Geomaterials
Vol. 2 No. 4 (2012) , Article ID: 24012 , 4 pages DOI:10.4236/gm.2012.24012
Optimization of Chromate Reduction by Whole Cells of Arthrobacter sp. SUK 1205 Isolated from Metalliferous Chromite Mine Environment
Microbiology Laboratory, Department of Botany, University of Calcutta, Kolkata, India
Email: dey1919@gmail.com, amalk_paul@yahoo.co.in
Received September 11, 2012; revised October 17, 2012; accepted October 25, 2012
Keywords: Arthrobacter sp.; Chromate Reduction; Chromite Mine Overburden; Detoxification; Hexavalent Chromium; Metal Resistance; Bioremediation; Environmental Pollution
ABSTRACT
Arthrobacter sp. SUK 1205 isolated from metalliferous chromite mine environment of Orissa, India showed wide degree of tolerance to heavy metals including Cr(VI), variety of antibiotics and was also capable of reducing Cr(VI) during growth. Freshly grown whole cells of this bacterium were evaluated for chromate reduction under batch culture using Vogel Bonner (V. B.) broth as the base. Cells of SUK 1205 were capable of completely reducing 100 µM Cr(VI) in V. B. broth within 48 h of incubation. Reduction of chromate increased with increase in cell density which attained maximum at 1010 cells/ml, however, reverse was the phenomenon when the concentration of Cr(VI) increased gradually. Glycerol, glycine and glucose promoted chromate reduction efficiency of the cells when used as electron donors. Optimum pH and temperature were found to be 7.0 and 35˚C respectively. The process of reduction was inhibited by Ni(II), Mn(II), Zn(II) and Co(II), but Cu(II) and Fe(III) was promotive in nature. On the other hand, 2,4-dinitrophenol was found to be neither promotive nor inhibitory for the reduction process, but carbonyl cyanide-m-chloro phenyl hydrazone, sodium azide, sodium fluoride and N,N,-dicyclohexyl carboiimide were inhibitory. Cells of SUK 1205 when permeabilized with toluene, triton X-100 and tween 80 showed an enhancement of the process and thereby indicated that reduction of Cr(VI) was mainly associated with soluble component of the cells. Arthrobacter sp. SUK 1205, therefore, showed great promise for use in Cr(VI) detoxification under a wide range of environmental conditions.
1. Introduction
Environmental pollution of chromium due to industrial operations such as metallurgical, refractory and chemical manufacturing is common occurrence. In addition, weathering and leaching of chromium from overburdens dumped in chromite mining areas along with accumulation of seepage water in quarries also play a significant role in contaminating the environment. In most cases, chromium is represented by both trivalent [Cr(III)] and hexavalent [Cr(VI)] forms as they are the most stable oxidation states. Mobilization of Cr(III) is slow unless dissolved in acidic environment or complexed by organic compounds [1] and is less bioavailable in natural environment, whereas Cr(VI) is highly toxic, carcinogenic and mutagenic [2] due to its high degree of solubility and membrane permeability leading to oxidative stress, DNA damage and altered gene expression.
Bioreduction of toxic Cr(VI) to less toxic Cr(III) and its precipitation in aquatic environment is considered as a cost effective and eco-friendly strategy for treatment of Cr(VI) contaminated wastes in contrast to the traditional physico-chemical treatment process [3], which are not environment friendly. A wide variety of indigenous microbial cultures as well as microbial consortium have been tested for reduction of Cr(VI) under both aerobic [4, 5] and anaerobic conditions [6] and proved to effective for environment management.
Members of the genus Arthrobacter capable of surviving in various chromium contaminated industrial areas such as tannery, chromite mining area and Department of Energy (DOE) waste sites have been explored for their chromate reducing potential by several authors [4,7-10]. These isolates are able to reduce chromium during growth, by whole cells and also by cell-free extracts. Camargo et al., [10] have reported the chromate reducing efficiency of Arthrobacter crystallopoites ES 32 during growth. Similarly Arthrobacter sp. in a consortium was able to reduce nearly 94.3% of 100 mg/l Cr(VI) in 24 h of incubation [7]. On the other hand, Asianti et al., [9] and Meghraj et al., [4] have demonstrated that Arthrobacter strains were able to reduce nearly 35 and 30 µg/ml of Cr(VI) in 10 day and 46 h respectively.
During the course of our survey for bacterial strains capable of tolerating and reducing high concentration of Cr(VI) from metalliferous chromite mine environments, we have isolated an efficient chromite reducing bacterium, Arthrobacter sp. SUK 1205 (MTCC 8731) from overburdens of Sukinda, Orissa, India. The strain has been shown to reduce 64% of initial 2 mM Cr(VI) [11]. The present study confirmed the taxonomic identity, phyllogenetic analysis of the strain and optimized the cultural conditions for Cr(VI) reduction by whole cells of the strain under batch culture.
2. Materials and Methods
2.1. Source and Maintenance of Bacterial Isolate
Chromate reducing bacterial isolate SUK 1205 (MTCC 8731) was isolated from metalliferous chromite mine overburden samples collected from chromite mining environment of Orissa, India. The strain was grown on slopes of peptone yeast-extract and glucose (PYEG) agar medium [12] supplemented with 2 mM Cr(VI) and maintained at 4˚C after 48 h growth in the same medium.
2.2. Phyllogenetic Analysis of the Strain
While the morphological, physiological and biochemical characteristics along with tentative identity of the isolate has been reported in Dey and Paul [11], the identity of the isolate SUK 1205 was confirmed based on 16S rDNA analysis. The DNA was isolated and purified by phenol/ chloroform extraction and precipitated by adding 3 M potassium acetate and isopropanol.
