Open Journal of Genetics
Vol.4 No.3(2014), Article
ID:47055,8
pages
DOI:10.4236/ojgen.2014.43021
Low Genetic Polymorphism at the Cytochrome C Oxidase I in Silkworm Strains of the Brazilian Germplasm Bank
Verônica Aureliana Fassina1, Thaís Souto Bignotto1, Roxelle Ethienne Ferreira Munhoz1, Bruno Fulan1, Juliana Pereira Bravo1, Laura B. Garay2, Rafaela Bespalhuk1, Claudia Regina das Neves Saez1, Naiara Climas Pereira1, Graziele Milani Pessini1, Maria Aparecida Fernandez1*
1Departamento de Biotecnologia, Genética e Biologia Celular, Universidade Estadual de Maringá, UEM, Paraná, Brasil
2Seda y Fibras S.R.L., Hernandarias, Paraguay
Email: *mafernandez@uem.br
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 23 April 2014; revised 13 May 2014; accepted 5 June 2014
ABSTRACT
Nucleotide sequences have been used to distinguish species and specimens for many years. More recently, the use of a partial sequence of 650 bp of the cytochrome c oxidase I, COI mitochondrial gene, has been proposed for species identification, known as DNA barcodes. In this work, a short sequence of the DNA barcode is described—approximately 250 bp, named as “DNA mini-barcode” —to molecularly identify different silkworm strains maintained at the unique public Germplasm Bank of Bombyx mori, at the Universidade Estadual de Maringá, UEM, Brazil. Analysis revealed no significant differences among the silkworm strains. The phylogenetic tree obtained by the neighbor-joining method and K2P distance, in which specimens of B. mandarina were used as outgroup, clustered all the specimens of B. mori in a unique clade. Genetic variability detect within B. mori was low or nonexistent. In conclusion, the partial region of 250 bp of the mitochondrial gene COI herein analyzed may not be efficient to discriminate silkworm strains from the UEM Germplasm Bank of Bombyx mori.
Keywords:Bombyx mori, Mitochondrial Gene, COI, DNA Mini-Barcode

1. Introduction
The silkworm Bombyx mori is a completely domesticated insect and is no longer found in nature. Currently, several strains of B. mori geographically distributed and genetically improved are available only in germplasm banks distributed around the world, especially where sericulture presents expressive activity [1] . In Brazil, the unique public germplasm bank of B. mori is located at the Universidade Estadual de Maringá, UEM, Paraná, Brazil. The conservation and maintenance of the original characteristics of the species and its different strains in germplasm banks are, therefore, very important, as well as their correct identification.
Despite external morphological aspects of larvae and silk moths are often very similar among silkworm strains, B. mori strains have been described based on morphological, biological and/or productive characteristics [2] -[4] . However, these characteristics are highly variable and influenced by the environment. Discrimination of silkworm strains based on molecular tools provides more accurate analysis of this genetic diversity than morphological traits [5] [6] .
Genetic characterization of silkworm strains maintained at the UEM Brazilian Germplasm Bank, UBGB, based on molecular markers is, therefore, essential. Mitochondrial DNA (mtDNA) is an efficient and commonly used marker for genetic diversity studies. Several mtDNA sequences could be employed, such as the gene cytochrome c oxidase subunit I (COI). COI has recently been a frequently used gene in molecular taxonomy for characterization and identification of species, a methodology named as “DNA barcode” [7] .
The 5’ region of the COI gene can be easily sequenced and provides resolution to the species level of approximately 98% for the major taxonomic groups such as birds, mammals, fish and various arthropods. The use of a short barcode, with approximately 100 - 300 base pairs (bp), herein referred as “DNA mini-barcode”, has proved to be an effective tool in the identification at the species level of most specimens in museums, formaldehyde-preserved samples and processed biological material [8] . Studies have shown that sequences of 250 bp-long can reach more than 95% of species identification [9] , which can be explained by the fact that the COI region shows small intraspecific variation, a possible reflection of the selection process in the coevolution of the mtDNA [10] .
Since 2005, UEM has the only public germplasm bank of silkworm of Brazil, which has strains from Chinese, Japanese and Indian origins. The genetic characterization of these silkworm strains is important not only for conservation and maintenance but also for the correct use of these genetic resources in breeding programs. Accordingly, the present study aimed the development of DNA mini-barcodes for the different Bombyx mori strains that are maintained at the UEM Brazilian germplasm bank, in order to perform their molecular characterization.
