Open Journal of Genetics
Vol. 2 No. 4 (2012) , Article ID: 26310 , 6 pages DOI:10.4236/ojgen.2012.24024
Combined effect of CYP2C9 and VKORC1 polymorphisms on warfarin maintenance dose in Omani patients
![]()
1Department of Hematology, Sultan Qaboos University Hospital, Muscat, Oman
2College of Medicine & Health Sciences, Muscat, Oman
3INSERM, Université Paris Diderot, Paris, France
4INSERM, Ecole Pratique des Hautes Etudes, Evry, France
Email: *pathare@squ.edu.om
Received 28 September 2012; revised 29 October 2012; accepted 16 November 2012
Keywords: CYP2C9; CYP4F2; VKORC1; Pharmacogenetics; Polymorphism; Omani
ABSTRACT
Excepting host genetic factors, other influences on the pharmacokinetic and pharmacodynamic behavior of warfarin are subject to variations during the treatment despite attempts to stabilize the INR. In 214 Omani patients on warfarin therapy, we evaluated the extent of influence of known genetic predictors of warfarin dose variability, namely CYP2C9, CYP4F2 and VKORC1 gene polymorphisms in a genetically heterogeneous patient population. When patients were stratified according to their daily warfarin maintenance dose (to maintain INR between 2 and 3) into “low dose” (sensitive), “medium dose” (intermediate) and “high dose” (resistance) groups, overall, seven patients with three or four mutant alleles fell in the sensitive group and consequently 25% (7 out of 28) of at risk patients for over anticoagulation can be recognized by prospective pharmacogenetic testing in this patient population. Pre-prescription genotyping of these loci prior to therapy initiation will therefore benefit a small fraction of this population.
1. INTRODUCTION
Warfarin is one of the most widely used anticoagulants with a narrow therapeutic index; small warfarin dose variations in a subset of individuals could have adverse drug reactions (ADRs) and result in either hemorrhage or failure to prevent thrombotic complications. [1] In order to avoid ADRs, the traditional warfarin dose adjustment relied on a trial-and-error basis by administering an initial dose of 5 mg or 10 mg, [2,3] with subsequent dose adjustment depending upon the INR response until the desired stable INR values were obtained on three consecutive visits. Inter-patient variability in warfarin dose requirement is multifactorial involving clinical, demographic, life style, environmental and genetic factors [1-3].
Studies involving genome-wide scan have consistently shown that the genetic contribution to warfarin dose requirement is restricted to three polymorphic loci, namely CYP2C9, CYP4F2 and VKORC1 [4,5]. Polymorphisms in these three loci vary in frequency across populations, [6,7] and consequently, the relative contribution of the CYP2C9, CYP4F2 and VKORC1 genotypes to the estimates of warfarin dose will also vary. Hence the proportions of individuals at risk for ADRs will differ across population groups.
The variant “warfarin sensitive” alleles of CYP2C9 and VKORC1 account for only 10% of the warfarin dose variability in African Americans as opposed to 30% in Caucasians [8-10]. Similarly, performance is quite variable even within the oriental population groups [9,10]. In the present study we analyzed the frequency distribution of warfarin dose-predictive (warfarin sensitive as well as warfarin resistant) genetic polymorphisms in the CYP2C9, CYP4F2 and VKORC1 loci in a micro-geographicallydefined, ethnically admixed Omani patients on warfarin. We also assessed the proportion of Omani patients who will benefit directly from prospective pharmacogenetic testing especially as a locally developed pharmacogenetic algorithm in patients on warfarin could explain 63% dose variability [11].
2. MATERIALS AND METHODS
2.1. Patients
The study cohort was recruited after approval by the institutional review board and after obtaining written informed consent from the participants. Subjects included are consecutive patients aged 13 years or more who were attending the medical and hematology clinics at the Sultan Qaboos University Hospital and were started on warfarin for arterial or venous thrombosis or non-ischemic heart disease viz. atrial fibrillation or dilated cardiomyopathy or had cardiac prosthesis. Exclusion criteria included the presence of severe comorbidities like liver cirrhosis, uremia, or malignant disease.
