins related to the response to various therapies. Once tools for conducting comprehensive proteome analysis became available, much of the interest turned towards analyzing proteins for the purpose of finding novel biomarkers of diseases, such as cancer. In the future, the ability to routinely identify thousands of proteins in the body fluids will be available. Because of the unique protein content of these samples, strategies for removing highly abundant proteins needed to be developed [33] . Several highly expressed proteins, particularly serum albumin, transferin, and immunoglobulins, often mask lower abundance proteins. Thus purification measures to fractionate and better resolve protein population have to be undertaken. One of the biggest challenges in finding biomarkers in clinical samples was throughput. To obtain the identification of thousands of proteins in a sample a high throughput technique is needed. Unless there is a major breakthrough in technology, biomarker discovery studies using comprehensive proteomic identification will remain low?throughput compared to genomic microarray analysis.
Initial proteomic studies relied on 2D-gel electrophoresis, which separates proteins based on isoelectric point and molecular weight [31] . This process has limited reproducibility, is complicated and not robust. Moreover weakly soluble proteins cannot be easily resolved and only a tiny portion of the proteome can be effectively stained. Another shortcoming of this method is that low-level expressed proteins can be masked by greater expression within a similar molecular weight or isoelectric point, or both. Edgar et al. applied this approach to the hippocampal proteome of schizophrenia patients [34] .
Proteomics advanced dramatically with the advent of mass spectrometric analysis for peptides (MALDI) [35] [36] . There are four steps in mass spectrometry. First, the ion source generates ionized proteins from the sample. Second, the mass analyzer sorts and resolves proteins based on their mass/charge ratio. Third, the ion detector spots the ions and composes data on the ion mass/charge ratio, quantity, and time of flight (TOF), or the time it took to reach the detector. Finally, bioinformatic analysis allows interpretation of the raw data. After a mass spectrometry run, in a process called peptide mass fingerprinting, the peptides are arranged into several databases to allow protein identification. Although peptide mass fingerprinting is a method of protein identification, it often requires extensive and often complex purification, as it tenders to interpret protein match by peptide masses rather than by sequences. Mass spectrometry has evolved to incorporate tandem mass spectrometric technology that permits effective sequencing. The MALDI?TOF is an advanced technology, a cutting-edge proteomic tool with direct amino acid sequencing and characterization capabilities [29] [35] . As mass spectroscopy continues to improve, it may replace immunoassays as the best method for measuring specific analytes in biologic samples.
Surface Enhanced Laser/Desorption Ionization (SELDI), a variation of MALDI, is a new generation of mass spectrometric analysis, and offers better accuracy with built in chromatography [29] . The central technology platform is a protein chip mass spectrometer, which uses a powerful new approach (surface-enhanced laser desorption/ionization time-of-flight mass spectrometry, or SELDI-TOF MS) to the analysis of complex protein mixtures such as serum and tissue extracts by profiling hundreds of proteins in the same time, thus creating characteristic patterns related to various cellular states or disease conditions. Each chip contains a unique chromatographic surface for selective protein capture [37] .
Functional study employed the profiling and sequencing properties of tandem mass spectrometric analysis. However, several studies have demonstrated the potential of this technology in determining the complexities of the dynamic proteome. Sequence analysis can detect important post?translation protein modifications such as methylation, acetylation, sulfation, phosphorylation, ubiquitylation and glycosylation [31] [38] . Protein detection can be performed on microarrays, however heterogeneity and relative instability of proteins is a challenge. Current research focus both on protein microarray construction and molecular strategies for specific and sensitive detection Antibody arrays can be used for protein expression studies and as diagnostic and discovery tools in autoimmunity [39] .
One of the important tools in clinical proteomics is tissue microarrays. They allow analyzing hundreds of tissue specimens in the same time. Tissue arrays investigate the distribution of proteins directly at the disease site [40] . The obtained results can be assessed manually or automatically and can be analyzed together with clinical data.
The number of diagnostic tools has been steadily expanding since the advent of modern medicine. New omics technologies will allow thousands of results per sample to be generated. Novel clinical tests could be used to determine therapy type, duration, and dose; and its efficiency. Genomics and proteomics may also reveal distinct etiologies and subtypes for better classification of the disease. Applying genomic and proteomic methods to clinically accessible body fluids (e.g., serum, cerebrospinal fluid, and urine) would place valuable objective analytical power in the hands of the clinician [41] . With personalized medicine, therapy will be based on individual patient characteristics that become known through bioinformatics. The expected results will give the response rates close to 100%, as well as increased survival rates, improved quality of life, cost savings, and reduced morbidity and mortality. For several years proteomics research has been expected to lead to the finding of new markers that will translate into clinical tests applicable to numerous clinical samples such as serum, plasma and urine [42] [43] . Attempts to implement technologies applied in proteomics, in particular protein arrays and surface-enhanced laser desorption ionization time?of?flight mass spectrometry, as diagnostic tools have initiated constructive discussions on opportunities and challenges. Genome and proteome-based research offer the promise of more effective diagnostic tools, greater understanding of an individual’s healthcare needs, and targeted treatments for diseases that affect a vast majority of the population such as cancer, diabetes and cardiovascular disease [44] [45] . The role of genomics and proteomics in health care is increasingly driven by the need for integrated approaches to disease prevention, earlier diagnosis and overall response to therapy―the new era of personalized medicine [46] .
2.1. Techniques and Methods Used for the Detection of Auto Antibodies
Screening tests for autoantibody detection has been performed for a variety of systemic and organ specific autoimmune diseases. Requests for these tests have risen remarkably, mainly due to the increased understanding of the nature of auto antibodies. A simultaneous development of new methods and analytical systems in clinical immunology has involved a constantly increasing expenditure of economic resources for the assay of antibodies. Inadequate use of laboratory tests is one of the most frequent problems in autoimmunity, leading to incorrect diagnoses and inadequate treatment [47] .
2.1.1. Immunofluorescence
The indirect immunofluorescence (IIF) technique, which uses various tissue sections or the human tumor cell line (HEp-2) as an antigenic source, has had major implications for the diagnosis of autoimmune diseases in a routine laboratory setting. In IIF, undefined antigens are recognized by auto antibodies from the patient’s sera and yield specific patterns which must be interpreted in their relation to disease association (Table 3) [47] .
The IIF staining pattern of a positive sample can be used to evaluate which appropriate antigen specificities to look for. It is known that the HEp-2 cells used for the detection of autoantibodies do not have a satisfactory ability to give positive IIF results for antibodies to SS-A/Ro-52 and Jo-1 (histidil-tRNA synthetase). Many serum samples give speckled or grainy homogenous staining patterns which cannot be clearly identified as one of the known patterns. The presence of various antigenic targets on the tissue section results in an excellent overall sensitivity. Antinuclear antibodies which have undefined specificities can be seen in the serum from patients with a wide variety of autoimmune diseases, infectious diseases and in some healthy individuals. The lack of specificity may result in misleading interpretation and demonstrate the limitations of the IIF technique for screening purposes. Indirect immunofluorescence microscopy is a sensitive method; yet, it has some limitations like substrate variations, manual performance, subjective result interpretation, low reproducibility and a lack of standardization. IIF
Table 3. Autoantibody patterns and their relation to specific auto antibodies and disease.