2. Materials and Methods
2.1. Biological Material
Twenty B. mori strains of Chinese, Japanese and Indian origins were obtained at the UEM Brazilian germplasm bank, UBGB. In addition, a commercial hybrid of the state of Paraná was included in the analyses, totaling twenty one genotypes (Table 1). Bombyx mori larvae of each strain were reared separately in hygienic and controlled environments, in breeding boxes, with temperature of 28˚C and relative humidity of 50%. Larvae were subjected to artificial lighting with a photoperiod of 14 hours of light and 10 hours of dark. Silkworm larvae were fed with fresh mulberry leaves.
2.2. DNA Isolation and Quantification
At the third day of the fifth instar, silkworms were stored at −20˚C to induce dormancy. Then, the medial part of the silk glands was dissected and used for DNA isolation. The medial portion of the silk gland was used to reduce the possibility of heteroplasmy, which may interfere in the DNA analysis.
DNA isolation of three specimens of each silkworm strain was performed according to [11] , with modifications. The medial portion of the silk gland was homogeneized in 750 μL of extraction buffer (10 mM Tris-HCl pH 7.5, 60 mM NaCl, 10 mM EDTA pH 8.0, 5% sucrose, 0.15 mM spermine and 0.15 mM spermidine), centrifuged at 7,826 g for 15 minutes and transferred to a new tube containing Proteinase K buffer (0.2 M Tris-HCl pH 9.0, 30 mM EDTA pH 8.0, 5% sucrose, 2% SDS and 100 μg/ml Proteinase K). Samples were incubated at
Table 1. Bombyx mori strains of the UEM Brazilian germplasm bank, UBGB, according to their geographical origin.
50˚C for 2 hours, centrifuged at 11,269 g for 15 minutes and extracted with chloroform and phenol/chloroform. The DNA-containing phase was ethanol-precipitated in the presence of 0.2 M NaCl and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA pH 8.0) containing 10 mg/ml RNase. The DNA was re-extracted with phenol/chloroform and ethanol-precipitated as described above.
The DNA concentration was determined by electrophoresis in 0.8% agarose gels and by UV-visible spectrophotometry (Shimadzu UV-1650PC). DNA was then diluted to 60 ng/μL.
3. Sequencing and Data Analysis
The partial COI segment of the twenty silkworm strains and the commercial hybrid were PCR amplified (Polymerase Chain Reaction) with the pair of primers COI F (Forward, 5’GAG CAC CAG ATA TAG CAT TCC C3’) and COI R (Reverse, 5’AAA TTG GGT CTC CTC CTC CA3’). The amplification reactions were performed in a total volume of 25 µL containing 120 ng of DNA, 0.1 mM of dNTP mix, 10 μM of each primer, 1X PCR Buffer, 0.001 mM triton, 1.5 mM of MgCl2 and 1 unit of Taq DNA polymerase (Invitrogen). PCR was performed in Thermal Cycler programmed for 1 min at 94˚C and 36 cycles of 40 s at 94˚C and then 40 s at 58˚C and lastly 1 min at 72˚C. PCR products were visualized on 1.5% agarose gel stained with EthBr in 1X TBE buffer (45 mM Tris-borate, 1 mM EDTA, pH 8.0) and compared to a molecular marker 100 pb ladder (Invitrogen). The gel was photographed under ultraviolet light in the BioImaging system.
Before sequencing, PCR products were purified with PEG (polyethylene glycol), according to Rosenthal et al. (1993), to remove impurities of the PCR reaction. The final product of each PCR reaction was used directly in the sequencing reaction with the DYEnamic ET Dye Terminator Kit (Amersham-Ge). The purified PCR products, 10 pmol of the COI F primer, 4 μL of DYEnamic ET Dye Terminator and ultra-pure water to complete 10 μL volume, were used in the sequencing reaction. The sequencing reaction was performed in Thermal Cycler programmed for one cycle of 30 s at 95˚C followed by 20 - 25 cycles of 15 s at 50˚C and 1 min at 60˚C. Sequencing was conducted in the automatic sequencer DNA MegaBACE 1000 (Amersham Biosciences), according to the manufacturer’s instructions.