A total of 214 subjects (114 males and 100 females) consented to participate in the study. Their mean age was 49.9 (±17.8, SD) years. Their demographic data, indication for anticoagulant therapy, target INR range, concomitant disease, concomitant medications, and dietary information were collected from the medical charting system and recorded. All patients were individually monitored and warfarin dose adjusted based on the results of international normalized ratio (INR) for prothrombin time (PT). The warfarin maintenance dose for stable anticoagulation was adjusted to achieve a target INR between 2 to 3 and modifications were made based on the ACCP guidelines for warfarin therapy [12,13]. A patient was considered to have a stable INR when his/her INR was between 2 to 3 on at least 3 consecutive assessments, three months after initiating the therapy. The mean warfarin dose in patients with wild type for both loci (CYP2C9*1/*1 and VKORC1-1639GG genotypes ±2 standard deviations) was used to define the warfarin sensitive and relative resistant groups.
2.2. DNA Studies
A 5 ml blood sample was collected in tubes containing EDTA. Genomic DNA was isolated using the semi-automated ABI PRISM™ 6100 Nucleic Acid Prep Station, (Applied Biosystems, Foster city, CA, USA) and samples were stored at –20˚C pending analysis. All DNA sequence polymorphisms were studied by direct sequencing of the relevant genome segments amplified by polymerase chain reaction (PCR) on an ABI PRISM™ 3100 Genetic Analyzer (Applied Biosystems, Foster city, CA, USA) using home-designed primers (available on request).
2.3. Genotyping: CYP2C9 and CYP4F2
The DNA segments encompassing the exons 3 and 7 of CYP2C9 and exon 11 of CYP4F2 genes were amplified by PCR and the PCR products were submitted for DNA sequencing to explore the following single nucleotide polymorphisms (SNPs); CYP2C9 *2 (430C > T; rs 1799853), *3 (1075A > C; rs 1057910), *4(1076T > C; rs 56165452), *5 (1080C > G; rs 28371686), *8 (449G > A; rs 7900194), *11 (1003C > T; rs 28371685) and CYP4F2*3 (1297G > A; rs 2108622).
VKORC1: The DNA segments containing the SNPs –1639G > A or 3673G > A (rs 9923231) and +1173C > T or 6484 C > T (rs 9934438); [10] of the VKORC1 locus (GeneBank accession number AY 587020) were PCR amplified and analyzed by DNA sequencing of the PCR product as above. In subjects exhibiting warfarin resistance phenotype, all the three exons of the VKORC1 gene, including the splice-site junctions, were amplified by PCR using intronic primers. Sequence variations, if any, were reconfirmed by replicate sequencing of both the sense and anti-sense strands.
2.4. Patient Stratification
Our patients were stratified into three warfarin doserequirement groups, namely “low dose” (sensitive), “medium dose” (intermediate) and “high dose” (resistance), based on their mean daily dose requirement (≤2.5 mg, >2.5 to <8.5 mg, and ≥8.5 mg respectively) to maintain their therapeutic target INR values between 2 and 3 for stable anticoagulation. The cut off value of 2.5 corresponds to the two standard deviation difference from the median value observed in subjects with the wild type genotype simultaneously for CYP2C9 *1/*1 and VKORC1 –1639GG (polymorphism at 1173C > T was in complete linkage disequilibrium with –1639G > A in our study population). Interestingly, a similar cut-off value was reported for a Brazilian population, which also consists of an ethnically admixed group [14].
2.5. Statistical Analysis
Continuous variables, expressed as mean (range), were used for age, INR, and daily warfarin doses. Frequency distribution of the CYP2C9, CYP4F2 and VKORC1 –1639 G > A polymorphisms were studied. The observed allele frequencies were used in Hardy-Weinberg’s equation for analyzing the degree of deviation, if any, between the observed and expected genotype frequencies by using weighted least square estimates of allele frequencies and chi-square goodness-of-fit tests, using SPSS software (ver.15) for all statistical analysis. The differences in the proportion of patients with 1, 2, 3 and 4 mutant alleles were analyzed by chi square or Fisher exact test as appropriate.
3. RESULTS
The demographic data, indications for anticoagulant therapy, concomitant co-morbidities and concomitant medications are tabulated in Table 1.
