Advances in Biological Chemistry
Vol.3 No.3(2013), Article ID:33565,9 pages DOI:10.4236/abc.2013.33033
Rad51 ATP binding but not hydrolysis is required to recruit Rad10 in synthesis-dependent strand annealing sites in S. cerevisiae
![]()
Department of Chemistry and Biochemistry, California State University Northridge, Northridge, USA
Email: paula.fischhaber@csun.edu
Copyright © 2013 Justin Karlin, Paula L. Fischhaber. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 22 April 2013; revised 20 May 2013; accepted 30 May 2013
Keywords: Rad1; Rad10; Rad51; Synthesis Dependent Strand Annealing; Yeast; Double Strand Break Repair
ABSTRACT
Several modes of eukaryotic of DNA double strand break repair (DSBR) depend on synapsis of complementary DNA. The Rad51 ATPase, the S. cerevisiae homolog of E. coli RecA, plays a key role in this process by catalyzing homology searching and strand exchange between an invading DNA strand and a repair template (e.g. sister chromatid or homologous chromosome). Synthesis dependent strand annealing (SDSA), a mode of DSBR, requires Rad51. Another repair enzyme, the Rad1-Rad10 endonuclease, acts in the final stages of SDSA, hydrolyzing 3( overhanging single-stranded DNA. Here we show in vivo by fluorescence microscopy that the ATP binding function of yeast Rad51 is required to recruit Rad10 SDSA sites indicating that Rad51 pre-synaptic filament formation must occur prior to the recruitment of Rad1- Rad10. Our data also show that Rad51 ATPase activity, an important step in Rad51 filament disassembly, is not absolutely required in order to recruit Rad1- Rad10 to DSB sites.
1. INTRODUCTION
DNA double strand breaks (DSB) occur when both sugar phosphate backbones of the DNA double helix are severed. They may be instigated by both exogenous agents (e.g. ionizing radiation) and endogenous by-products of normal cellular metabolism (e.g. hydroxyl radicals) [1]. If left unrepaired prior to cellular replication, a single DSB can result in permanent loss of genetic material and can even cause death of the organism [1]. DSBR pathways are thus among the most essential of the repair pathways, as evidenced by the myriad repair pathways exhibiting redundant function [1-3].
In S. cerevisiae, DSB that occur during cell division can be repaired by a mode of homologous recombination (HR) known as synthesis-dependent strand annealing (SDSA). This process begins with 5¢ ® 3¢ resection of regions flanking the DSB and invasion of a resultant 3¢ overhanging end into a sister chromatid or homologous chromosome (i.e. “strand invasion”). Strand invasion and subsequent identification of homologous DNA (i.e. “homology searching”) are facilitated by RPA, Rad51 and Rad52 [4-7]. Rad51 forms a helical, filamentous multimer (i.e. “pre-synaptic filament”) surrounding the single-stranded portion of the resected chromosome in a process requiring ATP. The pre-synaptic filament couples to the sister chromatid or homologous chromosome and moves distally until homologous DNA has been located. Once complementary DNA is found, a conformational change occurs that enables hybridization of homologous regions (i.e. synapsis). Template-directed DNA synthesis commences from the 3¢ end of the invading strand catalyzed by DNA Polymerase δ (i.e. post-synapsis) [1,8]. The nascent DNA then dissociates from the template and reanneals with the resected end at the break site in a process that is promoted by Rad52 [9]. The reannealing process may leave a 3¢ DNA overhanging flap which must be removed prior to the ligation. This flap is hydrolyzed by the structure-specific Rad1- Rad10 endonuclease in a process that also requires the Msh2-Msh3 complex from the mismatch repair pathway [10-13]. The result is a repaired segment of DNA identical in sequence to that found on the original, unbroken chromosome.
Our previous work showed that deletion of RAD51 abrogates Rad1-Rad10 recruitment to SDSA sites in S. cerevisiae [14]. The mechanism by which Rad51 is required for Rad1-Rad10 recruitment, however, remains unknown. It has previously been shown that Rad51 binds ATP to form the pre-synaptic filament, and hydrolyzes ATP to catalyze filament dissociation following homology searching. In the absence of ATP, the Rad51 filament rapidly dissociates from DNA [3,15,16]. The extent to which Rad51 filament formation and dissociation are carried out in advance of recruitment of downstream processing factors remains an open question. Our previous study shed light on the temporal overlap of Rad51 and Rad1-Rad10 by showing in vivo that a significant proportion of SDSA sites are occupied simultaneously by Rad51 and Rad10. Herein we use a fluorescence microscopy assay to investigate whether Rad51 ATP binding, hydrolysis or both are required to recruit Rad1-Rad10 to sites of SDSA.
2. MATERIALS AND METHODS
2.1. Preparation of Yeast Strains Containing the DSB Induction System and rad51K191A or rad51K191R
S. cerevisiae strains containing the required genetic markers were prepared by genetic crosses using standard procedures. Parent strain PF027-34D (Table 1) was transformed with plasmid pWJ554 [17] and crossed with strains LSY0983 and LSY0979 to produce strains PF075-8B and PF073-10D, respectively.