PCR amplification was performed using the 8 F (5’- AGAGTTTGATCCTGGCTCAG-3’) and 1492 R (5’- TACGGYTACCTTGTTACGACTT-3’) as forward and reverse primers respectively. Reactions were carried out using BDT v 3.1 cycle sequencing kit. The reaction mixture consisted of 2 µl BDT v 3.1 (ABI, cat #4337455), 1 µl sequencing buffer (ABI, cat #4336697), 2 µl primer (@ 4 µM), 4 µl template and 1 µl PCR water. The 16S rRNA gene was amplified using a 26 cycle PCR (96˚C, for 10 sec; annealing temperature, 55˚C, for 5 sec; extension temperature, 60˚C for 4 min) and hold at 4˚C. The PCR amplification products were analyzed by electrophoresis on a 1% agarose gel and purified. DNA sequencing was performed using the dideoxy chain termination method with an ABI 3730 × 1 Genetic Analyzer. A consensus sequence of 801 b.p. of 16S rDNA gene was generated from forward and reverse sequence data using aligner software. The 16S rDNA gene sequence was analyzed using BLAST programme with NCBI GenBank database. Based on maximum indentity score, first ten sequences were selected and aligned using multiple alignment software program Clustal W. Distance matrix was generated using RDP database and the phylogenetic tree was constructed using MEGA 4. The sequence was deposited at NCBI Gen Bank with accession No. JQ 312666.
2.3. Heavy Metal Tolerance
Heavy metal tolerance of the isolate SUK 1205 was evaluated by broth dilution method [13]. The Vogel Bonner (V. B. broth) supplemented with increasing concentration of heavy metal was inoculated with overnight grown culture and incubated at 35˚C for 24 - 48 h under continuous shaking (120 rpm). Optical density was recorded at 540 nm using uninoculated broth as control. Minimum inhibitory concentration (MIC) of the metal was determined as the lowest concentration responsible for complete inhibition of growth of the bacterium.
2.4. Antibiotic Susceptibility
Susceptibility of the isolate to different antibiotics was evaluated following standard disc-diffusion method. Antibiotic impregnated discs (6 mm dia. HIMEDIA) were placed on freshly prepared lawns of the isolate on PYEG agar medium and incubated at 35˚C for 24 h. The diameter of inhibition zone against each of the antibiotic was measured to nearest mm and isolates were identified as sensitive, resistant and intermediate following standard antibiotic sensitivity testing method [14]. Disc containing the following antibiotics were used: streptomycin (25 mg/disc), tetracycline (30 mg/disc), neomycin (30 mg/ disc), kanamycin (30 mg/disc), chloramphenicol (30 mg/ disc), doxycycline (30 mg/disc), ampicillin (10 mg/disc), polymixin B (50 units/disc), penicillin G (10 units/disc), erythromycin (15 mg/disc), methicilin (5 mg/disc), nalidixic acid (30 mg/disc), gentamycin (10 mg/disc), rifamcipin (30 mg/disc), netilin (30 mg/disc), novobiocin (30 mg/disc) and norfloxacin (30 mg/disc).
2.5. Reduction of Cr(VI) by Whole Cells
The isolate SUK 1205 was grown in PYEG medium at 35˚C under continuous shaking for 24 h and cells were harvested by centrifugation (10,000 × g) for 10 min at 4˚C. The cell pellet was thoroughly washed with sterile Tris HCl buffer (pH 7.0) and resuspended in the same buffer. The reduction of Cr(VI) by suspended whole cells was carried out in V. B. broth (25 ml/100ml flask) supplemented with 100 µM Cr(VI) and the cell density was adjusted at 109 cells/ml. The flasks were incubated at 35˚C under continuous shaking (120 rpm). Samples were withdrawn aseptically and analyzed for residual Cr(VI) following standard diphenyl carbazide method [15].
2.6. Reduction by Permeabilized Cells
To obtain permeabilized cells, overnight grown cultures were harvested (centrifugation at 10000 × g for 10 min), and washed with Tris-HCl buffer (pH 7.0) and suspended in the same buffer. Toluene, Triton X100 and Tween 80 were added to the cell suspension at a desired concentration and vortexed for 15 min to permeabilized the cells. Chromate reduction assay with these permeabilized cells was performed in the same way as with untreated whole cells as describe before.
Each experiment was performed in triplicates and the mean of triplicate readings ± Standard Error were represented.
3. Results and Discussion
3.1. Phyllogenetic Analysis of the Strain
The chromium reducing Gram-positive bacterial isolate SUK 1205 was obtained from chromite mine overburden samples of Sukinda Valley, Orissa, India. It showed characteristic rod to cocci cell cycle during growth, identified as Arthrobacter sp. based on phenotypic characteristics [11] and deposited to Microbial Type Culture Collection Institute of Microbial Technology, Chandigarh (MTCC 8731). The identity of the isolate was further confirmed by 16S rDNA analysis. The consensus sequence of 801 b.p. for 16S rDNA of the isolate SUK 1205 was generated to carry out BLAST with database of NCBI GenBank and confirmed as Arthrobacter sp. having 98.0% similarity with Arthrobacter sp. 3 - 4 A [16]. A phyllogenetic tree has been drawn (Figure 1) using Neighbour-joining programme in MEGA 4 software between the reported Arthrobacter species and the present isolate. The nucleotide sequence has been deposited to GenBank with an accession number JQ 312666. While a number of species of Arthrobacter have been documented to survive in diverse metal stressed environments [7,9] and exhibited the exceptional property of detoxifying the hexavalent chromium by reducing it to non-toxic Cr(III) [4,10,17], Arthrobacter 3 - 4 A reported by O’Niell et al., [16] from Anthrosol of Brazil was capable of surviving at higher pH, phosphorus and calcium contents, but was not reported to reduce hexavalent chromium.