The frequencies of the allelic variants and genotypes for CYP2C9 (n = 208), CYP4F2 (n = 191) and VKORC1 loci (n=199), are shown in Table 2 along with the daily warfarin dose (Mean ± SD). The mean daily warfarin dose (mg) was 1.0, 3.5, 3.8, 3.8, 4.5 and 5.5 in the CYP2C9*3 homozygotes, *2/*3 compound heterozygotes, *8 heterozygotes, *3 heterozygotes, *2
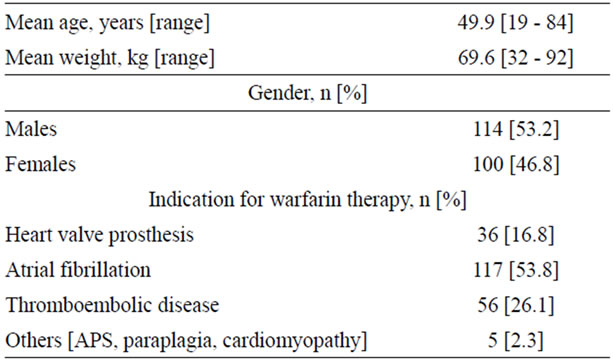
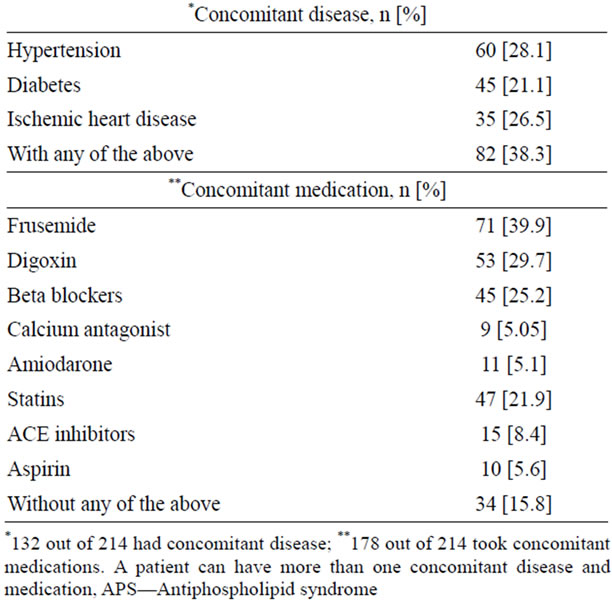
Table 1. Demographic and clinical characteristics of the study cohort [n = 214].

Table 2. Genetic characteristics of the patients and their mean daily warfarin maintenance dose (mg) [n = 214].
heterozygotes and *1 homozygotes (wild type) respectively regardless of genotype combinations with other loci. Similarly, the independent mean warfarin daily dose (mg) was 5.8, 5.0 and 4.6 mg in CYP4F2 *3 homozygotes, *1/*3 heterozygotes, and *1 homozygotes (wild type) respectively. The mean daily warfarin dose was significantly lower with the VKORC1 –1639AA genotype (2.5 mg) as compared to the VKORC1 –1639GA (4.3 mg), and –1639GG (6.2 mg) genotypes (p < 0.05), again independent of genotypes of other loci.
The association between the CYP4F2 V433M allele and warfarin dose requirement in our Omani patients is similar to the values reported in the literature with a significantly higher warfarin dose requirement in the CYP4F2 V433M homozygotes. However, the influence of CYP4F2*3 allele in warfarin dose requirement in combined analysis with other loci was weak and hence was not included in further analysis. Only the CYP2C9 and VKORC1 genotype combinations were relevant with respect to variability in the warfarin dose requirement in our study group.
Table 3 shows the mean warfarin dose range in relation to combined profile of CYP2C9 and VKORC1 genotypes [n = 192 each] in the study cohort, stratified into low warfarin dose (sensitive) [n = 28], medium dose (intermediate) [n = 158] and high warfarin dose (resistance) [n = 6] groups. The range of values of daily dose requirement and genotype combinations within the three groups show considerable overlap irrespective of the number of “sensitive” alleles (i.e. VKORC1 –1639A, CYP2C9 *2 or *3). Nevertheless, the lowest maintenance dose (0.5 mg/day) was noted in a subject who was homozygous for mutant alleles of both loci. An increased number of mutant alleles seemed to segregate in the warfarin low dose (sensitive) group rather than the warfarin medium dose (intermediate) group (p < 0.05, chi square).