2.2. Preparation of a Yeast Strain Containing DSB Induction System and rad2(
A rad2D transplacement knockout DNA cassette was prepared by PCR amplification of a segment of chromosomal DNA from a diploid hybrid of the SX46a and BJ1991 strains, WS9091. The primers used were Rad2- F1 5¢-CCTACGATACTTTATGGCTTTGGCTC-3¢ and Rad2-B16 5¢-GCCATAGCAAGAGCGTTTGTG-3¢which corresponded to the regions immediately 5¢ and 3¢ of the rad2D::TRP1 knockout cassette. This PCR product was ligated into the pGEM-T-easy vector (Promega Corporation) using manufacturer’s instructions. Approximately 40 µg of the resulting plasmid was linearized (EcoRI, Invitrogen) and transformed into W303-1A yeast using standard procedures. Transformants were selected on SC-Trp agar, and the presence of the deletion cassette was verified by PCR. The resulting rad2D strain (PF094) was crossed to strain PF027-34D using standard methods to produce strain PF099-31A that was used in fluorescence microscopy experiments. Strain PF094 was used in UV survival experiments.
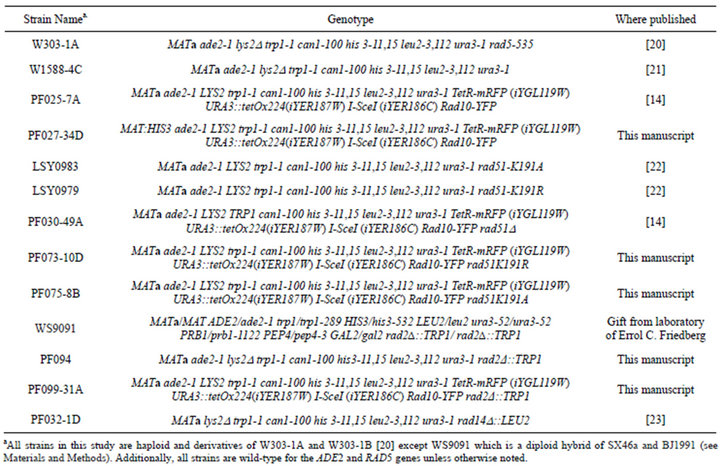
Table 1. Strains used in this study.
2.3. General Microscopy
Microscopy was performed on a Zeiss AxioImager M1 microscope with a Plan-Apochromat 100×, 1.46 numerical aperture (NA) objective oil immersion lens, a motorized Z-drive and automated shutters. The fluorescent light source was an X-Cite 120 IRIS Fluorescent Light Source. Images were captured with a Hamamatsu ORCAER digital camera (CA4742-80-12AG). For 11-slice Zstacks, images at different focal planes through the entire thickness of the cells were captured at 0.3 mm intervals along the Z-axis (a Z-stack); for 3-slice Z-stacks images were also captured at 0.3 mm intervals but only imaging the 3 slices bounding the center focal plane of the cell. Filter sets employed were Yellow GFP, Chroma # 41028, (lex = 500/20, ldichroic = 515, lem = 535/30), CFP, Chroma # 31044V2 (lex = 436/20, ldichroic = 455, lem = 480/40) and NAR/EX DsRed, Chroma # 41035, (lex = 546/11, ldichroic = 560, lem = 605/75). Integration time for image acquisition was 800 ms for Rad10-YFP and 400 ms for TetR-RFP. Microscope and camera were controlled by the Volocity software package (Improvision v3.7.0 for the Zeiss). Foci were counted by inspecting images from each focal plane of the Z-stack and contrast enhanced as previously described [14]. Images were prepared for publication using the Adobe Photoshop and Adobe Illustrator software packages (Adobe Systems, Mountain View, CA).
2.4. Induction of DSB by I-SceI
Strains for DSB induction experiments bear chromosomally integrated copies of the Tetracycline repressor protein (TetR) fused to RFP (TetR-RFP) and 224 copies of the Tetracycline operator (tetO) repressor binding site abutted with one copy of the I-SceI cut site at the iYER- 186 intergenic region on chromosome V as described [18]. The site is cut by the I-SceI endonuclease with 60% - 70% efficiency in asynchronously growing cells [18,19]. The strains used in I-SceI DSB induction experiments were PF025-7A, PF030-49A, PF073-10D, PF075-8B and PF099-31A.
Cultures for microscopy experiments were initially propagated in SC medium supplemented with 200 mg/mL adenine (SC + ade) at 23˚C. Strains were then transformed with plasmid pWJ1320, containing the I-SceI gene under control of the GAL1 promoter and an ADE2 selectable marker as described [24]. Colonies were isolated from Synthetic Complete agar lacking adenine, to ensure the presence of the pWJ1320 plasmid, and containing 2% raffinose (SC – ade + raff) to suppress I-SceI expression. Liquid cultures for DSB induction experiments were then propagated in SC – ade + raff at 23˚C overnight. Cultures were diluted to 0.1 OD600 and incubated for 3 h to allow cells to return to log phase growth. At that point, half of the culture was incubated in the presence of galactose (final concentration of 2%) and the other half was allowed to grow untreated (i.e. uninduced controls). Following growth for an additional 4 h, cells were processed for microscopy by brief, gentle centrifugation (1 min, 7000 rpm) to concentrate cells from 1 mL of culture into a small volume followed by mounting of the cell concentrates on microscope slides by mixing aliquots (1 mL) with an equal volume of mounting medium (SC + ade + 2% low melting agarose, 42˚C) as described previously [14].
2.5. Figure Preparation and Statistical Analyses
At least 2 independent trials were carried out for each experiment and at least 100 cells were examined and counted for each condition. The data were analyzed and graphs were prepared using Prism 6 for Mac OS X (GraphPad Software). The mean and standard deviation were calculated, and an unpaired two-tailed Student’s t-test was used to compare the mean colocalized focus percentages between different strains statistically (e.g. focus induction following enzyme induction in a RAD51 strain compared to a rad51-K191R strain). P < 0.05 was set as the cut-off value for significance in all experiments.
