Open Journal of Animal Sciences
Vol. 2 No. 4 (2012) , Article ID: 23936 , 6 pages DOI:10.4236/ojas.2012.24031
Isolation and characterization of superior rumen bacteria of cattle (Bos taurus) and potential application in animal feedstuff
![]()
1National Research Centre on Mithun, Indian Council of Agricultural Research, Medziphema, India
2Biorefining Research Institute, Department of Biology, Lakehead University, Thunder Bay, Canada; *Corresponding Author: wqin@lakeheadu.ca
Received 16 August 2012; revised 20 September 2012; accepted 28 September 2012
Keywords: Rumen; Bacteria; Feed Additive; Characterization
ABSTRACT
Rumen of cattle harbors many microorganisms responsible for bioconversion of nutrients into a source of energy for the animals. In recent years many rumen microbes have been isolated and characterized by sequence analysis of 16S ribosomal RNA gene. Some of the microbes have also been recommended as feed additives for improving the overall growth or production of animals. Rumen bacteria which have potential application in animal feed stuffs were isolated and characterized in this experiment. Isolation was carried out from the rumen of cattle (Bos taurus) using techniques of serial dilutions and repeated tubing of the selectively enriched microbial cultures by using the specific media for rumen bacteria. All the isolates were then screened for in vitro gas production and cellulase enzyme activity and four superior isolates were selected and characterized. There were 18.00% to 23.00% increases in gas production on addition of these isolates to the rumen fluid of cattle and there was better cellulase enzyme activity. Two isolates were identified as Butyrivibrio fibrisolvens, one isolate as Streptococcus species and one isolate as Clostridium aminophilum. This indicated that, these isolates are superior and may have potential to be used as microbial feed additive in ruminants if fed in higher quantity.
1. INTRODUCTION
Rumen microbes are of great importance for livestock and they are able to utilize plant nutrients efficiently as source of energy. Microbes survive in the rumen under different constraints which may be either natural or feed associated as some of the feeds contain a significant amount of anti-nutritional factors [1]. Any contaminant which cannot survive these constraints is eliminated. Therefore, the microbes which harbor the rumen are unique and for better understanding of rumen environment, identification and molecular characterization of microbes are highly essential. Moreover, the rumen manipulation by improving the digestibility of plant nutrient is a priority area for improving the growth or production of livestock.
In recent years many rumen microbes have been isolated and characterized by sequence analysis of 16S ribosomal RNA gene. Prokaryote diversity in the rumen of yak (Bos grunniens) and Jinnan cattle (Bos Taurus) were estimated by 16S rDNA homology analysis [2]. Some of the microbes have also been recommended as feed additive for improving the overall growth or production of animals [3]. The tropical forage Leucaena leucocephala contains mimosine, a non-protein amino acid. When consumed by ruminants in Australia and some parts of India, DHP causes goitrogenic effects. The detoxification of the 3-hydroxy-4(1H)-pyridone (DHP) by Synergistes jonesii, isolated from Hawaiian cattle, is one of the examples in manipulating rumen fermentation with bacteria [4]. Similarly addition of Megasphaera elsdenii has also experimentally prevented acidosis in steers [5]. The enzymes produced by rumen bacteria may contribute to the breakdown of switch grass, a renewable biofuel energy source [6]. Cows in early lactation produced 5.9% more milk when enzyme mixture was added to the concentrate feed [7]. The average increase in Dry Matter Intake (DMI) was 1.0 ± 1.3 kg/day and the average increase in milk yield was 1.1 ± 1.5 kg/d. In our study some of the superior bacteria of cattle were isolated and characterized which has got potential to be used as feed additive.
2. MATERIALS AND METHODS
Isolation of bacteria was done from the rumen of cattle (Bos Taurus, Simmental breed) of Canada. The slaughter house of Thunder Bay (Thunder Bay Meat Processing Plant, Murille, Thunder Bay) Canada was selected for sample collection and samples were collected immediately after slaughtering of animals (5 to 6 no) in the slaughter house. About 3 to 4 samples of rumen content were collected from rumen of each animal in the slaughter house and then mixed together to form a composite sample which was brought to the laboratory in strict anaerobic conditions for further processing.
2.1. Isolation Procedure
The isolation was carried out using conventional techniques of serial dilutions and repeated tubing of the selectively enriched microbial cultures by using the specific media for rumen bacteria (Figure 1). Strict anaerobic conditions were maintained during the isolation procedure.