In the six warfarin resistant patients (daily warfarin dose > 8.5 mg), in whom the entire VKORC1 coding region was sequenced, we identified two patients with Val66Met mutation, (g.1331G > A, GTG > ATG) and one patient with Asp36Tyr mutation, (g.107A>G; GAC> GGC) in heterozygous state. These mutations have previously been described in the literature and were shown to be associated with warfarin resistance [15,16]. In the remaining three warfarin resistant patients, we did not find any mutation in the coding region of the VKORC1 gene. Furthermore, in none of these six patients we could find CYP4F2*3 variant linked to warfarin resistance.
4. DISCUSSION
Predictive host genetic biomarkers that explain the interpatient warfarin dose variance are not necessarily predic-
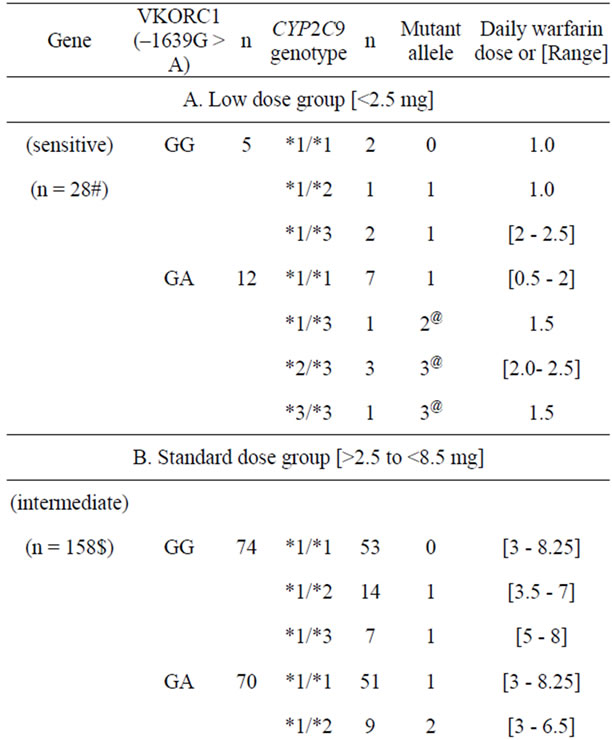
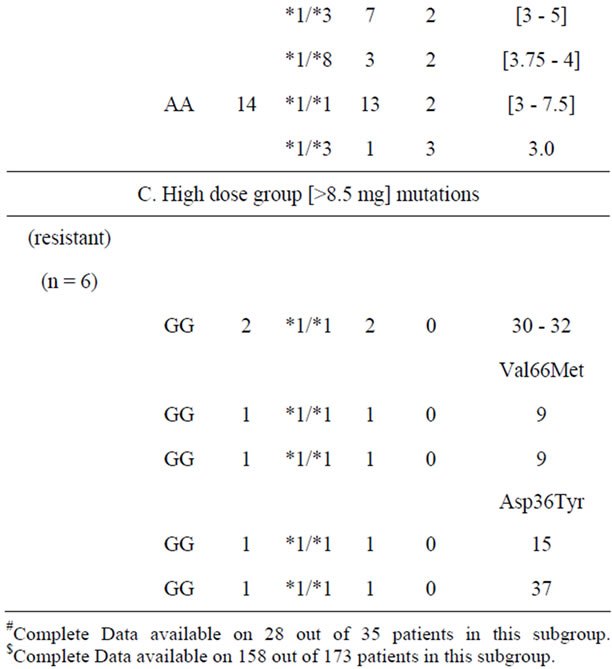
Table 3. Warfarin dose/range in subjects with combined genotype of both VKORC1 and CYP2C9 loci [n = 192].
tors of dose requirement in individual patients as a multitude of external factors do influence the pharmacokinetic and pharmacodynamic behavior of the drug. Hence algorithms that include the variables such as age, sex, bodyweight/surface area, co-medications, concurrent illness and the genetic factors, were developed to improve the dose predictability and even then, we and others have shown, that only around 60% of the inter-patient dose variance can be explained by such algorithms [11,17,18]. The remaining 40% of variance is believed to be influenceed by non-genetic life style factors which are rather difficult to monitor, control and quantify. Contrasting with these unstable and non-quantifiable factors, genetic factors can be reliably assessed in an objective manner. Understanding the extent of exclusive contribution of genetic factors to dose variability in a given patient population may unveil the proportion of patients who warrant stringent monitoring and management protocol. Hence in the present study we have essentially focused our attention to evaluate the extent of influence of genetic factors in the Omani patients on maintenance therapy with warfarin. This allowed us to determine the proportion of patients who are particularly vulnerable to the oscillating external influence on the clinical warfarin phenotype.