2.6. UV Survival Studies
UV survival experiments were carried out in duplicate by standard methods [14]. The strains used were W1588-4C (RAD2 RAD14), PF094 (rad2D RAD14) and PF032-1D (RAD2 rad14D). Briefly, cultures of cells were grown to mid-log phase (OD600 = 0.5) and cell densities counted using a hemocytometer. Appropriate dilutions of mid-log cultures were made onto YPD plates and cells distributed evenly on plate surfaces. Plates were incubated (30˚C, 1 h) to allow plating liquid to soak into plates and dry. Plates were irradiated with a germicidal UV-C lamp at the appropriate dosage (0 - 40 J/m2), wrapped in aluminum foil prior to exposure to room light and incubated (30˚C, 2 days). Colonies were counted and compared to unirradiated control plates to determine the fraction surviving.
3. RESULTS
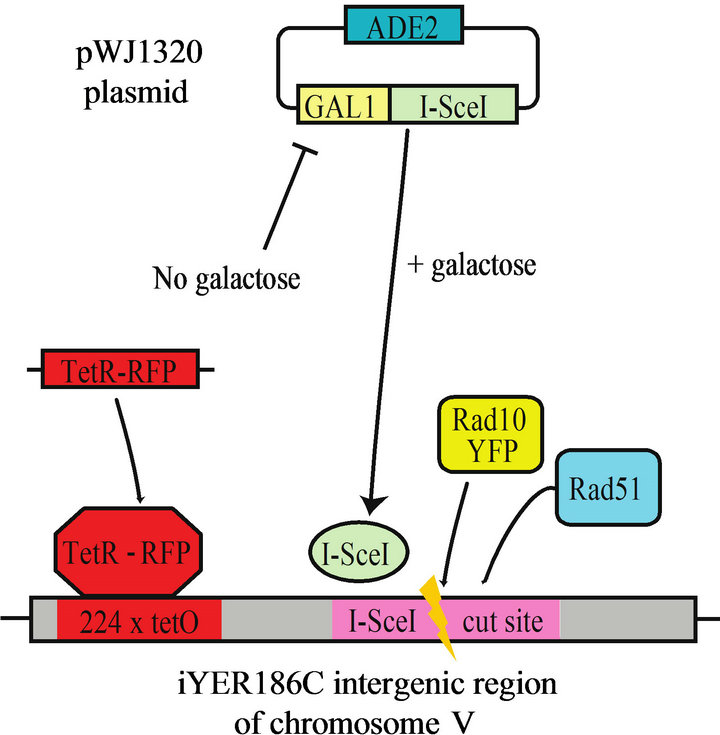
We set out to determine the extent to which Rad51 ATP binding, hydrolysis or both are required for recruitment of the Rad1-Rad10 complex to DSB sites in SDSA. To do so, we prepared a panel of RAD51 mutant S. cerevisiae strains chromosomally expressing Rad10 labeled with yellow fluorescent protein (Rad10-YFP), and a galactose-inducible, red-fluorescent-protein-labeled DSB site (DSB-RFP). The following alleles were used: RAD51 deletion (rad51D), point mutant Lys191Ala (rad51K191A, deficient in ATP binding), or point mutant Lys191Arg (rad51K191R, binds ATP but hydrolyzes extremely poorly exhibiting <5% residual ATPase activity) [25]. The inducible DSB site in these strains is not flanked by DNA repeats, and thus is repaired by SDSA and not single strand annealing (SSA) [14]. Addition of galactose derepresses the promoter of plasmid-encoded I-SceI gene (Figure 1(a)). I-SceI-induced DSB are situated proximal to numerous TetR-RFP binding elements, which labels the DSB site with a red fluorescent focus (DSB-RFP) (Figure 1(a)). An example of fluorescent images containing a colocalized Rad10-YFP/ DSB-RFP focus is shown (Figure 1(b)).
Examining cells in all phases of cell cycle, the data
 (a)
(a)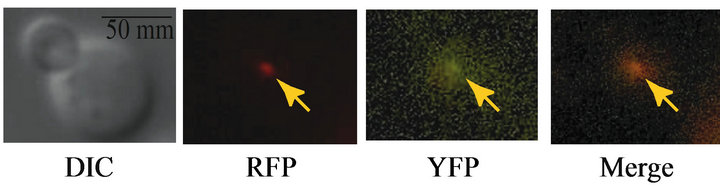 (b)
(b)
Figure 1. (a) Scheme showing experimental design. The I-SceI gene is introduced to the cells on an adenine selectable (ADE2) plasmid under the control of the galactose-inducible (GAL1) promoter. When propagated in medium containing raffinose, the GAL1 promoter is repressed; upon addition of galactose, the I-SceI gene is derepressed. A single I-SceI recognition sequence is installed within an intergenic region (iYER186C) of the S. cerevisiae chromosome V. This I-SceI recognition site lies adjacent to 224 tandem copies of the Tet Repressor Protein binding site (224 × tetO). The TetR gene fused to monomeric Red Fluorescent Protein (TetR-RFP) is constitutively expressed from an intergenic region on chromosome VII (iYGL119W). TetR-RFP binds to the array of 224 Tet repressor binding sites, thereby labeling the site of the induced DSB (DSB-RFP). (b) Example of a Rad10-YFP focus colocalized with TetR-RFP (DSB-RFP) focus (arrows). Fluorescence and DIC images are shown for strain PF025-7A transformed with plasmid pWJ1320 in the presence of galactose.