3.2. Heavy Metal Tolerance
Arthrobacter sp. SUK 1205 was also screened for its tolerance to chromium along with other heavy metals like Ni(II), Fe(III), Cu(II), Co(II), Mn(II), Zn(II), Cd(II) and Hg(II). It showed a high degree of tolerance to chromium (MIC 11.8 mM) like other species of Arthrobacter [4,8,17] where as MICs for Ni(II), Fe(III), Cu(II), Co(II) and Mn(II) were 7.8, 6.0, 4.8, 4.2 and 3.1 mM respectively. However, the isolate was comparatively sensitive to Zn(II) (MIC 2.8 mM) and Cd (MIC 2.1 mM). Mercury, on the other hand was most toxic for the isolate showing a MIC value as low as 0.001 mM (Table 1). The high degree of tolerance to hexavalent chromium by Arthrobacter SUK 1205, could be attributed to its long exposure as well as adaptation in highly Cr polluted environment. In addition, the isolate also showed considerable tolerance to metals like Ni(II), Fe(III), Cu(II) and Co(II), the most common constituents of chromite mine overburden.
3.3. Antibiotic Sensitivity
Since metal resistance of bacteria is often linked with resistance to antibiotics, the antibiotic sensitivity profile of the isolate SUK 1205 was determined by disc-diffusion
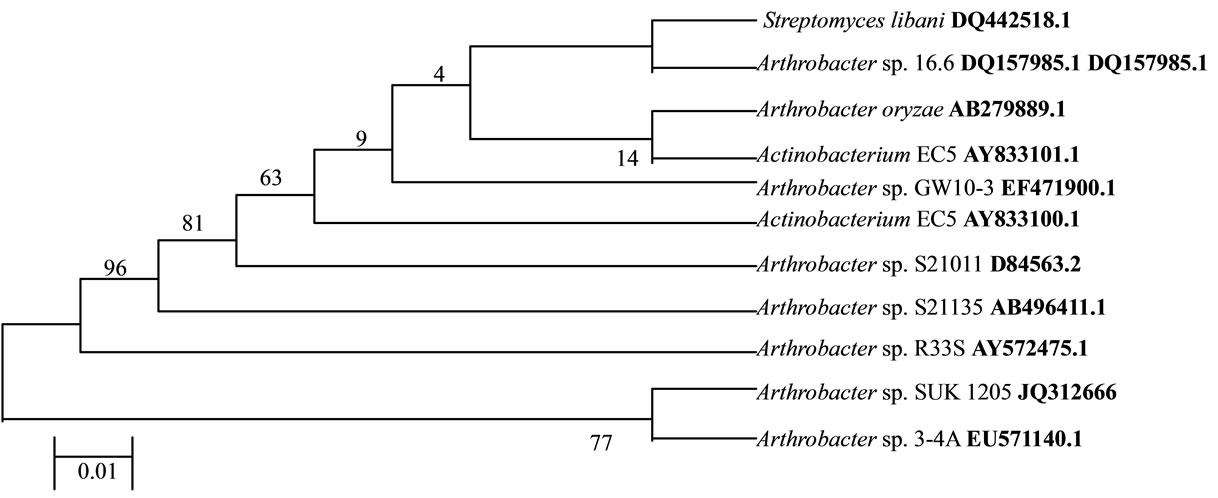
Figure 1. Phyllogenetic tree derived from 16S rRNA gene sequence of SUK 1205. The evolutionary history was inferred using the Neighbor-Joining method. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of base substitutions per site.
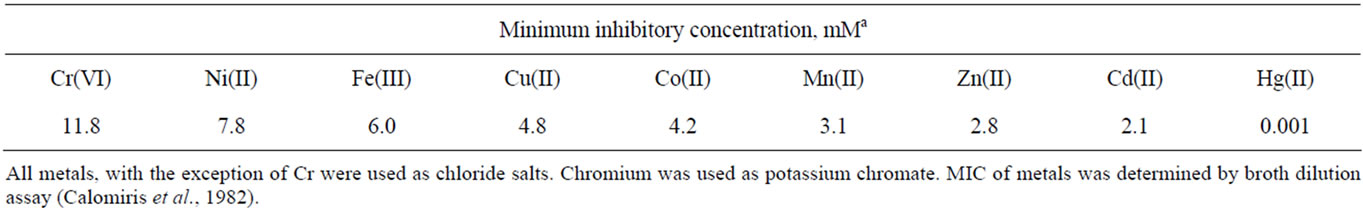
Table 1. Heavy metal tolerance profiles of bacterial isolate SUK 1205.
method. This clearly indicated that out of 17 different antibiotics tested, the isolate was resistant to at least 11 antibiotics including penicillin G, methicilin, ampicilin, doxycycline, polymyxin B, erythromycin, neomycin, norfloxacin, rifampicin, netilin and novobiocin (Table 2). However, it was sensitive to chroramphenicol, gentamycin and streptomycin. Response of the isolate to tetracycline, kanamycin and nalidixic acid was of intermediate nature.
3.4. Reduction by Whole Cells
Time course of chromate reduction by whole cells of Arthrobacter sp. SUK 1205 was determined under batch culture in V. B. broth containing 100 µM Cr(VI) and an initial cell density of 109 cells/ml. Complete reduction of 100 µM Cr(VI) was achieved in 48 h of incubation at 35˚C under continuous shaking (120 rpm). Reduction was accomplished by gradual discolouration of the medium, however, no significant increase in cell number/ ml of medium was recorded during the course of reduction process (Figure 2). The inability of the cells to grow during chromate reduction could be attributed to the poor nutritional status of the V. B. broth as well as the inhibitory effect of toxic hexavalent chromium.
3.5. Effect of Cell Density
The initial cell density of the reduction medium greatly influenced the Cr(VI) reduction by the isolate SUK 1205. Cr(VI) reduction increased proportionally with increase in cell density ranging from 106 to 1010 cells/ml. At the highest cell density (1010 cells/ml), reduction of 100 µM Cr(VI) was completed in 30 h, but at low cell concentration (106 cells/ml) only some 20% of the chromate was reduced in 48 h (Figure 3).