AA―autoantibody; AH―autoimmune hepatitis; APS―anti phospholipid syndrome; CENP―centromere protein; dcSS―diffuse cutaneous SSc; DIL―drug induced lups; DM―dermatomyositis; GW―glycin, tryptophan containing hUBF; lcSSc―limited cutaneous SSc; MCTD―mixed connective tissue disease; NLS―neonatal lupus syndrome; NOR―nuclear organizer; NuMA―nuclear mitotic apparatus; PBC―primary biliary cirrhosis; PCNA―proliferating cell nuclear antigin; PM―polymyositis; RNAP―RNA polymerase; RNP―ribonucleoprotin; RP―Raynauds phenomenon; SCLE―subacute cutaneous lupus erythematosus; SjS―sjogren syndrome; SLE―systemic lupus erythematosus; SRP―signal recognition particle; SSc―systemic sclerosis. (Source; Ilza Salamunić. Laboratory diagnosis of autoimmune diseases―new technologies, old dilemmas. Biochemia Medica 2010; 20 (1): 45-56.)
is time-consuming, resulting in a low throughput and increased personnel costs. In order to overcome this limitation, fully automated IIF interpretation systems with pattern-recognition software have been introduced recently. Standardization of autoantibodies testing by IIF remains a critical issue in and between routine laboratories and may be improved by automated interpretation systems.
The analysis of autoantibodies by IIF remains the hallmark of diagnosis, but some investigators have claimed that this technique is becoming out-of-date and that it could/should be replaced by enzyme immunoassays or multiplexed assays in the routine laboratory diagnostics [48] .
2.1.2. Enzyme Immunoassay
The enzyme-linked immunosorbent assay (ELISA) based either on antigens prepared from human tumor cell line (HEp-2) nuclear extracts and highly purified nuclear antigens or from recombinant antigens, has been the most promising, but the main differences, in terms of positively results, among various enzyme immunoassay methods have been described. The isolation of autoantigens from natural sources such as human tissues has major limitations with respect to reproducibility and purity. Many proteins are present only in limited amounts and their purification requires the removal of other potentially antigenic targets. The specificity of ELISAs for autoantibody measurements is strongly dependent on the quality of antigens used, and it is important that an antigen should have exactly the same sequence, conformation and post-translational modifications as the human antigen [49] .
EIA is now widely used for identifying specific auto antibodies to nuclear or cytoplasmic antigens of different group of organ-specific disorders, such as Grave’s disease, primary biliary cirrhosis, insulin-dependent diabetes mellitus or systemic affecting different organs like systemic sclerosis, Sjogren’s syndrome, mixed connective tissue disease or rheumatoid arthritis.
The HEp-2 antinuclear antibody EIA (HEp-2 ANA EIA) is an automated method with high reproducibility and internal calibration as a basis for standardization. However, the evaluation of clinically well-defined samples in case of scleroderma patients with the use of HEp-2 ANA EIA for example yielded a lower rate of positive results compared to ANA IIF. Many studies conducted under standardized conditions showed the analytical variability of different test systems.
2.1.3. Multiplexed Immunoassays
Multiplexed immunoassays support the identification of multiple auto antibodies from a single determination in the same time.
2.1.4. Microarray Based Assays
The line-blot immunoassay is a multiplexed immunoassay which allows the parallel analysis of different types of auto antibodies. Line assays use recombinant antigens almost exclusively which are immobilized in straight lines on a nylon test strip. When incubated with serum, auto antibodies that are present in the sample bound to the auto antigen lines on the strip. Bound auto antibodies are visualized with a color-detection system that relies on alkaline phosphatase activity. Results are interpreted by comparing the color intensities of the antigen lines with those of the cut off lines. Some publications have demonstrated that a few percent of IIF ANA negative sera will be positive using assay line assay, especially for anti-SS-A/Ro [50] .
The planar microarray technology was developed and applied for the simultaneous detection of different autoantibodies using the sandwich immunoassay format. Different autoantigens are immobilized on a microarray along with control proteins. The arrays are subsequently incubated with patient sera and bound auto antibodies are detected by a labeled secondary antibody. Most of the microarray assays formats applied chemiluminescence or fluorescence based detection methods.
2.1.5. Bead-Based Assay
As an alternative to planar microarrays, flow cytometry for the analysis of bead-based immunoassays has been developed [50] . Recently a commercially available microsphere-based fluorescent assay has been introduced for the detection of ANA. The potential low cost and time saving may be a reason for the routine use of these assays in the research and clinical laboratories. Immunoassay systems with micro-bead technology and flow cytometry detection (xMAP technology) have been applied to autoantibody measurement. The system uses polystyrene microspheres labeled internally with different ratios of two different fluorochromes. Each fluorochrome can have any of the 10 possible levels of fluorescence intensity, thereby creating a family of 100 spectrally addressed bead site. The antigens corresponding to auto antibodies are bound to the microspheres. Each of the 100 micro beads that can be differentiated by their fluorescence carries a specific immobilized antigen for a single autoantibody. At the same time, a green laser excites the external reporter fluorescence to quantify the specific reaction related to each autoantibody. Several companies supply commercial tests for simultaneous measurement of different auto antibodies by flow cytometry. The evaluation of a assay from different producers for the simultaneous quantitative determination in the same sample of nine antinuclear autoantibody specificities (dsDNA, SS-A/Ro, SS-B/La, Sm Sm/RNP, Scl-70, Jo-1, ribosome and centromere B) yielded good results. Clinical evaluation of a microsphere bead-based flow cytometry assays for simultaneous determination of anti-thyroidperoxidase and anti-thyroglo- bulin antibodies showed good agreement with ELISA [47] .
One problem in the detection of auto antibodies from patients is the lack of true quantitative calibration because of the different affinities of the antibodies to antigens. However, the major questions remain regarding whether the quantitative data obtained by multiplex bead-base assays are identical to, or at the least similar, to data obtained using other methods. Sensitivity, reliability and accuracy are similar to those observed with ELISA procedures.
2.1.6. Proteomic’s Technologies for the Study and Diagnosis of Autoimmune Diseases
Clinical proteomics offers opportunities to identify new disease biomarkers in body fluids, cells and tissues. The focus of clinical proteomics in on the analytical and clinical validation and implementation of novel diagnostic or therapy related markers [51] .
Protein microarrays represent a validated platform for profiling protein levels and their post-translational modifications. Proteomic technologies including antigen microarray platforms enable the large-scale characterization of immune responses against foreign and self antigens that may be involved in the development and progression of autoimmune disease. Antigen microarrays allow the comprehensive analysis of auto antibodies directed against hundreds to thousands of antigens, including proteins, peptides, nucleic acids, macromolecular complexes.
Molecular probes can be monoclonal or polyclonal antibodies. Arrays are probed with cell culture, supernatant, cell lysate or serum. Depending on the molecule probe used, proteins or antibodies in the sample are bound to the planar array. The bound molecules are detected by a secondary antibody marked with fluorescent dye, or directly if the sample has been fluorescently labeled. The incubated chips can be read by a variety of scanners based on planar guide technology.
Although progress toward the understanding the immune function has been made, the understanding of the underlying dysregulation and autoimmune response specificity remains limited. Alterations in genes that control pathways regulating self tolerance are critical in the pathogenesis of these diseases. The DNA microarray technologies are now available and providing a large number of information regarding the underlying pathophysiology of autoimmune diseases.