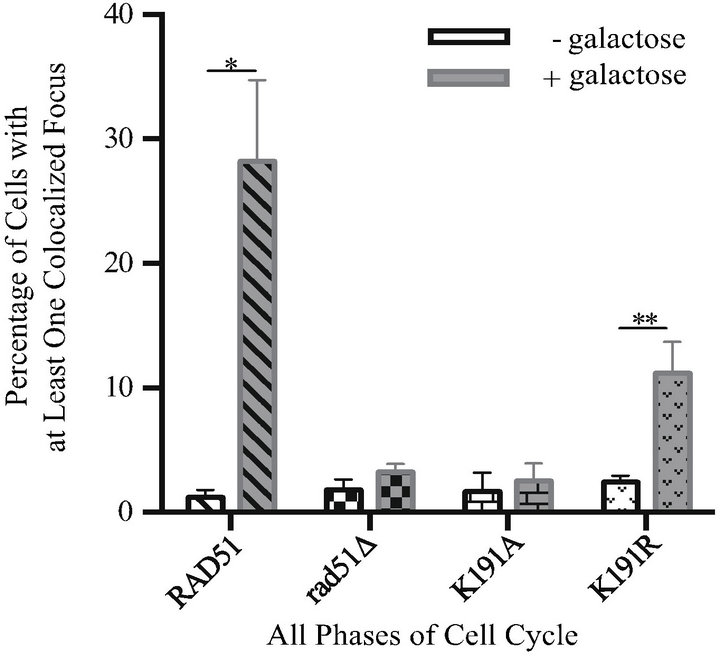
show that Rad10-YFP/DSB-RFP colocalization increases 24-fold in DSB induced (i.e. galactose-treated) samples over uninduced controls (mean 1.2% uninduced vs 28% induced) in a RAD51 strain (Figure 2(a)). In contrast, the rad51D cells exhibit only background levels of colocalization in response to DSB induction, with a 1.8-fold induction (mean 1.8% uninduced vs 3.2% induced) (Figure 2(a)). The rad51K191A mutant gave similar results to rad51D, exhibiting only a 1.5-fold induction (mean 1.7% uninduced vs 2.5% induced) indicating that the ATP binding activity of Rad51, and therefore formation of the Rad51 nucleoprotein filament, is necessary
 (a)
(a)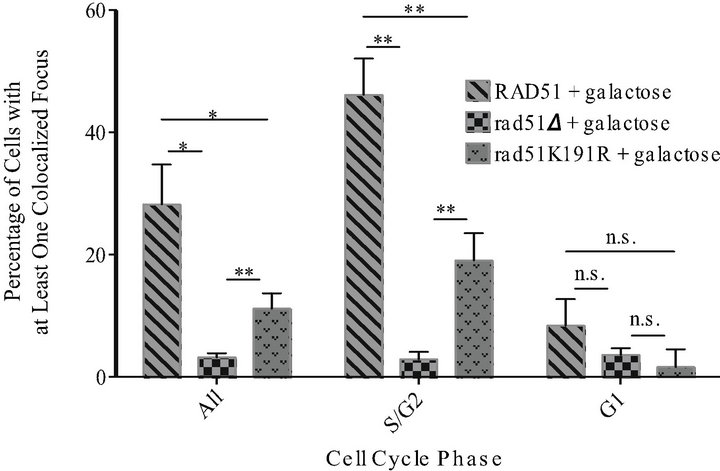 (b)
(b)
Figure 2. Rad10-YFP/DSB-RFP colocalized foci are not induced in rad51K191A cells but are partially induced in rad51K- 191R cells in response to DNA DSBs. White bars show results from uninduced control cells. Gray bars show results from cells induced with galactose. Diagonal hatched bars, RAD51 strain (PF025-7A); checked bars, rad51D strain (PF030-49A); bricked bars, rad51K191A strain (PF075-8B); stippled bars, rad51K- 191R strain (PF073-10D). Mean values are shown and error bars indicate the standard deviation of three independent experiments in each of which at least 100 cells were counted and classified. Unpaired, two-tailed T-tests were performed. “**” indicates 0.001 < P < 0.01; *indicates 0.01 < P < 0.05; n.s. indicates P values > 0.05. (a) Results from all cells without regard to phase of cell cycle. (b) Results from RAD51, rad51D and rad51K191R following galactose induction and organized by cell cycle phase.
for the recruitment of Rad10 (Figure 2(a)). This result was not unexpected since Rad51 nucleoprotein filament formation, homology searching, strand exchange and 3¢ extension of the broken chromosome presumably are required to create the DNA flap subsequently cleaved by Rad1-Rad10.
Interestingly, the ATPase-deficient rad51K191R mutant revealed an intermediate phenotype, exhibiting a 4.6 fold induction of Rad10-YFP/DSB-RFP colocalization (mean 2.4% uninduced vs 11% induced) (Figure 2(a)). Although the magnitude of this induction was not as robust as that observed in the RAD51 strain, it was statisticcally significant (P = 0.0040) (Figure 2(a)) when compared to rad51Δ cells. Together, these data indicate that, although the ATP binding activity of Rad51 is crucial to downstream recruitment of Rad10, the ATP hydrolysis activity is not absolutely required.
When results were further analyzed with respect to cell cycle, induction of Rad10-YFP/DSB-RFP colocalization was found almost entirely in dividing cells. In S phase or G2 phase (S/G2) cells, RAD51 and rad51K191R strains treated with galactose were found to have colocalized foci in 46% and 19% of cells, respectively, a statistically significant difference (P = 0.0032). In contrast, G1 phase cells exhibited 8.4 % and 1.7 %, respectively, a non-significant difference (P = 0.087) (Figure 2(b)). In rad51D both S/G2 and G1 cells exhibited only background levels of YFP/RFP colocalized foci (mean 2.9% and 3.6%, respectively) (Figure 2(b)). These data indicate that SDSA in haploid yeast proceeds in the presence of a sister chromatid, which is only available for template repair during S/G2 and early in M phase.