Freshly grown viable whole cells of the isolate SUK 1205 was capable of completely reducing 100 µM Cr(VI) in V. B. broth within 48 h of incubation. Such stimulation of Cr(VI) reduction process by increase in cell density has also been established with Microbacterium [18] and Bacillus sphaericus AND 303 [19].
3.6. Effect of Cr(VI) Concentration
Chromate reduction by suspended cells of bacteria has been demonstrated to be influenced by the initial Cr(VI) concentration [12,18,20,21] of the reduction medium.
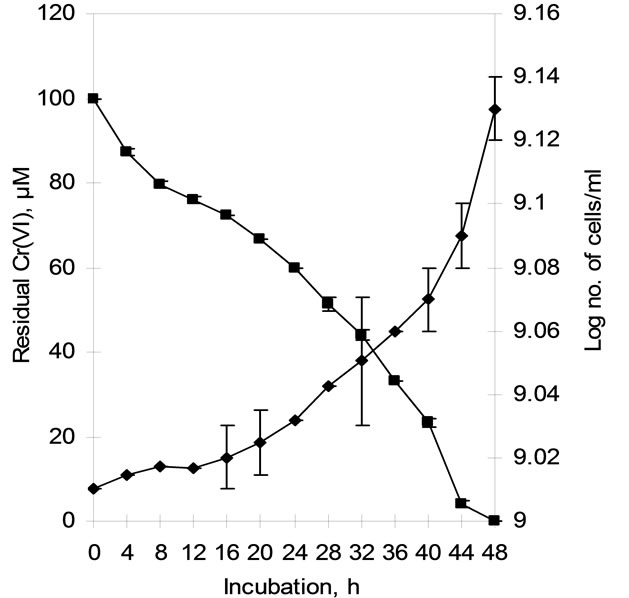
Figure 2. Time course of hexavalent chromium reduction by whole cells of isolate Arthrobacter sp. SUK 1205 in Vogel Bonner broth under batch culture (-♦- cell count, -■- Residual hexavalent chromium).
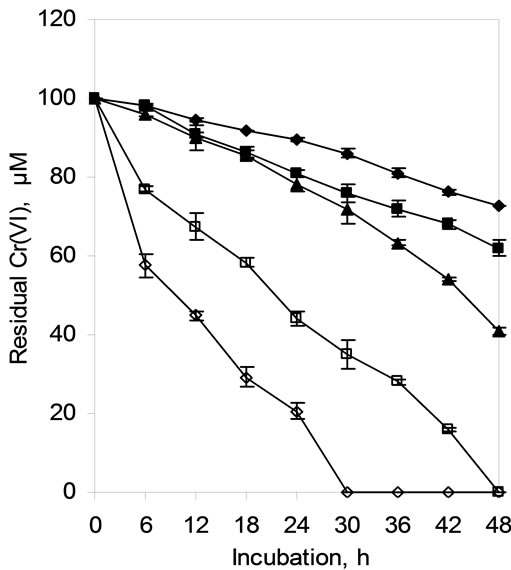
Figure 3. Hexavalent chromium reduction by cells of Arthrobacter sp. SUK 1205 as influenced by initial cell density (-♦- 106, -■- 107, -▲- 108 -□- 109, -◊- 1010 cells/ml).
Chromium reducing ability of the whole cells of SUK 1205 was monitored at Cr(VI) concentrations ranging from 50 to 800 µM and presented in Figure 4. Concentration of
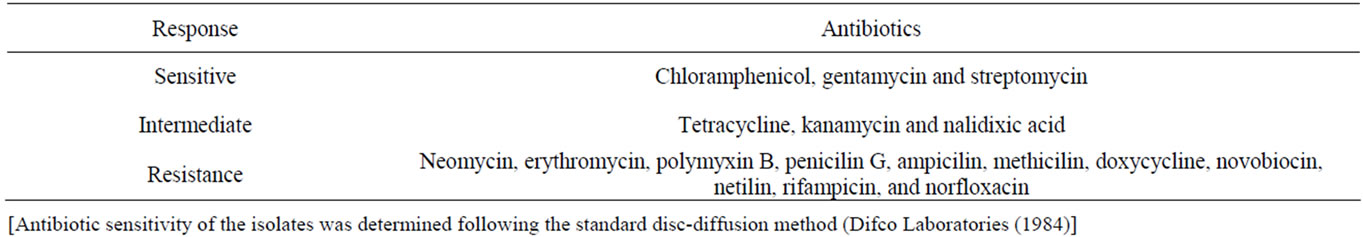
Table 2. Antibiotic sensitivity profile of the bacterial isolate SUK 1205.
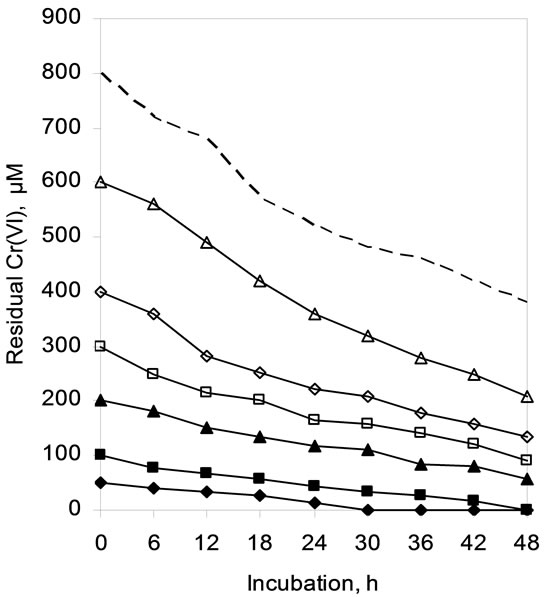
Figure 4. Hexavalent chromium reduction by whole cells of Arthrobacter sp. SUK 1205 as influenced by concentration of Cr(VI) (-♦- 50, -■- 100, -▲- 200, -□- 300, -◊- 400, -∆- 600, --- 800 µM).