The application of proteomic techniques in diagnosing autoimmune diseases, predicting a disease course, treating with the proper therapy and monitoring the impact of the therapy will change the currently valid diagnostic procedure in the future [19] .
2.2. Proteomics and Biomarkers in Autoimmune Diseases
Biomarkers are Bio-molecules that can be used to distinguish an abnormal from normal process, or a disease from condition. They can also be used as an indicator for a particular process such as drug interaction. These bio-molecules are usually found in blood, other body fluids, or tissues [52] . Proteomic technologies hold the potential to revolutionize clinical care by providing tools for the discovery of biomarkers for diagnosis, prediction of disease course, guiding therapeutic selection, and monitoring response to therapy. Nevertheless, tremendous work remains to develop, refine, validate and apply proteomics technologies to identify biomarkers in autoimmune disease. Several proteomics technologies and their application to autoimmune diseases, including (i), 2-DE and MS for autoantigen and biomarker discovery; (ii), autoantigen microarrays to characterize autoantibody responses; (iii), antibody array technologies to profile cytokines and other biomolecules; (iv), RP protein array studies to analyze phosphoproteins; and (v), flow cytometric analysis of phosphoproteins will be highlighted.
Biomarkers are valued tools used across the biological science spectrum, from research to diagnosis. Proteomics has the potential for advanced biomarker discovery. A number of proteomics platforms are available today for diagnostic and prognostic aspects of autoimmune diseases. The classical example of such biomarker of high specificity is anti-Citrullinated enolase autoantibodies and its diagnostic and prognostic implication in RA and SLE [53] . Both 2-D gel electrophoresis and mass spectrometry can be used to analyze complex mixtures of proteins and/or peptides in such systems. Newer approaches to address the limitations of 2-D gel electrophoresis, by detecting less abundant proteins through the use of protein chips arrayed with specific capture molecules like antibodies, are being pursued actively. Latest proteomic technologies for autoantibody profiling include multiplex analysis using protein chips/microarrays of antigen specific immunobeads, addressable tags, nanoparticles and arrays of living cells. Peptide-MHC tetramer arrays, reverse-phase protein microarrays, cDNA and oligonucleotides microarrays, antibody microarrays, bead-based multiplex assays, autoantigen microarrays, whole- proteome microarrays are such types of assays.
The completion of the human genome project and the availability of highly sensitive and accurate mass spectrometers have led to the ongoing proteomics revolution. Future advances in proteomics depend heavily on the quality and possibilities of related software. The required software can be seen as consisting of two segments. The first one is the analysis of the proteome population of proteins, i.e. proteome status. Unlike the genome, the compositions of the proteome change dynamically with the developmental state of the cell. Proteome analysis requires elaborated software to identify individual proteins by mass and mobility in 2-D electrophoresis and matrix assisted laser desorption/ionization experiments. The second software segment is focused on the proteome structural analysis. The primary amino acid sequences of proteins provide important advances in analysis of individual proteins of the proteome and making predictions regarding the functional status of the proteome. Exclusive prognosis can be based on estimating changes in the primary sequences and related 3-D structures of a set of proteins, i.e. protein markers [54] . Considering that on-chip procedure is quite easy to use, much less time consuming than Western blotting, and much more sensitive at least in the low molecular weight range, the SELDI-TOF technology is a very promising approach for the screening of auto-antibodies in autoimmune diseases. Due to its versatility, this on-chip technology could allow the large-scale screening for complex auto- antibody distributions for diagnostic purposes and early detection of autoimmune diseases could be made possible [55] .
Biomarkers of drug efficacy and toxicity are becoming a key need in the drug development process. Mass spectral-based proteomic technologies are ideally suited for the discovery of protein biomarkers in the absence of any prior knowledge of quantitative changes in protein levels. The success of any biomarker discovery effort will depend upon the quality of samples analysed, the ability to generate quantitative information on relative protein levels and the ability to readily interpret the data generated [4] .
A wealth of information available in bioinformatics includes a description of protein function, its domain structure, sub cellular localization, post-translational modifications, variants, similarities to other proteins, struc- tural classification, etc. Computational methods to define changes in the 3-D structure and stability of proteome proteins versus sequence variations and to make predictions about the proteome functional status are an integral part of proteomics. Newer methods of protein 3-D structure prediction, including homology modeling, molecular dynamics, protein 3-D structure visualization software (spdvb, Rasmol) are gaining impetus. The ongoing structural genomic project with its goal of solving the 3-D structures of all proteins encoded by the human genome serves as the main initiative to annotate basic proteome proteins.
A biomarker is a molecule that indicates an alteration of the physiological state of an individual in relation to health or disease state, drug treatment, toxins and other challenges of the environment. Marker discovery by proteomics is based on the assumption that “disease” can be defined as an altered flow of information in a biological system. One group of information carriers is in fact proteins. Consequently, the type and concentration of a protein (or a group of proteins) at any given time in a proteome and the correlation of those patterns present in a “disease state” as compared with a “healthy state” can be of high diagnostic value. These disease specific proteins can be used as a screening marker, a prognostic marker, a stratification marker, and an efficacy marker. The screening marker discriminates the “healthy” state from a beginning “disease” state, preferentially in the asymptomatic phase. The prognostic marker predicts a likely course of the disease and thus influences the aggressiveness of the therapy once the disease status is established. The stratification marker predicts the likely response to a drug before starting treatment classifying individuals as “responders” as compared with likely “non-responders”. The efficacy marker can be used to monitor the efficacy of drug treatment once the responder status is established [56] .
More recently, de Seny et al. used surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS) technology to identify serum protein biomarkers specific for RA. A total of 103 serum samples from patients and healthy controls were analysed. SELDI-TOF-MS data were analysed by a machine learning algorithm called decision tree boosting. Several peaks were highlighted as the biomarker candidates. The result of depletion test using anti-MRP-8 antibody suggested that the peak with mass/charge (m/z) value of 10,832 could be myeloid related protein 8. Another research used the same proteomics approach in detection of disease biomarkers in tear of patients with Sjögren’s syndrome (SS). They found that seven biomarkers were down-regulated, while three were up-regulated in primary SS group. The advantage of SELDI-TOF-MS technology combined decision tree boosting analysis is that it serves rapid analysis of numerous samples and detection of the biomarkers with discriminative m/z value. However, identification of the interested peaks seems a big problem. It has also investigated biomarkers at protein level using DIGE as well as at small peptide level using magnetic beads and filters combined MS and ClinPro Tools software analysis. Several proteins and peptides were found as candidate biomarkers in different diseases, such as RA, SLE, systemic sclerosis (SSc) and OA. The biofunction of these molecules are now on the way to be further studied [5] .