The growth media contained: 15 ml Mineral Solution I (KH2PO4 3.0 g; (NH4)2SO4 6.0 g; NaCl 6.0 g; MgSO4 0.6 g; CaCl2·2H2O 0.795 g per Litre), 15 ml Mineral Solution II (K2HPO4 3 g/litre), 0.25 g Yeast Extract, 1 g Tryptone, 0.1 ml Resazurine (0.1%), 0.2 ml Hemin (0.05%), 0.5 g Microcrystaline Cellulose, 0.1 g Cellobiose, 0.4 g Sodium Carbonate, 20 ml Clear Rumen Fluid, 50 ml Distilled Water and 50 mg Cysteine Hydrochloride. Fifty bacterial isolates were obtained and screened for superior isolates.
2.2. In Vitro Gas Production
The pure isolates were screened for in vitro gas production [8]. 100 ml calibrated glass syringes (haeberle Labortechnik, Germany) were used for this purpose and 24 hours gas production was recorded in triplicate for each isolate. The amount of gas produced during the incubation of isolates with rumen liquor is closely related to degradability and energetic value of isolates.
Rumen fluid was obtained from three adult Simmental breed of cattle. The rumen liquor was collected into the
Figure 1. Bacteria colonies in roll tube.
thermo flask that had been pre warmed to a temperature of 39˚C. Incubation procedure was done by using special type of calibrated transparent glass syringes with fitted silicon tube. Inoculums (30 ml) containing strained rumen liquor, buffer and mineral solution under continuous flushing with CO2 was dispensed using 50 ml plastic calibrated syringe after which 2 ml of isolates were immediately pushed by using another 10 ml syringe. The glass syringe was tapped and pushed upward by the piston in order to completely eliminate air in the inoculums. The silicon tube in the syringe was then tightened by a metal clip so as to prevent escape of gas. Incubation was carried out at 39˚C ± 1˚C and the volume of gas production was measured after 24 hours of incubation. The average of the volume of gas produced from the blanks was deducted from the volume of gas produced per sample against the incubation time.
2.3. Carboxy Methyl Cellulase (CMC) Activity
The pure isolates were also screened for enzymatic activity. The CMC enzyme activity was determined by measuring the amount of reducing sugar released from carboxymethyl cellulose. The reaction mixture contained 1.0 ml phosphate buffer (0.1 M pH 6.8), 0.5 ml of 1% carboxymethyl cellulose solution prepared in 0.1 M phosphate buffer (pH 6.8), 0.5 ml enzyme and incubated for 60 minutes at 39˚C. The reaction was halted and reducing sugars were determined by the addition of 3.0 ml of Dinitrosalicylic acid reagent [9]. Glucose was used as standard for determination of reducing sugars. The enzyme activity is expressed as International Unit (IU) which is micromole of glucose released per ml per hour.
2.4. Total Cellulase Activity by Filter Paper
Microplate-based filter paper assay [10] was conducted to measure total cellulose activity. Total cellulose activity is measured by the amount of glucose released from filter paper after two hrs incubation and is referred to as glucose equivalent. One FPU is defined as an average of one micromole of glucose equivalents released per min in the assay reaction.
2.5. DNA Isolation and PCR Amplification
Genomic DNA was extracted from the microbes of five isolates by using the standard kit (Nucleospin soil, manufactured by Macherey-Nagel GmbH & Co. KG). The DNA and its concentration were checked by agarose gel electrophoresis. Polymerase chain reaction (PCR) amplification of bacterial 16S rDNA was performed using the universal primer (forward: 5’-AGAGTTTGATCCTCAGAACGAACGCT-3’ and reverse: 5’-TACGGCTACCTTGACGACTTCACCCC-3’) of bacteria. The PCR reaction was set up in 20 μl volumes containing 1 μl template (100 ng/μl), 2 μl 10× buffer, 1.6 μl 25 mM MgCl2, 1 μl of each primer (10 μM), 0.4 μl of 10 mM dNTP mix, 0.2 μl Taq DNA polymerase and 12.8 μl distilled water. The amplification conditions were standardized for universal primer. The amplification conditions were as follows: 3 min of initial denaturation at 95˚C, followed by 30 cycles of denaturation at 94˚C for 30 sec, annealing at 60˚C for 30 sec, extension at 72˚C for 2 min with the last cycle followed by a 10 min extension step at 72˚C.
The PCR product was then visualized on an agarose gel (Figures 2 and 3). The bands were excised and DNA was purified from the gel slices using the standard kit (NORGEN Biotek Corporation, ON, Canada). The purified PCR product was sent for sequencing. The sequences were queried online in the NCBI BLAST database. The isolates were again confirmed by using the specific primer of bacteria.