Cr(VI) exceeding 100 µM could not be completely reduced in 48 h under the experimental conditions, about 50% of total Cr(VI) was reduced at a concentration of 800 µM (Figure 4). The extension of incubation period for complete reduction of Cr(VI) with increase in initial Cr(VI) concentration during present study is in well conformity with that of Meghraj et al., [4] with Arthrobacter sp.
3.7. Effect of Electron Donor
Chromate reducing organisms in general utilize a variety of organic compounds as electron donors for Cr(VI) reduction [22,23]. To facilitate the chromate reduction by whole cells of Arthrobacter sp. SUK 1205, organic substances such as acetate, benzoate, propionate, glucose, sucrose, glycerol, glycine, peptone, tryptone and yeast extract were added to the reduction medium at 0.1% level. Reduction of initial 100 µM Cr(VI) was completed within 48 h of incubation when glycerol, glucose and glycine was used individually as electron donor, whereas propionate, benzoate and sucrose were the least efficient as electron donors for Cr(VI) reduction (Figure 5). Marbrouk [24] also reported that peptone and yeast extract favoured chromate reduction by Streptomyces sp. MS-2. Ochrobactrum sp. strain CSCr-3 [20] and Bacillus cereus [25] were also found to utilize glucose as electron donor.
3.8. Effect of pH and Temperature
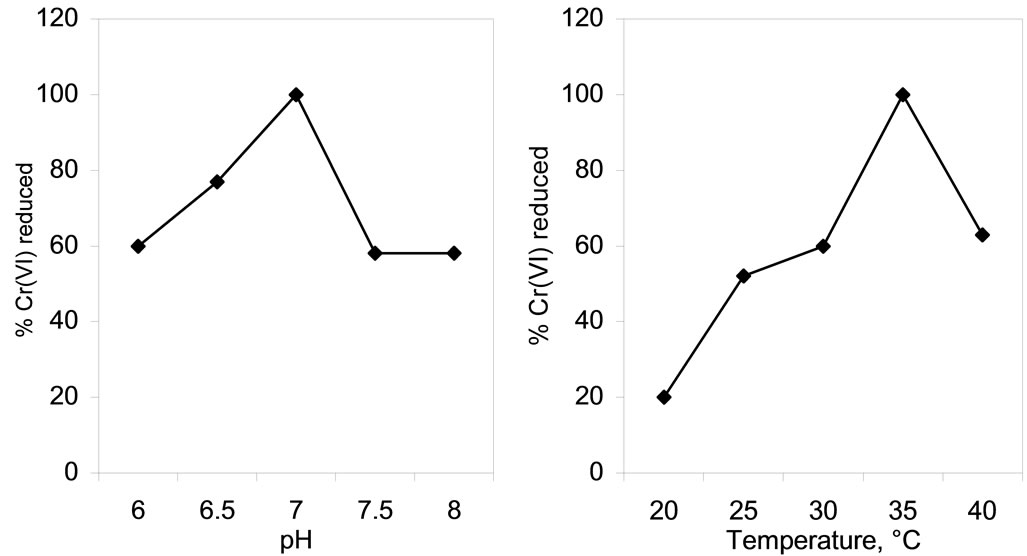
Both pH as well as temperature of the reduction medium were found to affect the chromate reducing potential of the isolate SUK 1205 (Figures 6(a) and (b)) as these two factors essentially interfere with metabolic activities of the cells. The optimum pH and temperature for Cr(VI) reduction were 7.0 (Figure 6(a)) and 35˚C (Figure 6(b)) respectively. On either side of the pH and temperature scale, the reduction process was negatively affected. Optimum temperature for chromate reduction was found to be 37˚C with Ochrobactrum intermedium Rb-2 [26] and Ochrobactrum intermedium SDCr-5 [27]. Deviation of these factors from their optimum conditions also altered chromate reductase activity possibly by altering the conformation of the enzyme. Farrell and Ranallo [28], postulated that the pH of the reaction medium affects the degree of ionization of the enzyme and changes in the protein conformation.
3.9. Effect of Metal Ions
Chromate reduction by whole cells of SUK 1205 was severely affected in presence of different heavy metals such as Ni(II), Zn(II), Mn(II) and Co(II). This could be explained by the possible metal toxicity and slowing down or inhibition of the Cr(VI) reduction process [29]. Cr(VI) reducing capability of the isolate was enhanced when Cu(II) and Fe(III) was supplemented in the reduction medium. Complete reduction of 100 µM Cr(VI) occurred within 24 h of incubation (Figure 7). Such stimulatory effect of Cu(II) on chromate reductase activity of SUK 1205 cells was probably due to the fact that it is a prosthetic group for many enzymes and acts as an electron redox centre and help in the shuttle of electron between different subunits [3]. Similar enhancement of Cr(VI) reduction ability was also observed with Ochrobactrum sp. CSCr-3 [20], Amphibacillus sp. KSUCr3 [30] and Bacillus sp. KSUCr9a [31].