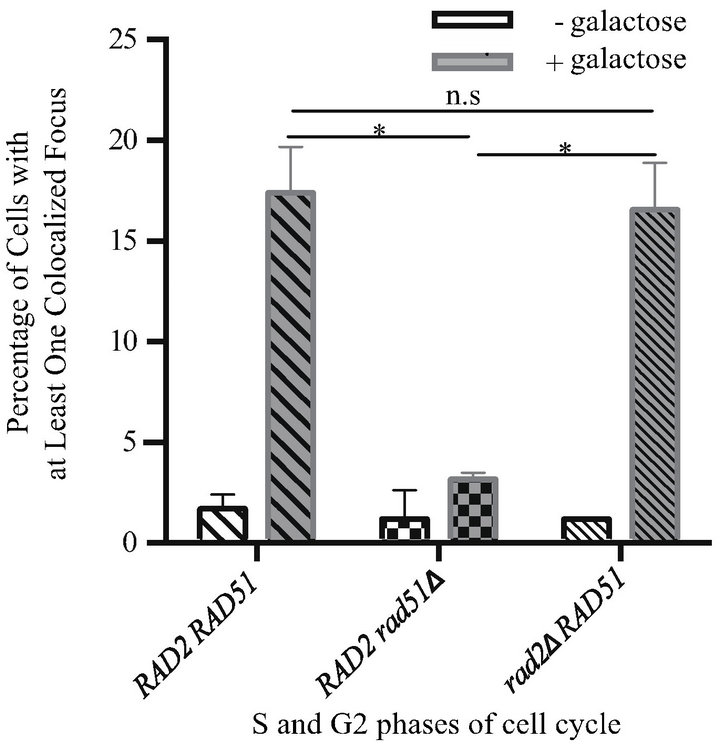
The Rad1-Rad10 endonuclease plays a central role in another repair pathway, nucleotide excision repair (NER). To ensure that the observed Rad10-YFP/DSB-RFP foci corresponded to sites of DSBR and not sites of NER, we carried out DSB induction experiments in an NER-null mutant (i.e. RAD2 deletion mutant, rad2D). Comparing strains harboring either the RAD2 or rad2D or alleles, we observed no statistically significant difference (P = 0.75) in Rad10-YFP recruitment to DSB-RFP sites in response to DSB induction, observing 12-fold and 16-fold induction of YFP/RFP colocalized foci in the RAD2 RAD51 and rad2D RAD51 strains, respectively (Figure 3(a)). As expected, a strain harboring the rad51D allele exhibited only 3.0-fold induction (Figure 3(a)).
To confirm that the rad2D strain was truly NER-null, we performed a UV sensitivity assay. Indeed, cells harboring the rad2D allele exhibit UV sensitivity comparable to another NER-deficient strain (rad14D) (Figure 3(b)). We thus conclude that Rad10-YFP recruitment to DSB-RFP sites in response to DSB induction corresponds to Rad1-Rad10 recruitment to sites of DSBR and not NER.
 (a)
(a) (b)
(b)
Figure 3. Rad10-YFP/DSB-RFP colocalized focus induction is not altered in an NERdeficient rad2D strain. (a) White bars show results from uninduced control cells. Gray bars show results from cells induced with galactose. Bars with wide diagonal hatches, RAD51 strain (PF025-7A); checked bars, rad51D strain (PF030-49A); bars with narrow diagonal hatches, rad2D (PF099-31A). Mean values are shown and error bars indicate the standard deviation from two independent experiments in each of which at least 100 cells in all phases of the cell cycle were counted and classified. Unpaired two-tailed T-tests were performed. *indicates 0.01 < P < 0.05; n.s. indicates P values > 0.05. (b) UV survival experiment showing that the rad2D strain exhibits similar UV sensitivity to a rad14D strain. Yeast strains W1588-4C (wild-type), PF032-1D (rad14D) and PF094 (rad2D) were cultured to log phase in YPD, diluted into water, plated on YPD agar and irradiated with UV-C according to standard methods. Irradiated plates were incubated in the dark 2 days and colonies of surviving cells counted. Mean values are shown and error bars indicate the standard deviation from three independent experiments.
4. DISCUSSION
The timing and order of the individual steps in SDSA are carefully orchestrated. Although Rad1-Rad10 and Rad51 have not been shown to form a direct physical complex, Rad51 binds Rad52 directly, and through this interaction forms a multiprotein complex involving Saw1 and Rad1 [7,26]. The point in the repair process when Rad1- Rad10 is recruited to the repair site has not been identified, except insofar as Rad10 presence at the SDSA site temporally overlaps with the presence of both Rad51 and Rad52 [14]. We set out to determine whether Rad1- Rad10 recruitment may occur very early during repair, perhaps as early as Rad51 pre-synaptic filament formation. Since ATP binding of Rad51 is prerequisite to filament formation, we examined the ATP binding-deficient rad51K191A mutant and found Rad10 recruitment was completely abrogated, indicating that initiation of filament formation is indeed required for Rad1-Rad10 recruitment. Likewise, we tested whether ATP hydrolysis was required for Rad10 recruitment by employing the ATP binding-proficient, ATP hydrolysis-deficient rad51- K191R mutant. We observed an intermediate phenotype in which recruitment of Rad10 was occurring, but at lower levels than those observed in the RAD51 wild-type strain. An illustration depicting our model explaining the possible molecular basis for these results and role of accessory proteins in displacing Rad51 is shown (Figure 4).