Nucleotide sequences of B. mori available in Genbank regarding the same segment of the COI gene were used in our analyses for comparison purposes (AB649182, BmoHap1A; AB649183, BmoHap1B; AB649184, BmoHap1D; AB649185, BmoHap2A; AB649186, BmoHap2C; AB649187, BmoHap2D; AB649188, BmHap2E; AB649189, BmoHap2F; AB649190, BmoHap3A; AB649191, BmoHap4D; AB649192, BmHap5D; AB649193, BmoHap6D; AB649194, BmoHap7A; AB649195, BmoHap8D). Specimens of Bombyx mandarina were used as outgroup, since it is a close species to B. mori (AF167280, Bma10_Yamanashi; AF167276, Bma12_Tsushima; AF167274, Bma1_Tsukuba; AF167272, Bma3_Fukuoka). The nucleotide sequences of the partial segment of the COI gene were aligned using ClustalW [12] and edited in the Chromas Lite 2.3.0.0 software. When necessary, the sequences were adjusted manually. The BlastN software (http://blast.ncbi.nlm.nih.gov) was used to verify the identity and homology of our nucleotide sequences. The neighbor-joining phylogenetic analysis based on the evolutionary model Kimura-2-parameters (K2P) as well as the K2P genetic distance values were obtained using the software MEGA 5.0 [13] . Bootstrap analyses were based on 10,000 replicates. The DNA mini-barcode sequences obtained in the present work are available at Genbank (Accession Numbers: KJ704283-KJ704326).
4. Results
Although genomic DNA of three specimens of each B. mori strain have been isolated, sequencing of the COI segment was successful for only one or two specimens, due to sequencing difficulties. A fragment of approximately 400 base pairs (bp) regarding a partial segment of the mitochondrial gene cytochrome c oxidase I, COI, was PCR amplified. However, a nucleotide sequence of only 252 bp was selected for the analysis due to the higher quality and reliability of sequencing after manual edition. The bases proportion for all samples was 35.3% of thymine, 15.1% of cytosine, 35% of adenine and 14.7% of guanine. Transition were more frequent, and the rates of transition/transversion were k1 = 9.06 (purines) and k2 = 49.612 (pyrimidines).
The ClustalW alignment revealed fifteen single polymorphic sites among specimens of B. mori and B. mandarina (Figure 1). Regarding B. mori specimens exclusively, five nucleotides substitutions were observed: two were identified in M11-A (2) and one substitution in the samples J1 (2), BmoHap6D (AB649193) and BmoHap7A (AB649194). However, except for BmoHap6D and BmoHap7A, no polymorphism was exclusive, which impeded the molecular characterization of the silkworm strains and/or the commercial hybrid based on the partial segment of the COI gene. Nucleotide substitutions were of one base only, with no indels (insertion/ deletion events).
According to the alignment, nine mitochondrial haplotypes were identified: five were unique to B. mori and four to B. mandarina (Table 2). Bmo-I of B. mori was the most frequent haplotype, grouping 32 specimens of B. mori, including the hybrid specimen (Hbd-Bratac). COI sequences of B. mori revealed few nucleotide differences, therefore low polymorphic haplotypes. In contrast, for specimens of B. mandarina, we observed high polymorphism and high number of haplotypes in the only four specimens analyzed (Bma1, Bma3, Bma10, and Bma12).
The average genetic distance based on the 252 bp COI sequences of B. mori was of 0.1%, whereas for B. mandarina was of 1.2%, suggesting a low genetic variability with respect to this mitochondrial region. However, comparisons between B. mori and B. mandarina revealed an average distance value of 3.7%. According to [7] , genetic distance values for Lepidoptera at or above 3% are indicative of inter-specific divergence. Thus, the partial sequence of the COI gene of 252 bp used in this study was sufficient to discriminate B. mori and B. manda-

Figure 1. ClustalW alignment of the haplotypes based on the DNA mini-barcode region (252 bp of the mitochondrial gene cytochrome c oxidase I) of the silkworm Bombyx mori and Bombyx mandarina.
Table 2. Species and strains of Bombyx mori and B. mandarina, analyzed in the present work, and their haplotypes regarding the 252 bp DNA mini-barcode region, consisting of 252 bp of the mitochondrial gene cytochrome c oxidase I, COI.
rina. However, the discrimination of B. mori strains based on the same region of the mitochondrial DNA was not possible.
The reconstruction of the phylogenetic tree (Figure 2) was based on the neighbor-joining method and the evolutionary model K2P, as suggested by [7] . Results revealed clear separation of specimens in two groups: one group including all specimens of B. mori, and another containing only specimens of B. mandarina, which was employed as outgroup. The phylogenetic tree also revealed that the genetic variability within B. mandarina was higher than within B. mori (Figure 2).