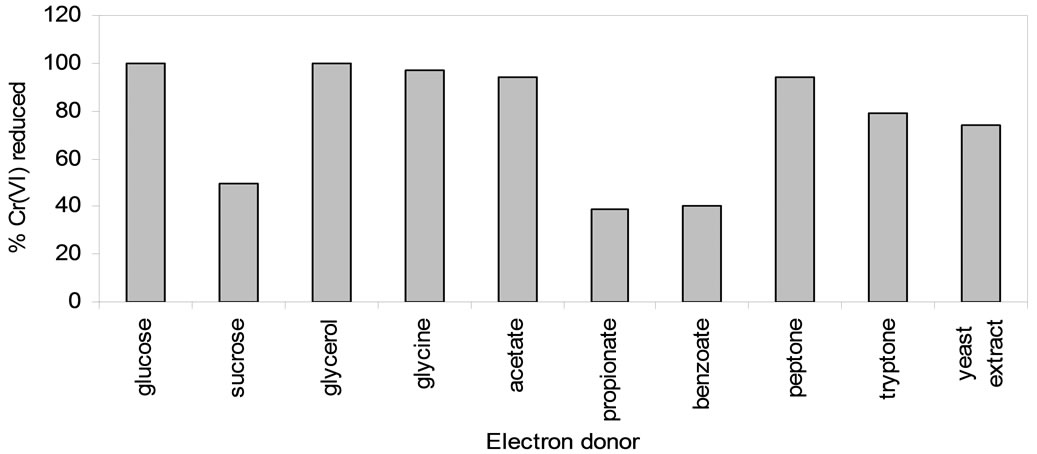
Figure 5. Effect of electron donor on Cr(VI) reduction by whole cells of Arthrobacter sp. SUK 1205.
 (a) (b)
(a) (b)
Figure 6. Effect of pH (a) and temperature (b) on chromate reduction by whole cells of Arthrobacter sp. SUK 1205.
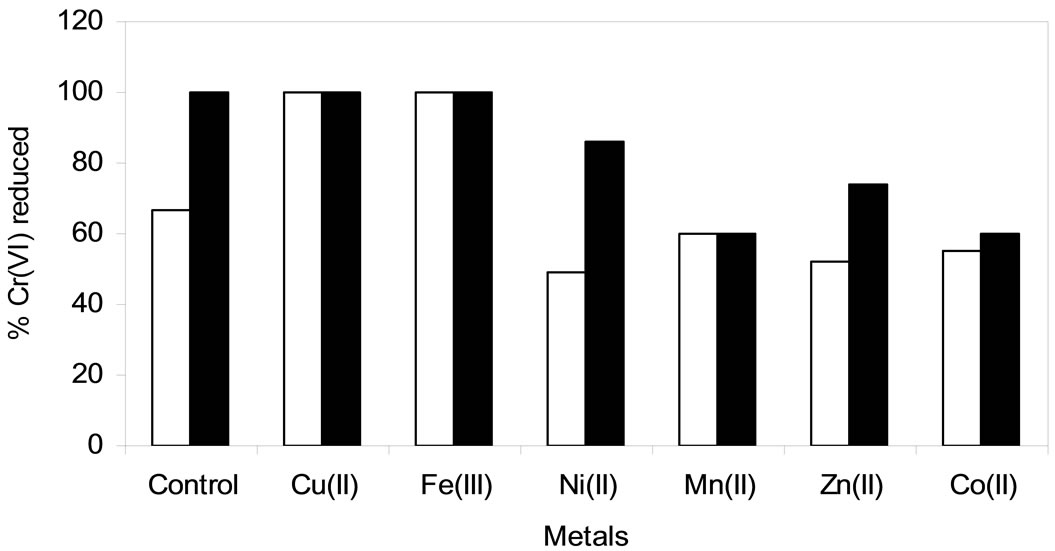
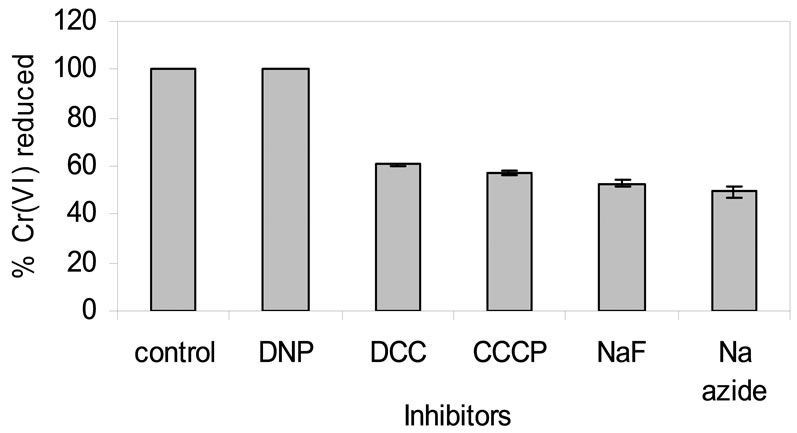
Figure 8. Effect of inhibitor on chromate reduction by whole cells of isolate Arthrobacter sp. SUK 1205. [DNP = 2,4-Dinitrophenol, DCC = N,N,-Dicyclohexyl carboiimide; NaN3 = Sodium azide; NaF = Sodium fluoride; CCCP = Carbonyl cyanidem-chloro phenyl hydrazone].
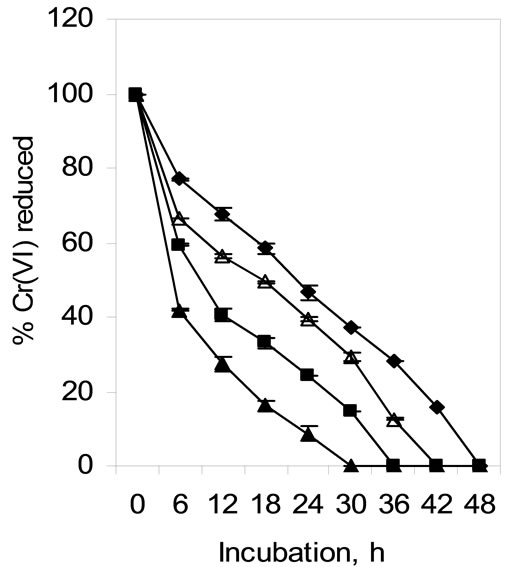
Figure 9. Hexavalent chromium reduction by permeabilized whole cells of isolate Arthrobacter sp. SUK 1205 (-♦- control, -■- triton, -▲- toluene, -∆- tween 80).