Given that Rad51 ATPase activity plays a central role in Rad51 filament disassembly, our experimental observations indicate that failure of Rad51 to catalyze its own dissociation significantly diminishes or delays the downstream steps of SDSA. This result seems to contradict prior studies which indicated that Rad51 and Rad1- Rad10 can be found at a DSB site at the same time [14]. Several mechanistic possibilities may be able to reconcile this potential contradiction. In one possible scenario, filament dissociation from the overhanging flap may initiate and proceed in the 3¢ ® 5¢ direction and the liberated flap may become reannealed to the broken chromosome prior to complete removal of the Rad51 filament. This mechanism would display a requirement for some measure of Rad51 removal (normally catalyzed by Rad51-ATPase activity) but also, ostensibly could result in a period of time between 3¢ flap hydrolysis by Rad1- Rad10 and dissociation of the final Rad51 molecule from the repair site, thereby explaining earlier observations of temporal overlap between Rad51 and Rad1-Rad10 occupancies at repair sites [14]. In support of this model, in a study by Antony et al., in vitro dissociation of Rad51 filaments was found to occur distal to the 5¢ end of the nucleoprotein filament [15]. Rad1-Rad10 recruitment may thus require only partial dissociation of Rad51 filaments. In another possible scenario, following DNA
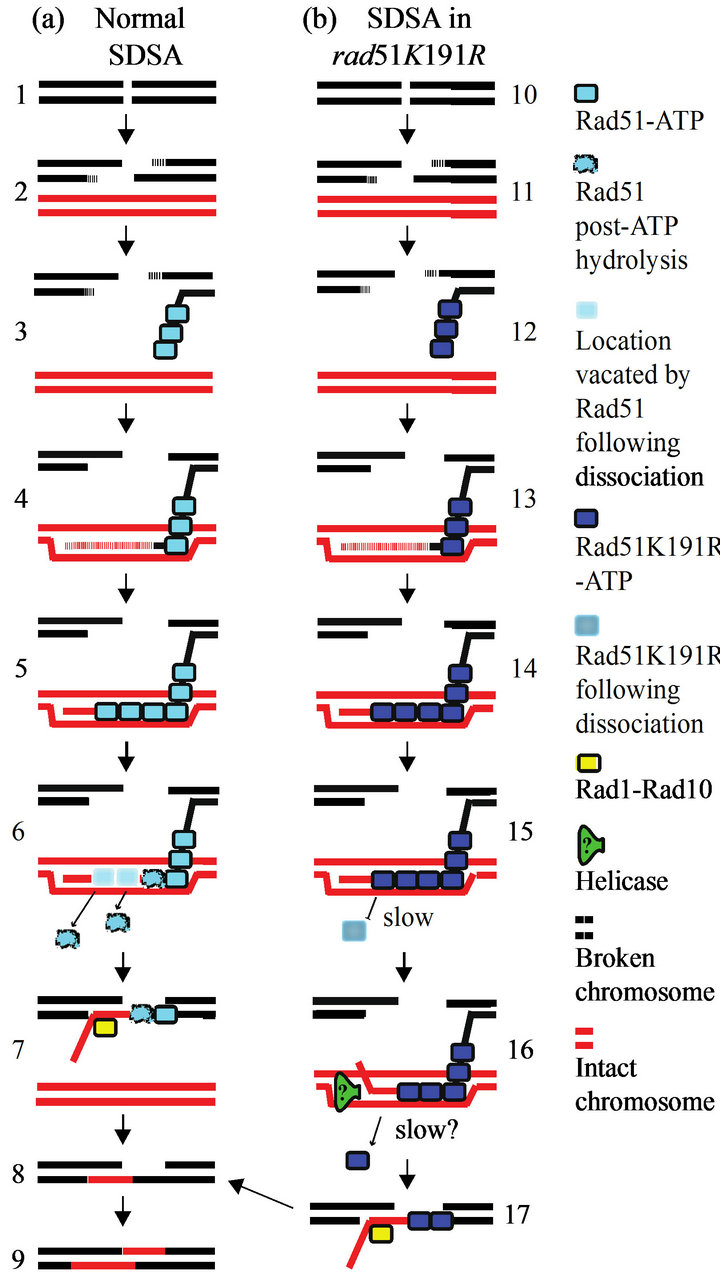
Figure 4. Model for Rad1-Rad10 recruitment to sites of SDSA in the presence of rad51K191R. (a) In wild-type cells, I-SceI-induced strand breaks (1) are resected (2) to give 3¢ singlestranded regions. ATPbound Rad51 (blue boxes) is recruited to form the presynaptic (3) and synaptic filaments (4), along with Rad52 (not shown). The invading strand is copied by DNA polymerase (4 and 5) templated from homologous base pairing on the sister chromatid (shown in red). After DNA synthesis, Rad51 hydrolyzes the bound ATP (6, distorted blue boxes) and rapidly dissociates. The newly synthesized strand anneals with the broken chromosome (7). Overlap between the newly annealed strands is removed by Rad1- Rad10 (7, yellow box), leading to ligation of the nick (8) and gap filling of the opposite strand (9). (b) In the rad51K191R strain, the initial steps of SDSA (10 - 14) are similar to wild-type. Following new DNA synthesis, however, Rad51-K191R protein cannot hydrolyze ATP and dissociates very slowly from the synapse (15). Other factors, possibly helicases (16, green triangle) may stimulate Rad51-K191R displacement by unwinding the newly synthesized DNA and liberating it so can anneal with the broken chromosome and be trimmed by Rad1-Rad10 (17, yellow box). The combination of slow spontaneous dissociation by Rad51-K191R and accessory factor displacement (if any) result in a lowered ability to recruit Rad1-Rad10.
strand invasion and synthesis using the sister chromatid as a template, the nascent strand may re-invade the original chromatid, forming a Holliday Junction, as has been proposed [27]. Subsequent resolution of this structure can involve Rad1-Rad10. If some measure of dissociation of Rad51 filaments must occur before the Holliday Junction is resolved, such a mechanism would explain both the previously observed temporal overlap of Rad51 and Rad1-Rad10 at repair sites, as well as the defect we observed in the Rad51-K191R mutant in the present study.