5. Discussion
The DNA mini-barcodes can significantly expand the application of the DNA barcodes in biodiversity studies, since that sometimes it is difficult to sequence fragments as long as 650 bp, especially of specimens in which DNA is degraded [9] . According to [14] , the DNA mini-barcodes generally provide measures of variability and genetic divergence in the same levels of the standard DNA barcodes. In the present work, the size of the sequenced fragments (~250 bp) showed the possibility of developing DNA mini-barcodes for B. mori strains of the UEM Brazilian germplasm bank, UBGB. Additionaly, the designed primers can be tested for other silkworm strains or phylogenetically related species of B. mori.
The low values of genetic distance and few haplotypes revealed low polymorphism among different strains of B. mori analyzed in this work. The low polymorphism found in B. mori of the UBGB can be attributed possibly to the very short segment analyzed. Studies including longer sequences of the COI region (standard DNA barcode) will be required to provide more information about the nucleotide sequence polymorphism and the level of conservation of B. mori of the UBGB. The amplification and characterization of 147 strains of B. mori based on COI fragments of 714 bp long were possible, according to [15] . The authors were able to identify eight haplotypes based on nucleotide differences in eight segregating sites. When non-coding and coding regions were analyzed together, 14 haplotypes were observed, eight haplotypes based on differences in nucleotide coding sites and six haplotypes segregating in non-coding sequences, revealing that DNA barcodes can be used to differentiate silkworm strains.
Based on the suggested requirements for data analyzes of DNA barcodes [7] , we reconstructed a neighborjoining phylogenetic tree based on the K2P evolutionary model (Figure 2). Specimens were divided into two groups, one group consisting of B. mori specimens, including the commercial hybrid, and other group formed exclusively by B. mandarina. Although the B. mori group consisted of Japanese, Chinese and Indian strains and of the hybrid specimen, it revealed no significant genetic differentiation. Most samples of B. mori showed little or no variation in the nucleotide sequence of the DNA mini-barcode analyzed.
However, it was clear that the genetic variability within B. mandarina was higher than in B. mori. Bombyx mori was domesticated and selected for many years, which has caused a strong genetic bottleneck; however, B. mandarina is a wild species that is exposed to different selective pressures. Strains of B. mori available in
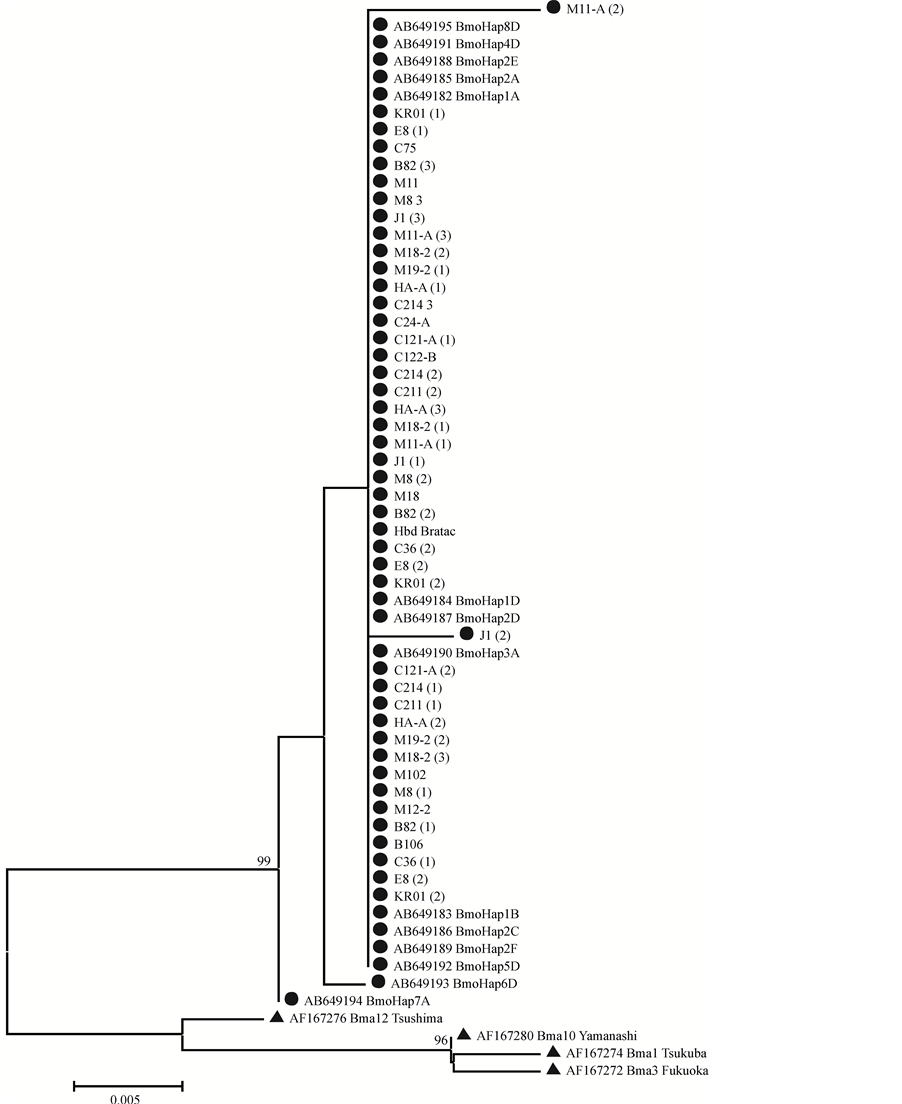
Figure 2. Neighbor-joining phylogenetic tree based on the evolutionary model Kimura-2-parameters (K2P) and on the DNA mini-barcode region (252 bp of the mitochondrial gene cytochrome c oxidase I) of specimens of B. mori (●, Bmo) and B. mandarina (▲, Bma). Values in parentheses represent the number of samples for each strain. The numbers above the branches refer to the bootstrap values based on 10,000 replicates.