2.6. Phylogenetic Analysis
The sequences of the isolates were compared with those available in the database. The reference sequences were downloaded from the nucleotide sequence databases and were used in further analysis for comparison. The obtained sequences were aligned using clustal V method of megAlign software (DNASTAR) and then phylogenetic tree was plotted.
3. RESULTS AND DISCUSSION
The isolation was carried out using conventional techniques of serial dilutions and repeated tubing of the selectively enriched microbial cultures by using the specific media for rumen bacteria. Strict anaerobic conditions were maintained during the isolation procedure.
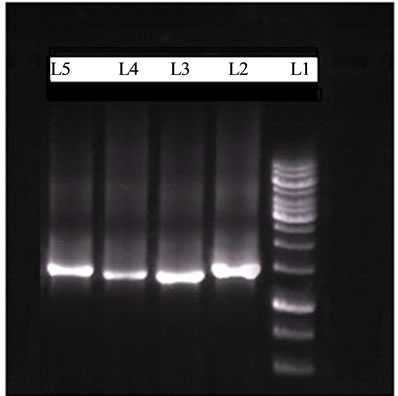
Figure 2. Agarose gel electrophoresis of PCR Products after amplification with Universal Primer (L1: 1 kb DNA Marker, L2-L5: isolates of BF12, BF14, SB13 and CA22).
3.1. In Vitro Gas Production
Four bacterial isolates i.e. BF12, BF14, SB13 and CA22 were observed in microscope and found to be cocci. On addition of these isolates to the rumen fluid of cattle, there was increase in gas production (Table 1). The gas production of isolates after 24 hours incubation varied from 8 to 10.5 ml/ml of isolate, highest in CA22 and lowest in BF14. It was observed that, there were 18.00% to 23.00% increases in gas production indicating release of energy on fibre-based mixed ration. In vitro gas production is the indicator of digestibility and nutritive value of feed stuffs [11,12]. Increased gas production in this experiment indicated that, these isolates may be termed as superior isolates and have potential to be used as microbial feed additives in ruminants (Table 1).
3.2. Enzymatic Analysis of Isolates
The isolates were subjected to enzymatic analysis like carboxymethyl cellulose (CMC), and total cellulose activity (filter paper activity). All the four isolates showed superior fibre degrading activity like CMC, and total cellulose activity, highest in CA22 and lowest in BF14

Figure 3. Agarose gel electrophoresis of PCR products amplification with specific primers of B. fibrisolvens. L1: 1 kb DNA Marker, L2-L3: B. fibrisolvens isolates (BF12, BF14).
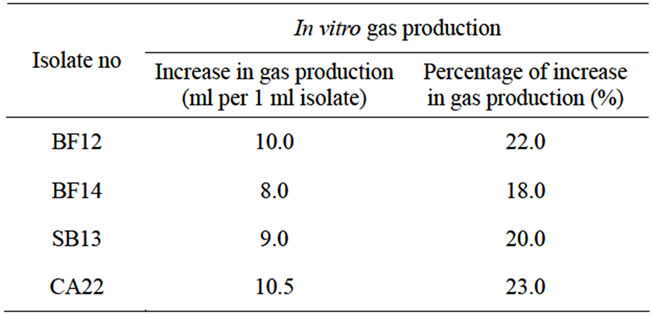
Table 1. In vitro gas production of bacterial isolates.
(Table 2). The in vitro gas productions were in same order as enzymatic analysis of the isolates.
3.3. Identification of Isolates
For identification of isolates, DNA was isolated from all pure isolates and DNA was found to be sufficient for PCR reaction as quantified by spectrophotometer. The DNA was amplified by universal and specific primers of bacteria and amplification product of required band size was obtained (Figures 2 and 3). The bands were excised and purified PCR products were sent for sequencing. One isolate (BF12) was homologus to Butyrivibrio fibrisolvens (Genbank Accession number EU684229) where as other isolate (BF14) was homologus to Butyrivibrio fibrisolvens (Genbank accession number EF 427365). Isolate SB13 was homologus to Streptococcus species (Genbank Accession number FJ611790) where as isolate CA22 was homologus to Clostridium aminophilum (Genbank Accession number DQ278862). This indicated that, these isolates have potential to be used as microbial feed additive in ruminant animal for better growth and production. There is also ample scope in using the enzymes of these bacteria as microbial feed additives.