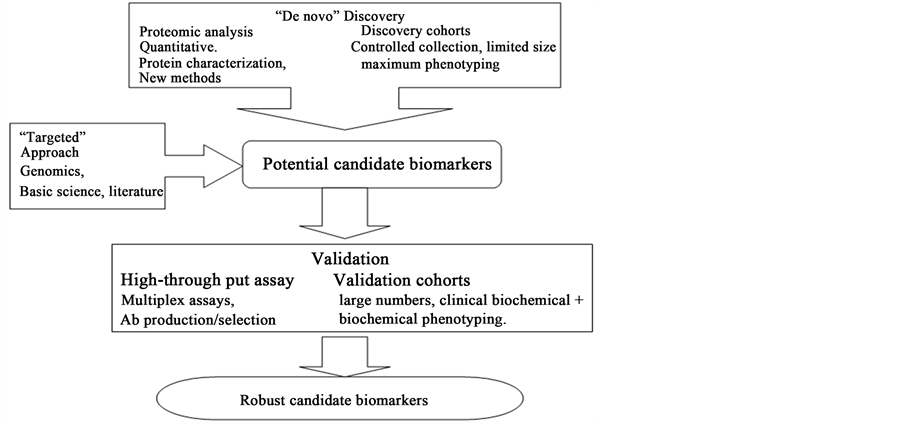
In general, two different strategies are being used to discover biomarkers using proteomic technologies (Figure 2). The first strategy is a “targeted” approach based on the more traditional hypothesis-driven evaluation of specific biomarker candidates, selected either on the basis of a biological rationale or from analysis of candidates derived from other sources. The second strategy is a “de novo” discovery approach that uses different proteomic techniques and finally validates potential biomarker candidates. Both strategies are complementary, have advantages and disadvantages, and may be performed in parallel. Regardless of the strategy used, it is believed that the discovery and development of a robust biomarker candidate, using proteomics, demands a systematic in-depth approach in which discovery and validation are coupled. Figure 3, illustrates such an approach, as adopted in the laboratory for serum/plasma and tissue samples.
2.3. Emerging Trends in Proteomics
A number of emerging concepts have the potential to improve current features of proteomics. Obtaining absolute quantification of proteins and monitoring post-translational modifications are the two tasks that impact the understanding of protein function in healthy and diseased cells. Advances in quantitative proteomics would clearly enable more in-depth analysis of cellular systems. For many cellular events, the protein concentrations do not change; rather, their function is modulated by post-transitional modifications (PTM). Methods of monitoring PTM are an underdeveloped area in proteomics. Selecting a particular subset of protein for analysis substantially reduces protein complexity, making it advantageous for diagnostic purposes where blood is the starting material. Another important aspect of proteomics, yet not addressed, is that proteomics methods should focus on studying proteins in the context of the environment. The increasing use of chemical cross linkers, introduced into living cells to fix protein-protein, protein-DNA and other interactions, may ameliorate this problem partially. The
Figure 2. Two strategies for biomarker discovery and validation: a “targeted” approach, based on hypothesis-driven evaluation of specific biomarker candidates, and a “de novo” discovery approach using different proteomic technologies followed by validation of potential biomarker candidates. (Source: Physiological Genomics; American Physiological Society, 2008).
Figure 3. The platform for biomarker discovery in serum and tissue samples combines multiple synergistic protein fractionation and separation methods including one- and two-dimensional gel electrophoresis (1-DE, 2-DE), differential in-gel electrophoresis (2D-DIGE), and one- and two-dimensional liquid chromatography (1DLC, 2DLC), coupled with mass spectrometry (MS) identification and validation methods, using multiplex arrays. RPLC, reversed phase liquid chromatography; VSMC, vascular smooth muscle cell; cTnI, cardiac troponin I; Igs, immunoglobulins. (Source: Physiological Genomics; American Physiological Society, 2008).
challenge is to identify suitable methods of preserving relevant interactions. Another goal for studying protein is to develop more sophisticated methods to image proteins and other molecules in living cells and real time [6] .
2.4. Proteomics in Diagnosis of Autoimmune Diseases
Profiling of autoantibody responses can be attempted using biological fluids derived from patients who are suffering from autoimmune diseases and proteomics technologies are found to be of great help in this process. Moreover, proteomics techniques provide a powerful tool to distinguish autoreactive B-cell resposnses in diseases such as rheumatoid arthritis, multiple sclerosis, autoimmune diabetis and systemic lupus erythematosus. Classsiification of individual patients based on their “autoantibody fingerprint”, autoantibody profiling and epitope spreading have lead to characterization of autoantigens for the benefit of antigen-specific therapy. The most prominent future challenges and trends is to concentrate on Proteomics technologies which will broaden the understanding of the basic mechanisms of diagnosing, prognosticate and treatment of autoimmune diseases [4] .
Several autoimmune diseases are characterized by specific auto antibodies that are important in diagnosis. These include anti-double stranded DNA (dsDNA) antibodies and anti-Smith antibodies in SLE, anti-U1- snRNP antibodies in MCTD, anti acetylcholine receptor antibodies in myasthenia gravis, and antithyroid stimulating hormone receptor antibodies in Graves’ disease. Measurement of serum antibodies specific for multiple epitopes of a particular antigen have proven useful in identifying dominant linear epitopes in SLE antigens and in increasing the sensitivity of detection of anticitrulline antibodies in RA by combining the frequencies of reactivity to a panel of citrulline-containing peptides [57] [58] . Studies using antigen arrays allow for the simultaneous detection of hundreds of auto antibodies on one array with 1 μl or less of patient serum and therefore are ideal for profiling auto antibodies in SLE and other autoimmune diseases.
An example of using first-generation antigen arrays to characterize the specificity of the autoantibody response in human SLE patients is presented in Figure 4. More recently, it has expanded the antigen arrays to



Figure 4. Antigen array detection of autoantibodies specific for common autoantigens in systemic lupus erythematosus (SLE). Protein arrays were fabricated by spotting common SLE antigens including DNA, histone 2-A protein, and serine/arginine (SR) proteins onto poly-L-lysine-coated glass microscope slides using a robotic microarrayer. Arrays were incubated with patient serum followed by secondary antibody covalently conjugated to a spectrally resolvable fluorescent label, and then scanned with a GenePix 4000B Array Scanner (Axon Instruments, Union City, CA). Shown are scanned images of arrays incubated with serum from a healthy control subject (a), Normal human serum (control) and 2 SLE patients ((b) SLE-1 and (c) SLE-2), followed by Cy3-labeled anti-human Ig secondary antibody. In SLE patient 1, autoantibodies specific for DNA (yellow circles) and SR proteins (orange circles) are detected. SLE patient 2 has autoantibody reactivity primarily against DNA (yellow circles) and histone 2-A protein (blue circles). The array-determined autoantibody profiles in these 2 SLE serum samples and the control sample are in exact concordance with the specificities previously determined using traditional methods including Western blot, enzyme-linked immunosorbent assay, and immunoprecipitation analysis. Antibodies specific for influenza A virus vaccine are detected in all samples (white circles). Spotted antibodies specific for human IgG detect the presence of IgG in each sample (red circles). (Source: Robinson, W. H., Steinman, L. and Utz, P. J. (2002), Proteomics technologies for the study of autoimmune disease. Arthritis & Rheumatism, 46: 885-893).
include ∼200 distinct protein, peptide, nucleic acid, and protein complex antigens targeted to humans with various autoimmune diseases including RA, SLE, polymyositis, limited and diffuse scleroderma, primary biliary sclerosis, and Sjögren’s syndrome. It has also generated “myelin proteome” arrays containing >500 proteins and peptides derived from the myelin sheath that are targeted in multiple sclerosis (MS) and its animal model (experimental autoimmune encephalomyelitis [EAE]). They are using these arrays to characterize specificity and epitope spreading and to guide selection of antigen-specific therapies in EAE. They are also using our myelin proteome arrays to characterize the specificity of the autoantibody response in CSF and brain plaque-eluted antibody from humans with MS [23] .