3.10. Effect of Inhibitor
Effect of 5 different inhibitors such as sodium azide (NaN3), sodium fluoride (NaF), 2, 4-Dinitrophenol (DNP), carbonyl cyanide-m-cholophenyl hydrazone (CCCP) and N,N,-Di-cyclohexyl carboiimide (DCC) was used at equimolecular concentration to evaluate their influence on chromate reduction. Reduction of Cr(VI) in presence of DNP was almost parallel with the control, whereas sodium azide was most inhibitory showing only 49% Cr(VI) reduction (Figure 8). Cr(VI) reduction by isolate Arthrobacter sp. SUK 1205 was not inhibited by DNP as it is an uncoupler and might have accelerated the respiratory chain linked electron transport mechanism [32]. Enhancement of Cr(VI) reduction by DNP has also been reported in Burkholderia cepacia [32] and Staphylococcus gallinarum [21]. But, sodium azide, sodiu fluorideCCCP and DCC inhibited the process of reduction as they are known to inhibit the activity of cytochrome oxidase, enolase [33], disrupts chemiosmotic gradient and inhibits the ATPase activity.
3.11. Effect of Permeabilized Cells on Reduction
Freshly grown cells of Arthrobacter sp. SUK 1205 were permeabilized in presence of triton, toluene and tween 80 and used for chromate reduction studies. Efficient reduction of hexavalent was achieved with toluene (in 30 h), followed by triton (in 36 h) and tween 80 (in 42 h) treated cells (Figure 9), which might indicate that the Cr(VI) reduction is mediated by soluble protein of the cell [34]. Similar enhancement in Cr(VI) reduction rate was observed with Providencia sp. [35].
4. Conclusion
Optimization of conditions for Cr(VI) reduction by whole cells of Arthrobacter SUK 1205 established its biotechnological potential for transformation of highly toxic and mutagenic Cr(VI) to less toxic Cr(III) and thus could be an effective tool in detoxification of chromium pollutants. The isolate also showed wide range of tolerance to different heavy metals and antibiotics supporting its application for Cr(VI) bioremediation in metal contaminated areas.
5. Acknowledgements
The authors acknowledge the financial support received from the Department of Biotechnology, Ministry of Science and Technology, Government of India vide Sanction number BT/PR/5766/NDB/51/061/2005. The authors are grateful to Tata Iron and Steel Company (TISCO) and Orissa Mining Corporation (OMC) and Ferro Alloys Corporation Limited (FACOR) for providing permission and logistic support in sample collection.
REFERENCES
- D. Rai, B. M. Sass and D. A. Moore, “Chromium (III) Hydrolysis Constants and Solubility of Chromium (III) Hydroxide,” Inorganic Chemistry, Vol. 26, No. 3, 1987, pp. 345-349. doi:10.1021/ic00250a002
- D. Bagchi, S. J. Stohs, W. O. Bernard, M. Bagchi and H. G. Preus, “Cytotoxicity and Oxidative Mechanism of Different Forms of Chromium,” Toxicology, Vol. 180, 2002, pp. 5-22. doi:10.1016/S0300-483X(02)00378-5
- F. A. O. Camargo, B. C. Okeke, F. M. Bento and W. T. Frankenberger, “In Vitro Reduction of Hexavalent Chromium by a Cell-Free Extract of Bacillus sp. ES 29 Stimulated by Cu2+,” Applied Microbiology and Biotechnology, Vol. 62, No. 5-6, 2003, pp. 569-573. doi:10.1007/s00253-003-1291-x
- M. Megharaj, S. Avudainayagam and R. Naidu, “Toxicity of Hexavalent Chromium and Its Reduction by Bacteria Isolated from Soil Contaminated with Tannery Waste,” Current Microbiology, Vol. 47, No. 1, 2003, pp. 51-54. doi:10.1007/s00284-002-3889-0
- U. Thacker, R. Parikh, Y. Shouche and D. Madamwar, “Reduction of Chromate by Cell-Free Extract of Brucella sp. Isolated from Cr(VI) Contaminated Sites,” Bioresource Technology, Vol. 98, No. 8, 2007, pp. 1541-1547. doi:10.1016/j.biortech.2006.06.011
- C. Cervantes, J. Campos-Garcia, S. Devars, F. G. Corona, H. Loza-Tavera, J. Carlos, T. Guzman and R. M. Sanchez, “Interactions of Chromium with Microorganisms and Plants,” FEMS Microbiology Reviews, Vol. 25, No. 3, 2001, pp. 335-347. doi:10.1111/j.1574-6976.2001.tb00581.x
- P. E. Molokwane, C. K. Meli and M. N. E. Chirwa, “Chromium (VI) Reduction in Activated Sludge Bacteria Exposed to High Chromium Loading: Brits Culture (South Africa),” Water Research, Vol. 42, No. 17, 2008, pp. 4538-4548. doi:10.1016/j.watres.2008.07.040
- R. N. Horton, W. A. Apel, V. S. Thompson and P. P. Sheridan, “Low Temperature Reduction of Hexavalent Chromium by a Microbial Enrichment Consortium and a Novel Strain of Arthrobacter aurescens,” BMC Microbiology, Vol. 6, 2006, p. 6.