A study by Fung et al. showed that the Rad51-K191R protein is defective in presynaptic filament formation in response to ionizing radiation [28]. In light of this data, it may seem that the observed attenuated, but not ablated, recruitment of Rad1-Rad10 to DSB sites in the presence of Rad51-K191R may be due to delayed kinetics of nucleoprotein filament formation rather than disrupted filament disassembly. Close inspection of data from that study, however, reveals that the defect in presynaptic filament formation occurs only up to 2 hours after induction of DNA damage. At 3 or more hours after DNA damage induction (a similar time point to that examined in our study), more Rad51-K191R is found to be bound to DNA compared to wild-type Rad51. So, although the kinetics of Rad51-K191R filament formation may be altered very early in the damage response, several hours after damage induction, the principal defect of Rad51- K191R is in filament disassembly.
If Rad1-Rad10 recruitment can occur in the absence of Rad51 ATPase catalyzed auto-dissociation, as our data suggest, then accessory factors, such as repair helicases, may be involved in removal or regulation of Rad51. One such helicase, Srs2, blocks Rad51-mediated S phase recombination and also inhibits Rad51-induced release from cell cycle checkpoint arrest. Srs2 has been shown to clear Rad51 filaments by binding Rad51 and allosterically triggering ATP hydrolysis [15]. It therefore seems unlikely that the modest Rad10 recruitment observed in rad51K191R experiments is due to Srs2 dissolution of filaments containing rad51K191R mutant proteins. Another helicase, Sgs1, regulates chromosome synapsis and may directly induce dissociation of presynaptic filaments [29-34]. Experiments in strains harboring Srs2 or Sgs1 deletions may shed light on the mechanism by which Rad51 participates in the recruitment of Rad1-Rad10.
We believe that spontaneous dissociation of Rad51- K191R (as opposed to helicase-mediated dissociation) does not contribute significantly to Rad10 recruitment to SDSA sites in rad51K191R cells. Experiments addressing the spontaneous dissociation of Rad51 filaments in vitro show that Rad51 dissociates from single-stranded DNA very slowly in the absence of ATP [15]. Furthermore, in our study, Rad10-YFP focus formation at SDSA sites in the rad51K191R strain amounted to approximately 20% that which we observed in the RAD51 strain (4.6-fold vs 24-fold, respectively), even though the residual ATP hydrolysis activity of Rad51-K191R has previously been shown to be less than 5% that of Rad51 [25]. Figure 4 illustrates our model—partial dissociation of Rad51, facilitated by other repair factors such as helicases, induces recruitment of Rad10 to SDSA sites.
5. ACKNOWLEDGEMENTS
The authors thank the laboratory of Lorraine Symington for yeast strains containing rad51K191A and rad51K191R mutants. This work was supported by the California State University Program for Education and Research in Biotechnology (CSUPERB) Don Eden Graduate Student Research Award (JK), National Institutes of Health grants GM081155 and GM093858 (PLF) and by the California State University Northridge Competition for Research Scholarship and Creative Activity Awards (PLF).
REFERENCES
- Friedberg, E.C., Walker, G.C., Siede, W., Wood, R.D., Schultz, R.A., et al. (2005) DNA repair and mutagenesis. 2nd Edition, ASM Press, Washington DC.
- Paques, F. and Haber, J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews, 63, 349- 404.
- Symington, L.S. (2002) Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiology and Molecular Biology Reviews, 66, 630-670. doi:10.1128/MMBR.66.4.630-670.2002
- New, J.H., Sugiyama, T., Zaitseva, E. and Kowalczykowski, S.C. (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature, 391, 407-410. doi:10.1038/34950
- Sugiyama, T., New, J.H. and Kowalczykowski, S.C. (1998) DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proceedings of the National Academy of Sciences of the United States of America, 95, 6049-6054. doi:10.1073/pnas.95.11.6049
- Sugiyama, T., Zaitseva, E.M. and Kowalczykowski, S.C. (1997) A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. The Journal of Biological Chemistry, 272, 7940-7945. doi:10.1074/jbc.272.12.7940
- Sung, P. (1997) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. The Journal of Biological Chemistry, 272, 28194-28197. doi:10.1074/jbc.272.45.28194
- Maloisel, L., Fabre, F. and Gangloff, S. (2008) DNA polymerase delta is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Molecular and Cellular Biology, 28, 1373-1382. doi:10.1128/MCB.01651-07
- Sugiyama, T., Kantake, N., Wu, Y. and Kowalczykowski, S.C. (2006) Rad52-mediated DNA annealing after Rad51- mediated DNA strand exchange promotes second ssDNA capture. The EMBO Journal, 25, 5539-5548. doi:10.1038/sj.emboj.7601412
- Bardwell, A.J., Bardwell, L., Johnson, D.K. and Friedberg, E.C. (1993) Yeast DNA recombination and repair proteins Rad1 and Rad10 constitute a complex in vivo mediated by localized hydrophobic domains. Molecular Microbiology, 8, 1177-1188. doi:10.1111/j.1365-2958.1993.tb01662.x
- Fishman-Lobell, J. and Haber, J.E. (1992) Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science, 258, 480-484. doi:10.1126/science.1411547
- Sugawara, N., Ira, G. and Haber, J.E. (2000) DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Molecular and Cellular Biology, 20, 5300-5309. doi:10.1128/MCB.20.14.5300-5309.2000
- Sugawara, N., Paques, F., Colaiacovo, M. and Haber, J.E. (1997) Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proceedings of the National Academy of Sciences of the United States of America, 94, 9214-9219. doi:10.1073/pnas.94.17.9214
- Moore, D.M., Karlin, J., Gonzalez-Barrera, S., Mardiros, A., Lisby, M., et al. (2009) Rad10 exhibits lesion-dependent genetic requirements for recruitment to DNA double-strand breaks in Saccharomyces cerevisiae. Nucleic Acids Research, 37, 6429-6438.