In addition to diagnosis of autoimmune disease, antigen microarrays have been designed to determine and monitor IgE reactivity profiles in patients with seasonal allergies. Hiller et al. developed a microarray containing 94 purified allergens. They demonstrated that the array results were consistent with patients’ known sensitization profiles based on skin testing or radio allergosorbent test (RAST)-based assays. Additional studies showed that these microarrays had a dynamic range comparable to RAST assays, that the sensitivity was similar to ELISA and exceeded that of RAST, and that no significant cross-reactivity was observed. Antigen arrays have also been applied for serodiagnosis of infectious diseases. Mezzasoma et al. designed arrays with antigens from multiple perinatal pathogens, including Toxoplasma gondii, cytomegalovirus, herpes simplex virus type 1 and 2, and rubella virus. Using a panel of characterized human sera, they validated the arrays, demonstrating a detection limit of 0.5 pg of antibody and sensitivity similar to ELISA. The arrays also included internal calibration curves for IgM and IgG, allowing quantification of individual immune responses. A comprehensive array for simian human immunodeficiency virus has also been used to dissect the B cell response in monkeys enrolled in an antigen-specific DNA vaccine trial [59] .
With each of these microarray systems, the advantages are that the time to run the assay, the cost, and the amount of serum remain the same regardless of the number of analytes. In contrast, with ELISA each analyte must be assayed individually, increasing time, cost, and amount of serum required. In fact, clinical laboratories typically request at least 0.5 mL of serum for autoantibody studies, with a bare minimum of 0.15 mL per assay. Therefore, the amount of serum needed to test all the antigens on an array could be prohibitive, especially in seriously ill or pediatric patients.
2.5. Application of Proteomics Technologies to Drive Development and Selection of Antigen-Specific Therapies for Autoimmune Disease
The use of autoantigen arrays helps for the development and selection of antigen specific therapies for human autoimmune disease. Conventional methods for determining the specificity of autoimmune responses, including T cell proliferation, ELISA, and radioimmunoassay analysis, do not enable large-scale determination of the specificity of autoimmune responses in individual patients. Antigen arrays allow simple parallel, multiplex determination of the specificity of autoimmune responses in individuals and cohorts of patients. Thus, they propose administration of antigen-specific therapeutic agents, defined by the binding specificity of patient autoantibodies. This could be accomplished utilizing a wide variety of different antigen-specific therapies. Antigen-specific therapies involve administration of the targeted autoantigen in a manner that induces immune tolerance to treat autoimmunity.
Such therapies include; oral administration of antigen to induce “oral tolerance”, administration of native peptides via intravenous or other routes, administration of altered peptide ligands, administration of whole protein antigens, administration of other biomolecules such as DNA, or proteins and peptides with posttranslational modifications, administration of DNA-tolerizing vaccines encoding the targeted self proteins. It is anticipated that future antigen-specific therapies based on this strategy will deliver multiple targeted epitopes or protein antigens as tolerizing agents. Such therapies could deliver a consensus dominant targeted epitope or antigen, and could also deliver a cocktail of 20 or more of the consensus targeted epitopes or antigens to treat patients with a specific disease or subset of that disease. Antigen arrays are used to guide development and selection of antigen- specific DNA-tolerizing vaccine therapy, a strategy that have been termed “reverse genomics.” Reverse genomics strategy is applied to develop and select antigen-specific DNA tolerizing vaccines to treat EAE and collagen induced arthritis, a model for RA [23] .
3. Future Perspectives and Challenges
Given the complex nature of proteins, optimal conditions for antigen arrays have not been established, and variation is seen using different slide surfaces and printing conditions. In addition, following attachment of antigens to a planar surface, epitopes may be altered, resulting in lack of detection of autoantigens on the array.
Proteomic approaches are constantly expanding the ability to quantify changes in protein expression and modification in an unbiased fashion for a given biological sample. Current limitations include lack of ability to extend comprehensive coverage to encompass the entire proteome to include even the low-abundance proteins with sufficient degrees of quantification and reproducibility.
The most prominent future challenges will be evaluation of depletion efficiency and reproducibility of each protein depletion kit using different serum samples [60] .
Encouragingly, the technology used has been steadily evolving over the past decade. For example, until recently, 2D gel electrophoresis was the most powerful proteomic profiling technique for both protein identification and quantitation in clinical samples. But this platform fails to detect proteins with extreme pH values, high and low molecular weights, proteins with low copy numbers, and those with hydrophobic domains. However, recently, liquid chromatography-based mass spectrometric (LC-MS) techniques have begun to exercise their dominance in the field, although they are more costly and technologically intensive. Present day technologies allow for more accurate quantitation of the differentially expressed proteins. Additionally, quantification of proteins in samples from different subjects, different stages of disease, or different treatment conditions can be achieved by using protein tags such as iTRAQ (isobaric tag for relative and absolute quantitation), ICAT (isotope-coded affinity tag) and cICAT (cleavable ICAT), which significantly reduce sample-to-sample variation and time-point variation. Consequently, the most effective strategy should combine the different techniques and therapeutic agents for evaluation of therapeutic effects in various disease states. Although most of the shortcomings of 2DGE can be alleviated, there is still room for improvement with LC-MS-based platforms as the field marches towards attaining total coverage of the entire proteome at an affordable cost [61] .
Although significant progress has been made in developing proteomics technologies, major hurdles and significant work remain. Extensive validation of array results, using thousands of sera already characterized for antibody specificities by standard methods, will be essential for regulatory approval and entry into routine clinical practice. Certain autoantigens are not amenable to detection using poly-L-lysine-coated glass slides [62] . This disadvantage can be addressed by using alternative surface chemical molecules, and linkers to orient and to serve as spacers between antigens and the surface, particle, or tag. Bead and tag systems are currently limited by the relatively small numbers of addressable elements available.
Autoantibody profiling using antigen microarray technology does not provide direct information about the specificity of the T cells that mediate autoimmunity. Although there are examples of discordance of the fine peptide epitope specificity of the autoreactive T-cell and B-cell responses, there is a high degree of concordance between autoreactive B-cell and T-cell responses at the macromolecular level [63] . It believed that specificity of the autoantibody response is predictive of the specificity of the overall autoimmune response at the level of whole autoantigens. Further studies will be necessary to determine whether this powerful and enabling hypothesis is, in fact, valid.
4. Conclusions
Proteome alterations in disease may occur in many different ways that are not predictable from genomic analysis, and it is clear that a better understanding of these alterations will have a substantial impact in medicine. A useful repertoire of proteomics technologies is currently available for disease-related applications, although further technological innovations would be beneficial to increase sensitivity, reduce sample requirement, increase throughput and more effectively uncover various types of protein alterations such as post-translational modifications. The use of these technologies will likely expand substantially, particularly to meet the need for better diagnostics and to shorten the path for developing effective therapy.