germplasm banks have undergone many evolutionary changes through natural mutations and artificial selection, thereby creating a large genetic diversity among strains, but this diversity becomes more homogeneous within the lineage [6] .
Although it was not possible the identification and characterization of different strains of B. mori from the UBGB based on the DNA mini-barcodes, previous studies based on morphological, molecular and/or productive characteristics revealed differentiation among strains. Conformation-sensitive gel electrophoresis (CSGE) was used to identify allelic variability in the third intron of the fibroin light chain gene in six B. mori strains of the Brazilian germplasm bank by [16] . Authors discussed that the Japanese strains HAA, M12B and M19-2 and the Chinese strains C25B and C24-2 revealed distinct heteroduplex profiles, except for the chinese strain C21A, that revealed only homoduplex DNA molecules.
Eight biological and productive characteristics of 16 B. mori strains from the UBGB were evaluated by [3] . It was possible to characterize these strains into three distinct groups. More recently, [4] evaluated and characterized 14 Chinese and Japanese strains (AS3, AS31, C75, C36, C37, E8, F6, JK, M8, M11-2, M18, M11, B82 and B106) of UBGB based on several biological and productive characters. Furthermore, the use of molecular data (RAPD) allowed the characterization of the strains, dividing them into three groups according to their geographical origin.
The nucleotide substitutions of the DNA mini-barcode region demonstrate that there is genetic variability between B. mori and B. mandarina species. Moreover, as no common haplotypes were detected between the two species, there is evidence that gene flow between species was discontinued long ago [15] . Thus, the partial sequence of the COI gene of only 252 bp used in this work was sufficient to discriminate these two species, but insufficient for the differentiation of B. mori strains. The possibility of species discrimination based on the DNA barcodes varies according to the group, for example, groups who have suffered recent speciation, or population with large size and reasonable stability, which is the case of many tropical insects, the identification of species is more complicated. DNA barcode can be less successful than suggested by its proponents, particularly with several tropical invertebrates with congeneric species and geographical strains [16] .
The DNA mini-barcode region, consisting of 252 bp of the mitochondrial gene COI, revealed insufficient polymorphism to characterize and discriminate different strains of B. mori that are maintained at the UEM Brazilian germplasm bank. COI sequences of larger sizes (approximately 650 bp) will probably reveal polymorphism among strains, which will allow the correct identification, maintenance and conservation of these genetic stocks of the silkworm B. mori.
Acknowledgements
This research was supported by CAPES, CNPq, Fundação Araucária and Secretaria de Estado da Ciência, Tecnologia e Ensino Superior—Fundo Paraná. Authors gratefully thank all the staff members of the Laboratório de Melhoramento Genético do Bicho-da-Seda for their assistance and the COMCAP from Universidade Estadual de Maringá, UEM, PR, Brasil.
References
- Nagaraju, J. and Goldismith, M.R. (2002) Silkworm Genomics—Progress and Prospects. Current Science, 83, 411-425.