- Antony, E., Tomko, E.J., Xiao, Q., Krejci, L., Lohman, T.M., et al. (2009) Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Molecular Cell, 35, 105-115. doi:10.1016/j.molcel.2009.05.026
- Sung, P. and Stratton, S.A. (1996) Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. The Journal of Biological Chemistry, 271, 27983-27986. doi:10.1074/jbc.271.45.27983
- Lisby, M., Rothstein, R. and Mortensen, U.H. (2001) Rad52 forms DNA repair and recombination centers during S phase. Proceedings of the National Academy of Sciences of the United States of America, 98, 8276-8282. doi:10.1073/pnas.121006298
- Lisby, M., Mortensen, U.H. and Rothstein, R. (2003) Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nature Cell Biology, 5, 572- 577. doi:10.1038/ncb997
- Barlow, J.H., Lisby, M. and Rothstein, R. (2008) Differential regulation of the cellular response to DNA double-strand breaks in G1. Molecular Cell, 30, 73-85. doi:10.1016/j.molcel.2008.01.016
- Thomas, B.J. and Rothstein, R. (1989) Elevated recombination rates in transcriptionally active DNA. Cell, 56, 619-630. doi:10.1016/0092-8674(89)90584-9
- Zhao, X., Muller, E.G. and Rothstein, R. (1998) A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Molecular Cell, 2, 329-340. doi:10.1016/S1097-2765(00)80277-4
- Morgan, E.A., Shah, N. and Symington, L.S. (2002) The requirement for ATP hydrolysis by Saccharomyces cerevisiae Rad51 is bypassed by mating-type heterozygosity or RAD54 in high copy. Molecular and Cellular Biology, 22, 6336-6343. doi:10.1128/MCB.22.18.6336-6343.2002
- Mardiros, A., Benoun, J.M., Haughton, R., Baxter, K., Kelson, E. P., et al. (2011) Rad10-YFP focus induction in response to UV depends on RAD14 in yeast. Acta Histochemica, 113, 409-415. doi:10.1016/j.acthis.2010.03.005
- Lisby, M., Barlow, J.H., Burgess, R.C. and Rothstein, R. (2004) Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell, 118, 699-713. doi:10.1016/j.cell.2004.08.015
- Chi, P., Van Komen, S., Sehorn, M.G., Sigurdsson, S. and Sung, P. (2006) Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amsterdam), 5, 381-391. doi:10.1016/j.dnarep.2005.11.005
- Li, F., Dong, J., Pan, X., Oum, J.H., Boeke, J.D., et al. (2008) Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Molecular Cell, 30, 325- 335. doi:10.1016/j.molcel.2008.02.028
- Mazon, G., Lam, A.F., Ho, C.K., Kupiec, M. and Symington, L.S. (2012) The Rad1-Rad10 nuclease promotes chromosome translocations between dispersed repeats. Nature Structural & Molecular Biology, 19, 964-971. doi:10.1038/nsmb.2359
- Fung, C.W., Fortin, G.S., Peterson, S.E. and Symington, L.S. (2006) The rad51-K191R ATPase-defective mutant is impaired for presynaptic filament formation. Molecular and Cellular Biology, 26, 9544-9554. doi:10.1128/MCB.00599-06
- Pfander, B., Moldovan, G.L., Sacher, M., Hoege, C. and Jentsch, S. (2005) SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature, 436, 428-433. doi:10.1038/nature03665
- Rockmill, B., Fung, J.C., Branda, S.S. and Roeder, G.S. (2003) The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Current Biology, 13, 1954- 1962. doi:10.1016/j.cub.2003.10.059
- Watt, P.M., Louis, E.J., Borts, R.H. and Hickson, I.D. (1995) Sgs1: A eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell, 81, 253-260. doi:10.1016/0092-8674(95)90335-6
- Yeung, M. and Durocher, D. (2011) Srs2 enables checkpoint recovery by promoting disassembly of DNA damage foci from chromatin. DNA Repair (Amsterdam), 10, 1213-1222. doi:10.1016/j.dnarep.2011.09.005
- Bugreev, D.V., Yu, X., Egelman, E.H. and Mazin, A.V. (2007) Novel proand anti-recombination activities of the Bloom’s syndrome helicase. Genes & Development, 21, 3085-3094. doi:10.1101/gad.1609007
- Ira, G., Malkova, A., Liberi, G., Foiani, M. and Haber, J. E. (2003) Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell, 115, 401-411. doi:10.1016/S0092-8674(03)00886-9