Proteomic profiling of autoantibody responses has the potential to determine the specificity of autoimmune responses in individuals and cohorts of patients. It has been observed that proteomic approach provides strong analytical tool for
Ø understanding the underlying mechanisms which lead to the development of various autoimmune disorders;
Ø finding out subsets of a disease which may need different approaches to therapy; and
Ø knowing more inclusive theory of pathogenesis of particular autoimmune phenomena depending on the proteomic evolution of the disease.
Proteomic techniques will pave the way for (a) new biomarker discovery as diagnostic or prognostic marker; (b) development of new ways to manage the disease, i.e. targeted intervention to modulate metabolic machinery in the cell or tissue early in the disease; and (c) many autoimmune diseases, which produce antibodies or reactive immuno competent cells that may damage, kill, remove or stimulate cellular proliferation. The range of auto antibodies and spectrum of their action in different autoimmune diseases at different stages of disease will guide a new dawn in understanding and managing these diseases which so far have been proved to be unsatisfactory and in certain cases elusive.
Proteomic analysis of biological fluids by 2DE-DIGE mass spectrometry is in its infancy and many commentators consider the roads leading to approval of biomarker assays long and difficult. Nonetheless, exploratory genomic/proteomic studies have already proven their remarkable ability to shed light on the mechanisms of autoimmune disease and identify new therapeutic targets.
At this time, these are exciting times for researchers and clinicians ready to embrace the vision of personalized medicine, and it is hoped that translating promising findings from genomic and proteomic studies into both innovative therapies and superior clinical predictors will ultimately benefit those for whom it matters most, the patients with debilitating autoimmune diseases.
In general, proteomics technologies in autoimmune diseases trend towards extensive utilization in clinical and basic researches. The antigen microarrays have been demonstrated effective to detect all the auto antibodies which are known in certain autoimmune disease in short time at low price, high accuracy and convenience. The commercial array kits are necessary in future for autoimmune diseases diagnosis, prognosis and treatment. The investigation of neoauantigen/antibodies, based on the high throughput of protein separation and identification, should continue to enrich the autoantigen/autoantibody contents, and highlight autoimmune pathogenesis of autoimmune diseases.
The developing proteomics technologies in searching disease biomarkers will eventually solve problems in pathogenesis and treatment of autoimmune diseases. Multiple proteomics technologies provide the potential to identify biomarkers with utility for personalized medicine and targeted therapy. Ultimately, the development and use of such proteomic biomarkers for diagnosis, assessing prognosis and guiding therapy will revolutionize the care for autoimmune disease patients.
The development of miniaturized proteomics technologies heralds the beginning of an era of multiplex, high- throughput analysis of autoantibody specificities and isotype usage. Spotted antigen arrays on derivatized microscope slides offer a fluorescence-based proteomics platform utilizing simple protocols and widely available equipment. In the future, fluid-phase arrays based on addressable particles and tags are likely to supplant planar arrays, due to their lower propensity to distort and to sterically interfere with immunologic epitopes. It is anticipated that proteomics monitoring of autoantibody responses will have a major impact on the diagnosis, monitoring, and therapy of autoimmune disease.
Autoimmunity laboratories use immunoassays as the basic technique for the determination of autoantibodies. The central and main procedure principle for all autoantibody diagnostic assays is the capture of autoantibodies from serum using immobilized autoantigens. However, there is an enormous variability in these tests that has led to differences in results, a variable degree of confidence in their utility and even misdiagnosis of the patient’s disease. There is no universal solution to resolve these problems, but it is possible to improve the standardization level for techniques and methods. Standardization is an international problem and it is desirable that one international organization in collaboration with all organizations responsible for quality assessment of assays should be responsible for the coordination between diagnostic developers, clinicians and clinical diagnostics companies. In practice, the laboratories are responsible to solve the dilemma on suitable applications of respective methods for detecting autoantibodies in cooperation with the laboratory experts, clinicians and manufacturers.
Multiple determination systems could play a crucial role in allowing the identification of autoantibody signatures in the individual patient. This, in turn, may lead to a more accurate diagnosis and a targeted therapy. The identification of proteins per se is not sufficient to understand biological function because most proteins are post-translationally modified. The promise of phosphoproteomics, or of glycoproteomics, which enables the study of important physiological PTMs of proteins, will revolutionize the understanding of the function of proteins.
Although significant progress has been made in developing proteomics technologies, major hurdles and significant work remains, because there are limitations such as, following attachment of antigens to a planar surface, epitopes may be altered, resulting in lack of detection of autoantigens on the array, lack of ability to extend comprehensive coverage to encompass the entire proteome to include even the low-abundance proteins with sufficient degrees of quantification and reproducibility and autoantibody profiling using antigen microarray technology does not provide direct information about the specificity of the T cells that mediate autoimmunity.
Cite this paper
GebrehiwotGebretsadik,M. K. C.Menon, (2016) Proteomics and Its Applications in Diagnosis of Auto Immune Diseases. Open Journal of Immunology,06,14-33. doi: 10.4236/oji.2016.61003
References
- 1. Anderson, N.L. and Anderson, N.G. (1998) Proteome and Proteomics: New Technologies, New Concepts, and New Words. Electrophoresis, 19, 1853-1861.
http://dx.doi.org/10.1002/elps.1150191103 - 2. Blackstock, W.P. and Weir, M.P. (1999) Proteomics: Quantitative and Physical Mapping of Cellular Proteins. Trends in Biotechnology, 17, 121-127.
http://dx.doi.org/10.1016/S0167-7799(98)01245-1 - 3. James, P. (1997) Protein Identification in the Post-Genome Era: The Rapid Rise of Proteomics. Quarterly Reviews of Biophysics, 30, 279-331.
http://dx.doi.org/10.1017/S0033583597003399 - 4. Abhilash, M. (2009) Applications of Proteomics. The Internet Journal of Genomics and Proteomics, 4, 1540-2630.
- 5. Xiang, Y. and Kato, T. (2006) Use of Proteomics in Analysis of Autoimmune Diseases. Lupus, 15, 431-435.
www.lupus-journal.com
http://dx.doi.org/10.1191/0961203306lu2329oa - 6. Weston, A.D. and Hood, L. (2004) Systems Biology, Proteomics, and the Future of Health Care: Toward Predictive, Preventative, and Personalized Medicine. Journal of Proteome Research, 3, 179-196.
http://dx.doi.org/10.1021/pr0499693 - 7. Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., et al. (2001) The Sequence of the Human Genome. Science, 291, 1304-1351.
http://dx.doi.org/10.1126/science.1058040 - 8. Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., et al. (2001) Initial Sequencing and Analysis of the Human Genome. Nature, 409, 860-921.
http://dx.doi.org/10.1038/35057062 - 9. Reboul, J., Vaglio, P., Tzellas, N., Thierry-Mieg, N., Moore, T., Jackson, C., et al. (2001) Open-Reading-Frame Sequence Tags (OSTs) Support the Existence of at Least 17 300 Genes in C. elegans. Nature Genetics, 27, 332-336.
http://dx.doi.org/10.1038/85913 - 10. Liebler, D.C. (2002) Introduction to Proteomics. Humana Press, New York.
- 11. Robinson, W.H., Steinman, L. and Utz, P.J. (2002) Protein and Peptide Array Analysis of Autoimmune Disease. BioTechniques, 33, 566-569.
- 12. US Department of Health and Human Services (2005) Progresses in Auto Immune Research.