In our patient cohort, we did not find a bias in the age distribution in the stratified sensitive and intermediate groups. We found a 2 - 3 fold increase in the number of patients on amiodarone along with warfarin in the sensitive group (14% in the sensitive group vs. 5% for the entire cohort). However, influence of amiodarone, was not found to be significant in our previous multivariate analysis [11].
Around 93% of the subjects of the sensitive group have at least one mutant allele as compared to 67% of those in the intermediate group. However, a minimum of two warfarin-sensitive mutant alleles was observed to be necessary for a significant lowering of the warfarin dose (p < 0.05; Chi square). These data suggest that the mutant alleles of CYP2C9 and VKORC1 loci, although contribute to the warfarin dose variability are not per se sufficient to explain the dose difference between the sensitive and the intermediate groups. Nevertheless, in only 7% of the patients, mutant allele of either locus was absent in the sensitive group as opposed to 33% in the intermediate group (p < 0.05; Chi square). However caution is warranted since our strategy of genotyping concerned essentially the common mutations and hence occurrence of rare functional variants affecting the warfarin metabolism/sensitivity cannot be excluded and need to be explored by a systematic DNA sequencing strategy for the entire genomic region of these two loci.
Although it is known that the sensitive and intermediate groups have mainly mutations in the structural gene of CYP2C9 (affecting the pharmacokinetics) and in the regulatory region of VKORC1 (affecting the pharmacodynamics), the resistant group distinguishes itself by having rare mutations in the structural gene of VKORC1 (exonic variations). Given the overriding effect of VKORC1 exonic mutations in conferring warfarin resistance in heterozygous state, warfarin sensitive alleles in the VKORC1 gene of homologous chromosome in warfarin resistant patients will less likely to have any impact on the observed warfarin phenotype [19].
The present study confirms that the combined CYP2C9 and VKORC1 polymorphisms have impact on warfarin dose phenotype in our microgeographically-defined, ethnically admixed Omani patient population with mixed Caucasian, Asian and African ancestries. However, given the fact that a subset of patients with three or four mutant alleles in CYP2C9 and VKORC1 loci are clearly at risk for overanticoagulation, clinicians can anticipate their potential fragility to the influence of external factors and accordingly can implement close monitoring and management including the prospects of potential drug interactions. The current ACCP recommendations for initiating anticoagulation insist on starting heparin in combination with warfarin. It is envisagable that algorithm-based warfarin dose adjustment can be conveniently initiated within 24 to 48 hours (allowing the necessary time for CYP2C9 and VKORC1 genotyping) that will minimize ADRs. Indeed, a recent pharmacogenetic-based dosing trial highlighted the importance of genotype-guided warfarin therapy in reducing significantly the ADRs [20].
5. ACKNOWLEDGEMENTS
This study was the result of a collaborative project between INSERM, UMR _S 763, Paris, France and Department of Haematology, Sultan Qaboos University, Muscat, Oman. It was supported by a research grant [IG/MED/HAEM/09/01] from the Sultan Qaboos University. We sincerely thank the College Research Committee of the Sultan Qaboos University for facilitating the study and the University Hospital administration for allowing the use of clinical material.
REFERENCES
- Guyatt, G.H., Akl, E.A., Crowther, M., Gutterman, D. and Schuünemann, H.J. (2012) Executive summary: Antithrombotic therapy and prevention of thrombosis, 9th ed: American college of chest physicians evidence-based clinical practice guidelines. Chest, 141, 7S-47S. doi:10.1378/chest.1412S3
- Kovacs, M.J., Rodger, M. and Anderson, D.R., et al. (2003) Comparison of 10-mg and 5-mg warfarin initiation nomograms together with low-molecular-weight heparin for outpatient treatment of acute venous thromboembolism. A randomized, double-blind, controlled trial. Annals of Internal Medicine, 38, 714-719.