3.4. Phylogenetic Characterisation
The phylogenetic tree displayed three main groups of bacteria i.e. Butyrivibrio, Streptococcus and Clostridium under the phylum Firmicutes (Figure 4). The isolates BF12 was closely related to Butyrivibrio fibrisolvens isolated from sheep rumen (Accession no EU684229) and isolate no BF14 was closely related to Butyrivibrio fibrisolvens strain of 16S ribosomal RNA gene (Acession no EF427365). The isolate SB13 was closely related to Streptococcus equinus isolated from sheep rumen (Acession no AB563264) and the isolate CA22 was closely related to Clostridium aminophilum isolated from cow rumen (Accession no DQ278862). This shows that, three bacteria isolated in the present experiment are inhabitant of rumen.

Table 2. Enzymatic analysis of bacterial isolates.
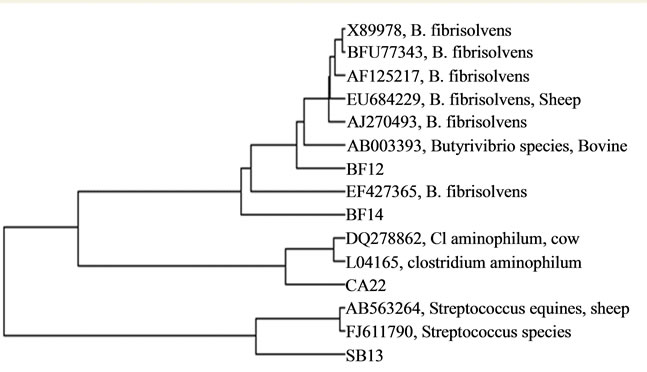
Figure 4. Phylogenetic characterization of isolates.
3.5. Potential of Rumen Bacteria as Microbial Feed Additives
Butyrivibrio species is the major cellulose and hemicellulose degrading bacteria in the rumen [13] and rapidly degrade xylan necessary for improving the overall efficiency of livestock production [14]. This may also be used as probiotics for reducing the aberrant crypt foci in the colon and rectum of mice [15]. Butyrivibrio species isolated in this experiment contained higher source of cellulolytic enzyme and in vitro gas production was increased on addition of these isolates to the rumen fluid of cattle.
Streptococcus species are the predominant cellulolytic micro-organisms that are associated with the possession of complex cellulose enzyme systems [16,17]. Some strains of streptococcus species are responsible for starch hydrolysis in the rumen [18]. Pulse dosing alone or pulse dosing with daily feeding of Streptococcus faecium M74 resulted in improved performance of incoming feedlot cattle [19]. Streptococcus species identified in this experiment are superior because of higher enzymatic activity and in vitro gas production. It has also been reported that, Streptococcus equinus are phylogenetically similar to Streptococcus bovis [20].
Clostridium strain IY-2 isolated from gut content of giant Panda possess cellulase activity towards untreated natural materials such as bamboo fibres, cellulose, rice straw and saw dust and comprises three major cellulases [21]. Clostridial isolate from swine were effective in degrading cell wall components from grasses [22]. As per the result of other worker, Clostridium aminophilum produces ammonia and is responsible for digestion of protein in the feed stuffs [23]. Though effectiveness depends on the strain of microbes, positive results were observed in this experiment.
4. CONCLUSION
Based on the findings of the result, it can be concluded that three species of microbes i.e. Butyrivibrio fibrisolvens, Steptococcus species and Clostridium aminophilus are superior and may have potential to be used as microbial feed additive as a source of microbial enzymes if fed in higher quantity. However, animal experiments are required to confirm the effectiveness of these three species of microorganism.
5. ACKNOWLEDGEMENTS
We are thankful to lab personnel Dr. Wenju Wan, Mehdi Dashtban, Miranda Maki, Bruce Rosa, Xi Chen and Yuanyuan Hong during the research work at Biorefining Research Initiative and Department of Biology, Lakehead University, Thunder Bay, Canada for providing help and cooperation for successful completion of this research work. This work was supported by NSERC-RCD funding of Canada to W. Q. and Indian Overseas fellowship of DBT to K. C. D.
REFERENCES
- Kamra, D.N. (2005) Rumen microbial ecosystem. Current Science, 89, 124-135.
- An, D.D., Dong, X.Z. and Dong, Z.Y. (2005) Prokaryote diversity in the rumen of yak (Bos grunniens) and Jinnan cattle (Bos taurus) estimated by 16S rDNA homology analyses. Anaerobe, 11, 207-215. doi:10.1016/j.anaerobe.2005.02.001
- Mamen, D., Vadivel, V., Pugalenthi, M. and Parimelazhagan, T. (2010) Evaluation of fibrolytic activity of two different anaerobic rumen fungal isolates for their utilization as microbial feed additive. Animal Nutrition and Feed Technology, 10, 37-49.