- 13. Oldstone, M.B.A. (1998) Molecular Mimicry and Immune-Mediated Diseases. The FASEB Journal, 12, 1255-1265.
- 14. Croxford, J.L., Anger, H.A. and Miller, S.D. (2005) Viral Delivery of an Epitope from Haemophilus influenzae Induces Central Nervous System Autoimmune Disease by Molecular Mimicry. The Journal of Immunology, 174, 907-917.
http://dx.doi.org/10.4049/jimmunol.174.2.907 - 15. Nissim, A., Winyard, P.G., Corrigall, V., Fatah, R., Perrett, D., Panayi, G. and Chernajovsky, Y. (2005) Generation of Neoantigenic Epitopes after Posttranslational Modification of Type II Collagen by Factors Present within the Inflamed Joint. Arthritis & Rheumatism, 52, 3829-3838.
http://dx.doi.org/10.1002/art.21479 - 16. Hahn, M., Nicholson, M.J., Pyrdol, J. and Wucherpfennig, K.W. (2005) Unconventional Topology of Self Peptide-Major Histocompatibility Complex Binding by a Human Autoimmune T Cell Receptor. Nature Immunology, 6, 490-496.
http://dx.doi.org/10.1038/ni1187 - 17. Carl, P.L., Temple, B.R. and Cohen, P.L. (2005) Most Nuclear Systemic Autoantigens Are Extremely Disordered Proteins: Implications for the Etiology of Systemic Autoimmunity. Arthritis Research & Therapy, 7, R1360.
http://dx.doi.org/10.1186/ar1832 - 18. James, J. and Harley, J. (1992) Linear Epitope Mapping of an Sm B/B’ Polypeptide. The Journal of Immunology, 148, 2074-2079.
- 19. Hueber, W. and Robinson, W.H. (2006) Proteomic Biomarkers for Autoimmune Disease. Proteomics, 6, 4100-4105.
http://dx.doi.org/10.1002/pmic.200600017 - 20. Prieto, S. and Grau, J.M. (2010) The Geoepidemiology of Autoimmune Muscle Disease. Autoimmunity Reviews, 9, A330-A334.
http://dx.doi.org/10.1016/j.autrev.2009.11.006 - 21. Davidson, A. and Diamond, B. (2001) Autoimmune Disease. The New England Journal of Medicine, 345, 340-350.
http://dx.doi.org/10.1056/NEJM200108023450506 - 22. Sherer, Y., Gorstein, A., Fritzler, M.J. and Shoenfeld, Y. (2004) Autoantibody Explosion in Systemic Lupus Erythematosus: More than 100 Different Antibodies Found in SLE Patients. Seminars in Arthritis and Rheumatism, 34, 501-537.
http://dx.doi.org/10.1016/j.semarthrit.2004.07.002 - 23. Robinson, W.H., Steinman, L. and Utz, P.J. (2002) Proteomics Technologies for the Study of Autoimmune Disease. Arthritis & Rheumatism, 46, 885-893.
http://dx.doi.org/10.1002/art.10129 - 24. Hueber, W., Utz, P.J., Steinman, L. and Robinson, W.H. (2002) Autoantibody Profiling for the Study of Treatment of Auto-Immune Disease. Arthritis Research & Therapy, 4, 290-295.
http://dx.doi.org/10.1186/ar426 - 25. Thadikkaran, L., Siegenthaler, M.A., Crettaz, D., Queloz, P.-A., Schneider, P. and Tissot, J.-D. (2005) Recent Advances in Blood-Related Proteomics. Proteomics, 5, 3019-3034.
http://dx.doi.org/10.1002/pmic.200402053 - 26. Vostry, M. (2009) Multiplex Immunoassays Chips and Beads. Electronic Journal of the IFCC, 20, No. 4.
- 27. Rados, C. (2005) Genomics and Medical Devices: A New Paradigm for Health Care. FDA Consumer, 39, 34-39.
http://findarticles.com/p/articles/mi_m1370/is_6_39/ai_n27869192/ - 28. Hueber, W. and Robinson, W.H. (2009) Genomics and Proteomics: Applications in Autoimmune Diseases. Pharmacogenomics and Personalized Medicine, 2, 39-48.
http://dx.doi.org/10.2147/PGPM.S4708 - 29. Ocak, S., Chaurand, P. and Massion, P.P. (2009) Mass Spectrometry-Based Proteomic Profiling of Lung Cancer. Proceedings of the American Thoracic Society, 6, 159-170.
http://dx.doi.org/10.1513/pats.200809-108LC - 30. Stoevesandt, O., Taussig, M.J. and He, M. (2009) Protein Microarrays: High-Throughput Tools for Proteomics. Expert Review of Proteomics, 6, 145-157.
http://dx.doi.org/10.1586/epr.09.2 - 31. Lakhan, S.E. (2006) Schizophrenia Proteomics: Biomarkers on the Path to Laboratory Medicine? Diagnostic Pathology, 1, 11.
http://dx.doi.org/10.1186/1746-1596-1-11 - 32. Walther, T.C. and Mann, M. (2010) Mass Spectrometry-Based Proteomics in Cell Biology. The Journal of Cell Biology, 190, 491-500.
http://dx.doi.org/10.1083/jcb.201004052 - 33. Issaq, H.J., Xiao, Z. and Veenstra, T.D. (2007) Serum and Plasma Proteomics. Chemical Reviews, 107, 3601-3620.
http://dx.doi.org/10.1021/cr068287r - 34. Edgar, P.F., Schonberger, S.J., Dean, B., Faull, R.L., Kydd, R. and Cooper, G.J. (1999) A Comparative Proteome Analysis of Hippocampal Tissue from Schizophrenic and Alzheimer’s Disease Individuals. Molecular Psychiatry, 4, 173-178.
http://dx.doi.org/10.1038/sj.mp.4000463 - 35. Master, S.R. (2005) Diagnostic Proteomics: Back to Basics? Clinical Chemistry, 51, 1333-1334.
http://dx.doi.org/10.1373/clinchem.2005.053686 - 36. Conrads, T.P., Zhou, M., Petricoin III, E.F., Liotta, L. and Veenstra, T.D. (2003) Cancer Diagnosis Using Proteomic Patterns. Expert Review of Molecular Diagnostics, 3, 411-420.
http://dx.doi.org/10.1586/14737159.3.4.411 - 37. Johann Jr., D.J. and Blonder, J. (2007) Biomarker Discovery: Tissues versus Fluids versus Both. Expert Review of Molecular Diagnostics, 7, 473-475.
http://dx.doi.org/10.1586/14737159.7.5.473 - 38. Hortin, G.L. (2005) Can Mass Spectrometric Protein Profiling Meet Desired Standards of Clinical Laboratory Practice? Clinical Chemistry, 51, 3-5.
http://dx.doi.org/10.1373/clinchem.2004.043281 - 39. Plebani, M., Pittoni, M., Celadin, M., Bernardi, D. and Mion, M.M. (2009) Recent Advances in Diagnostic Technologies for Autoimmune Diseases. Autoimmunity Reviews, 8, 238-243.
http://dx.doi.org/10.1016/j.autrev.2008.07.032 - 40. Kononen, J., Bubendorf, L., Kallioniemi, A., Bärlund, M., Schraml, P., Leighton, S., Torhorst, J., Mihatsch, M.J., Sauter, G. and Kallioniemi, O.P. (1998) Tissue Microarrays for High-Throughput Molecular Profiling of Tumor Specimens. Nature Medicine, 4, 844-847.
http://dx.doi.org/10.1038/nm0798-844 - 41. Pejcic, M., Stojnev, S. and Stefanovic, V. (2010) Urinary Proteomics—A Tool for Biomarker Discovery. Renal Failure, 32, 259-268.
http://dx.doi.org/10.3109/08860221003599759 - 42. Proteomics, K.J. (2002) Public-Private Group Maps out Initiatives. Science, 296, 827.