- Monkman, K., Lazo-Langner, A. and Kovacs, M.J. (2009) A 10 mg warfarin initiation nomogram is safe and effective in outpatients starting oral anticoagulant therapy for venous thromboembolism. Thrombosis Research, 124, 275-280. doi:10.1016/j.thromres.2008.12.001
- Cooper, G.M., Johnson, J.A., Langaee, T.Y., et al. (2008) A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood, 112, 1022-1027. doi:10.1182/blood-2008-01-134247
- Takeuchi, F., McGinnis, R., Bourgeois, S., et al. (2009) A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genetics, 5, 1-9. doi:10.1371/journal.pgen.1000433
- The Pharmacogenomics knowledge base [PharmGKB] home page. https://www.pharmgkb.org/
- Pathare, A.V., Al Zadjali, S.A., Misquith. R., et al. (2012) Warfarin pharmacogenetics: Polymorphisms of the CYP2C9, CYP4F2, and VKORC1 loci in a genetically admixed Omani population. Human Biology, 84, 67-77. doi:10.3378/027.084.0103
- Cavallari, L.H., Langaee, T.Y., Momary, K.M., et al. (2010) Genetic and clinical predictors of warfarin dose requirements in African Americans. Clinical Pharmacology & Therapeutics, 87, 459-464. doi:10.1038/clpt.2009.223
- Rieder, M.J., Reiner, A.P., Gage, B.F., et al. (2005) Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. The New England Journal of Medicine, 352, 2285-2293. doi:10.1056/NEJMoa044503
- Geisen, C., Watzka, M., Sittinger, K., et al. (2005) VKORC1 haplotypes and their impact on the inter-individual and interethnical variability of oral anticoagulation. Journal of Thrombosis and Haemostasis, 94, 773-779.
- Pathare, A.V., Al Khabori, M., Alkindi, S., et al. (2012) Warfarin pharmacogenetics: Development of a dosing algorithm for Omani patients. Journal of Human Genetics, 57, 665-669. doi:10.1038/jhg.2012.94
- Daly, A.K. and King, B.P. (2003) Pharmacogenetics of oral anticoagulants. Pharmacogenetics, 13, 247-252. doi:10.1097/00008571-200305000-00002
- Holbrook, A., Schulman, S., Witt, D.M., et al. (2012) Evidence-based management of anticoagulant therapy: Antithrombotic therapy and prevention of thrombosis, 9th ed: American college of chest physicians evidence-based clinical practice guidelines. Chest, 141, e152S-e184S. doi:10.1378/chest.11-2295
- Orsi, F.A., Bizzacchi, J.M.A., dePaula, E.V., et al. (2010) VKORC1 V66M mutation in African Brazilian patients resistant to oral anticoagulant therapy. Thrombosis Research, 126, 206-210. doi:10.1016/j.thromres.2010.06.008
- Loebstein, R., Dvoskin, I., Halkin, H., et al. (2007) A coding VKORC1 Asp36Tyr polymorphism predisposes to warfarin resistance. Blood, 109, 2477-2480. doi:10.1182/blood-2006-08-038984
- Harrington, D.J., Underwood, S., Morse, C., Shearer, M.J., Tuddenham, E.G. and Mumford, A.D. (2005) Pharmacodynamic resistance to warfarin associated with Val66Met Substitution in Vitamin K epoxide reductase complex 1. Thrombosis & Haemostasis, 93, 23-26.
- Vecsler, M., Loebstein, R., Almog, S., et al. (2006) Combined genetic profiles of components and regulators of the vitamin K-dependent gamma-carboxylation system affect individual sensitivity to warfarin. Journal of Thrombosis and Haemostasis, 95, 205-211.
- Herman, D., Peternel, P., Stegnar, M., Breskvar, K. and Dolzan, V., (2006) The influence of sequence variations in factor VII, gammaglutamyl carboxylase and vitamin K epoxide reductase complex genes on warfarin dose requirement. Journal of Thrombosis and Haemostasis, 95, 782-787.
- Rost, S., Fregin, A., Ivaskevicius, V., et al. (2004) Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature, 427, 537-541. doi:10.1038/nature02214
- Anderson, J.L., Horne, B.D., Stevens, S.M., et al. (2012) A randomized and clinical effectiveness trial comparing two pharmacogenetic algorithms and standard care for individualizing warfarin dosing (CoumaGen-II). Circulation, 125, 1997-2005. doi:10.1161/CIRCULATIONAHA.111.070920
NOTES
*Corresponding author.