- Allison, M.J., Mayberry, W.R., McSweeney, C.S. and Stahl, D.A. (1992) Synergistes jonesii, gen. nov., sp.nov: A rumen bacterium that degrades toxic pyridinediols. Systematic and Applied Microbiology, 15, 522-529. doi:10.1016/S0723-2020(11)80111-6
- Robinson, J.A., Smolenski, W.J., Greening, R.C., Ogilvie, M.L., Bell, R.L., Barsuhn, K. and Peters, J.P. (1992) Prevention of acute acidosis and enhancement of feed intake in the bovine by Megasphaera elsdenii 407A. Journal of Animal Science, 70, 310 (Abstract).
- Anonymous (2011) Cow rumen enzymes for better biofuels. Science Daily, 2011.
- Yang, W.Z., Beauchemin, K.A. and Rode, L.M. (2000) A comparison of methods of adding fibrolytic enzymes to lactating cow diets. Journal of Dairy Science, 83, 2512-2520. doi:10.3168/jds.S0022-0302(00)75143-5
- Menke, H.H. and Steingass, H. (1988) Estimation of the energetic feed value obtained from chemical analysis and in vitro gas production using rumen fluid. Animal Research and Development, 28, 7-55.
- Miller, G.L. (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry, 31, 426-428. doi:10.1021/ac60147a030
- Xiao, Z., Storms, R. and Tsang, A. (2004) Microplatebased filter paper assay to measure total cellulose activity. Biotechnol Bioeng, 88, 832-837. doi:10.1002/bit.20286
- Akinfemi, A., Adesanya, A.O. and Aya, V.E. (2009) Use of an in vitro gas production technique to evaluate some Nigerian feedstuffs. American-Eurasian Journal of Scientific Research, 4, 240-245.
- Das, K.C., Hundal, J., Mahapatra, P.S., Subudhi, P.K. and Sharma, K. (2010) Chemical composition and in vitro gas production of fodder tree leaves and shrubs. Indian Veterinary Journal, 87, 899-901.
- Krause, D.O., McSweeney, C.S. and Forster, R.J. (1999) Molecular ecological methods to study fibrolytic ruminal bacteria: Phylogeny, competition, and persistence. In: Bell, C.R., Brylinsky, M. and Johnson-Green, P., Eds., Microbial Biosystems: New Frontiers Proceedings of the 8th International Symposium on Microbial Ecology, Atlantic Canada Society for Microbial Ecology, Halifax, 1999.
- Cotta, M.A. and Zeltwanger, R.L. (1995) Degradation and utilization of xylan by the ruminal bacteria Butyrivibrio fibrisolvens and Selenomonas ruminantium. Applied and Environmental Microbiology, 61, 4396-4402.
- Ohkawara, S., Furuya, H., Nagashima, K., Asanuma, N. and Hino, T. (2005) Oral administration of Butyrivibrio fibrisolvens, a butyrate-producing bacterium, decreases the formation of aberrant crypt foci in the colon and rectum of mice. Journal of Nutrition, 135, 2878-2883.
- Schwarz, W.H. (2001) The cellulosome and cellulose degradation by anaerobic bacteria. Applied Microbiology & Biotechnology, 56, 634-649. doi:10.1007/s002530100710
- Oyeleke, S.B. and Okusanmi, T.A. (2008) Isolation and characterization of cellulose hydrolyzing microorganism from the rumen of ruminants. African Journal of Biotechnology, 7, 1503-1504.
- Dunican, L.K. and Seeley, H.W. (1962) Starch hydrolysis by Streptococcus equinus. Journal of Bacteriology, 83, 264-269. http://www.ncbi.nlm.nih.gov/pubmed/13888473
- Lee, R.W. and Botts, R.L. (1988) Evaluation of a single oral dosing and continuous feeding of Streptococcus faecium M74 (Syntabac) on the performance of incoming feedlot cattle. Journal of Animal Science, 66, 460.
- Saitou, N. and Nei, M. (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4, 406-425.
- Yu, I.K. (1996) Secretion of Clostridium cellulase by E. coli. United States Patent 5496725.
- Varel, V.H. and Pond, W.G. (1992) Characteristics of a new cellulolytic Clostridium sp. isolated from pig intestinal tract. Applied and Environmental Microbiology, 58, 1645-1649.
- Atiwood, G.T., Reilly, K. and Patel, B.K.C. (1996) Clostridium proteoclasticum sp. nov., a novel proteolytic bacterium from the bovine rumen. International Journal of Systematic Bacteriology, 46, 753-758. doi:/10.1099/00207713-46-3-753