- Porto, A.J., Okamoto, F., Cunha, E.A. and Otsuk, I.P. (2004) Caracterização de oito raças de bicho-da-seda (Bombyx mori). Ciência Rural, 34, 259-264. http://dx.doi.org/10.1590/S0103-84782004000100040
- Zanatta, D., Bravo, J.P., Barbosa, J.F., Munhoz, R.E.F. and Fernandez, M.A. (2009) Evaluation of Economically Important Traits from Sixteen Parental Strains of the Silkworm Bombyx mori L. (Lepidoptera: Bombycidae). Neotropical Entomology, 38, 327-331. http://dx.doi.org/10.1590/S1519-566X2009000300005
- Pereira, N.C., Munhoz, R.E.F., Bignotto, T.S., Bespalhuk, R., Garay, L.B., Saez, C.R.N., Fassina, V.A., Nembri, A. and Fernandez, M.A. (2013) Biological and Molecular Characterization of Silkworn Strains from the Brazilian Germplasm Bank of Bombyx mori. Genetics and Molecular Research, 12, 2138-2147. http://dx.doi.org/10.4238/2013.June.28.1
- Li, M., Shen, L., Xu, A., Miao, X., Hou, C., Sun, P., Zhang, Y. and Huang, Y. (2005) Genetic Diversity among Silkworm (Bombyx mori L., Lep., Bombycidae) Germplasms Revealed by Microsatellites. Genome, 48, 802-810. http://dx.doi.org/10.1139/g05-053
- Jingade, A.H., Vijavan, K., Somasundaram, P., Srinivasabaru, G.K. and Kamble, C.K. (2011) A Review of the Implications of Heterozygosity and Inbreeding on Germplasm Biodiversity and Its Conservation in the Silkworm, Bombyx mori. Journal of Insect Science, 11, 1-16. http://dx.doi.org/10.1673/031.011.0108
- Hebert, P.D.N., Ciwinska, A., Ball, S.L. and Dewaard, J.R. (2003) Biological Identifications through DNA Barcodes. Proceedings of the Royal Society B: Biological Sciences, 270, 313-321. http://dx.doi.org/10.1098/rspb.2002.2218
- Hajibabaei, M. and Mackenna, C. (2012) DNA Mini-Barcodes. http://link.springer.com/protocol/10.1007/978-1-61779-591-6_15#page-1.
- Meusnier, I., Singer, G.A.C., Landry, J.F., Hickey, D.A., Hebert, P.D.N. and Hajibabaei, M. (2008) A Universal DNA Mini-Barcode for Biodiversity Analysis. BMC Genomic, 9, 1471-2164. http://dx.doi.org/10.1186/1471-2164-9-214
- Hebert, P.D.N. and Gregory, T.R. (2005) The Promise of DNA Barcoding for Taxonomy. Systematic Biology, 54, 852-859. http://dx.doi.org/10.1080/10635150500354886
- Monesi, N., Jacobs-Lorena, M. and Paçó-Larson, M.L. (1998) The DNA Puff Gene BhC4-1 of Bradysia hygida Is Specifically Transcribed Early Prepupal Salivary Glands of Drosophila melanogaster. Chromosoma, 107, 559-569. http://dx.doi.org/10.1007/s004120050342
- Thompson, J.D., Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: Improving the Sensitivity of Progressive Multiple Alignment through Sequence Weighting, Positions-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Research, 22, 4673-4680. http://dx.doi.org/10.1093/nar/22.22.4673
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. and Kumar S. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution, 28, 2731-2739. http://dx.doi.org/10.1093/molbev/msr121
- Hajibabaei, M.; Janzen, D.H.; Burns, J.M.; Hallwachs, W. and Hebert, P.D.N. (2006) DNA Barcodes Distinguish Species of Tropical Lepidoptera. Proceedings of the National Academy of Sciences of the United States of America, 103, 968-971. http://dx.doi.org/10.1073/pnas.0510466103
- Yukuhiro, K., Sezutsu, H., Tamura, T., Kosegawa, E. and Kiuchi, M. (2011) Nucleotide Sequence Variation in Mitochondrial COI Gene among 147 Silkworm (Bombyx mori) Strains from Japanese, Chinese, European and moltinism classes. Genes Genetic Systems, 86, 315-323. http://dx.doi.org/10.1266/ggs.86.315
- Elias, M., Hill, R.I., Willmott, K.R., Dasmahapatra, K.K., Brower, A.V.Z., Mallet, J. and Jiggins, C.D. (2007) Limited Performance of DNA Barcoding in a Diverse Community of Tropical Butterflies. Proceedings of the Royal Society B, 274, 2881-2889. http://dx.doi.org/10.1098/rspb.2007.1035
NOTES

*Corresponding author.