- 43. Silva, J.C., Gorenstein, M.V., Li, G.Z., Vissers, J.P. and Geromanos, S.J. (2006) Absolute Quantification of Proteins by LCMSE: A Virtue of Parallel MS Acquisition. Molecular & Cellular Proteomics, 5, 144-156.
http://dx.doi.org/10.1074/mcp.M500230-MCP200 - 44. Manolio, T.A. (2010) Genomewide Association Studies and Assessment of the Risk of Disease. The New England Journal of Medicine, 363, 166-176.
http://dx.doi.org/10.1056/NEJMra0905980 - 45. Calzone, K., Wattendorf, D. and Dunn, B.K. (2010) The Application of Genetics and Genomics to Cancer Prevention. Seminars in Oncology, 37, 407-418.
http://dx.doi.org/10.1053/j.seminoncol.2010.05.005 - 46. Cowin, P.A., Anglesio, M., Etemadmoghadam, D. and Bowtell, D.D. (2010) Profiling the Cancer Genome. Annual Review of Genomics and Human Genetics, 11, 133-159.
http://dx.doi.org/10.1146/annurev-genom-082509-141536 - 47. Salamunic, I. (2010) Laboratory Diagnosis of Autoimmune Diseases—New Technologies, Old Dilemmas. Biochemia Medica, 20, 45-56.
http://dx.doi.org/10.11613/BM.2010.006 - 48. Dahle, C., Skogh, T., Åberg, A.K., Jalal, A. and Olcén, P. (2004) Methods of Choice for Diagnostics Antinuclear Antibody (ANA) Screening: Benefit of Adding Antigen-Specific Assays to Immunofluorescence Microscopy. Journal of Autoimmunity, 22, 241-248.
http://dx.doi.org/10.1016/j.jaut.2003.12.004 - 49. Binder, S.R. (2006) Autoantibody Detection Using Multiplex Technologies. Lupus, 15, 412-421.
http://dx.doi.org/10.1191/0961203306lu2326oa - 50. Gonzalez, C., Garcia-Berrocal, B., Talavan, T., Cassas, M.L., Navajo, J.A. and Gonzalez-Buitargo, J.M. (2005) Clinical Evaluation of a Microsphere Bead-Based Flow Cytometry Assay for the Simultaneous Determination of Anti Thyroid Peroxidase and Anti Thyroglobulin Antibodies. Clinical Biochemistry, 38, 966-972.
http://dx.doi.org/10.1016/j.clinbiochem.2005.08.004 - 51. Apweiler, R., Aslandis, C., Defuel, T., Gerstner, A., Hansen, J., Hochstrasser, D., et al. (2009) Approaching Clinical Proteomics: Current State and Future Fields of Application in Fluid Proteomics. Clinical Chemistry and Laboratory Medicine, 47, 724-744.
http://dx.doi.org/10.1515/CCLM.2009.167 - 52. http://lup.sagepub.com/content/15/7/431
- 53. Liu, W.L., Zhang, G., Wang, J.Y., Cao, J.Y., Guo, X.Z., et al. (2008) Proteomics-Based Identification of Autoantibody against CDC25B as a Novel Serum Marker in Esophageal Squamous Cell Carcinoma. Biochemical and Biophysical Research Communications, 375, 440-445.
http://dx.doi.org/10.1016/j.bbrc.2008.08.039 - 54. Wild, N., Karl, J., Grunert, V.P., Schmitt, R.I., Garczarek, U., et al. (2008) Diagnosis of Rheumatoid Arthritis: Multivariate Analysis of Biomarkers. Biomarkers, 13, 88-105.
http://dx.doi.org/10.1080/13547500701669410 - 55. Liotta, L.A. and Petricoin, E.F. (2000) Beyond the Genome to Tissue Proteomics. Breast Cancer Research, 2, 13-14.
http://dx.doi.org/10.1186/bcr23 - 56. Zolg, J.W. and Langen, H. (2004) How Industry Is Approaching the Search for New Diagnostic Markers and Biomarkers. Molecular & Cellular Proteomics, 3, 345-354.
http://dx.doi.org/10.1074/mcp.M400007-MCP200 - 57. Hueber, W., Kidd, B., Toomoka, B., Lee, B.-J., Bruce, B., et al. (2005) Antigen Microarray Profiling of Autoantibodies in Rheumatoid Arthritis. Arthritis & Rheumatology, 52, 2645-2655.
http://dx.doi.org/10.1002/art.21269 - 58. Schellekens, G.A., de Jong, B.A., van den Hoogen, F.H., van de Putte, L.B. and van Venrooij, W.J. (1998) Citrulline Is an Essential Constituent of Antigenic Determinants Recognized by Rheumatoid Arthritis-Specific Autoantibodies. Journal of Clinical Investigation, 101, 273-281.
http://dx.doi.org/10.1172/JCI1316 - 59. de Vegvar, H.E.N., Amara, R.R., Steinman, L., Utz, P.J., Robinson, H.L. and Robinson, W.H. (2003) Microarray Profiling of Antibody Responses against Simian-Human Immunodeficiency Virus: Postchallenge Convergence of Reactivities Independent of Host Histocompatibility Type and Vaccine Regimen. Journal of Virology, 77, 11125-11138.
http://dx.doi.org/10.1128/JVI.77.20.11125-11138.2003 - 60. Liu, B., Qiu, F.H., Voss, C., Xu, Y., Zhao, M., Wu, Y., Nie, J. and Wang, Z. (2011) Evaluation of Three High Abundance Protein Depletion Kits for Umbilical Cord Serum Proteomics. Proteome Science, 9, 24.
http://dx.doi.org/10.1186/1477-5956-9-24 - 61. Vanarsa, K. and Mohan, C. (2010) Proteomics in Rheumatology: The Dawn of a New Era. F1000 Medicine Reports, 2, 87.
- 62. Robinson, W.H., Di Gennaro, C., Hueber, W., Haab, B.B., Kamachi, M., Dean, E.J., Fournel, S., Fong, D., Genovese, M.C., Neuman de Vegvar, H.E., Skriner, K., Hirschberg, D.L., Morris, R.I., Muller, S., Pruijn, G.J., van Venrooij, W.J., Smolen, J.S., Brown, P.O., Steinman, L. and Utz, P.J. (2002) Autoantigen Microarrays for Multiplex Characterization of Autoantibody Responses. Nature Medicine, 8, 295-301.
http://dx.doi.org/10.1038/nm0302-295 - 63. Pietropaolo, M. and Eisenbarth, G.S. (2001) Autoantibodies in Human Diabetes. Current Directions in Autoimmunity, 4, 252-282.
http://dx.doi.org/10.1159/000060541
NOTES
*Corresponding author.