- N. V. Asianti, M. K. Abuladze, T. M. Kartvelishvili, N. G. Bakradze, N. A. Sapojnikova, N. Y. Tsibakhashvili, L. V. Tabatadze, L. L. Asanishvili and H. Holman, “Effect of Chromium (VI) Action on Arthrobacter oxydans,” Current Microbiology, Vol. 49, No. 5, 2004, pp. 321-326. doi:10.1007/s00284-004-4351-2
- F. A. O. Camargo, F. M. Bento, B. C. Okek and W. T. Frankenberger, “Hexavalent Chromium Reduction by an Actinomycetes, Arthrobacter crystallopoites ES 32,” Biological Trace Element Research, Vol. 97, No. 2, 2004, pp. 183-194. doi:10.1385/BTER:97:2:183
- S. Dey and A. K. Paul, “Hexavalent Chromium Reduction by Aerobic Heterotrophic Bacteria Indigenous to Chromite Mine Overburden,” Brazilian Journal of Microbiolgy, 2012, in Press.
- Y. T. Wang and C. Xiao, “Factors Affecting Hexavalent Chromium Reduction in Pure Cultures of Bacteria,” Water Research, Vol. 29, No. 11, 1995, pp. 2467-2474. doi:10.1016/0043-1354(95)00093-Z
- J. J. Calomiris, T. L. Armstrong and R. J. Seidler, “Association of Metal-Tolerance with Multiple Antibiotic Resistance of Bacteria Isolated from Drinking Water,” Applied and Environmental Microbiology, Vol. 47, No. 6, 1984, pp. 1238-1242.
- Difco Laboratories, “DIFCO Manual: Dehydrated Culture Media and Reagents for Microbiology,” 10th Edition, DIFCO Laboratories Inc., Detroit, 1984.
- C. H. Park, B. Keyhan, B. Wielinga, S. Fendorf and A. Matin, “Purification to Homogeneity and Characterization of a Novel Pseudomonas putida Chromate Reductase,” Applied and Environmental Microbiology, Vol. 66, No. 5, 2000, pp. 1788-1795. doi:10.1128/AEM.66.5.1788-1795.2000
- B. O’Neill, J. Grossman, M. T. Tsai, J. E. Gomes, J. Lehmann, J. Peterson, E. Neves and J. E. Thies, “Bacterial Community Composition in Brazilian Anthrosols and Adjacent Soils Characterized Using Culturing and Molecular Identification,” Microbial Ecology, Vol. 58, No. 1, 2009, pp. 23-35. doi:10.1007/s00248-009-9515-y
- S. Dey and A. K. Paul, “Optimization of Cultural Conditions for Growth Associated Chromate Reduction by Arthrobacter sp. SUK 1201 Isolated from Chromite Mine Overburden,” Journal of Hazardous Materials, Vol. 213-214, 2012, pp. 200-206. doi:10.1016/j.jhazmat.2012.01.078
- P. Pattanapipitpaisal, N. L. Brown and L. E. Macaskie, “Chromate reduction by Microbacterium liquefaciens Immobilized in Polyvinyl Alcohol,” Biotechnology Letters, Vol. 23, No. 1, 2001, pp. 61-65. doi:10.1023/A:1026750810580
- A. Pal and A. K. Paul, “Aerobic Chromate Reduction by Chromium-Resistant Bacteria Isolated from Serpentine Soil,” Microbiological Research, Vol. 159, No. 4, 2004, pp. 347-354. doi:10.1016/j.micres.2004.08.001
- Z. He, F. Gao, T. Sha, Y. Hu and C. He, “Isolation and Characterization of a Cr(VI)-Reduction Ochrobactrum sp. Strain CSCr-3 from Chromium Landfill,” Journal of Hazardous Materials, Vol. 163, No. 2-3, 2009, pp. 869-873. doi:10.1016/j.jhazmat.2008.07.041
- M. Z. Alam and S. Ahmad, “Toxic Chromate Reduction by Resistant and Sensitive Bacteria Isolated from Tannery Effluent Contaminated Soil,” Annals of Microbiology, Vol. 62, No. 1, 2012, pp. 113-121. doi:10.1007/s13213-011-0235-4
- L. Philip and C. Venkobachar, “Cr(VI) Reduction by Bacillus coagulans Isolated from Contaminated Soils,” Journal of Environmental Engineering, Vol. 124, No. 12, 1998, pp. 1165-1170. doi:10.1061/(ASCE)0733-9372(1998)124:12(1165)
- L. G. Liu, W. H. Xu and G. M. Zeng, “Experimental Study on Reduction by Pseudomonas aeruginosa,” Journal of Environment and Science, Vol. 16, No. 5, 2004, pp. 795-801.
- M. E. M. Mabrouk, “Statistical Optimization of Medium Components for Chromate Reduction by Halophilic Streptomyces sp. MS-2,” African Journal of Microbiology Research, Vol. 2, No. 5, 2008, pp. 103-109.
- Y. T. Wang and H. Shen, “Bacterial Reduction of Hexavalent Chromium,” Journal of Industrial Microbiology, Vol. 14, No. 2, 1995, pp. 159-163. doi:10.1007/BF01569898
- B. Rida, K. Yrjälä and S. Hasnain, “Hexavalent Chromium Reduction by Bacteria from Tannery Effluent,” Journal of Microbiology and Biotechnology, Vol. 22, No. 4, 2012, pp. 547-554. doi:10.4014/jmb.1108.08029
- S. Sultan and S. Hasnain, “Reduction of Toxic Hexavalent Chromium by Ochrobactrum intermedium Strain SDCr-5 Stimulated by Heavy Metals,” Bioresource Technology, Vol. 98, No. 2, 2007, pp. 340-344. doi:10.1016/j.biortech.2005.12.025
- S. O. Farrell and R. T. Ranallo, “Experiments in Biochemistry. A Hands-On Approach,” Saunders College Publications, Orlando, 2000.
- J. McLean and T. J. Beveridge, “Chromate Reduction by a Pseudomonad Isolated from a Site Contaminated with Chromated Copper Arsenate,” Applied and Environmental Microbiology, Vol. 67, No. 3, 2001, pp. 1076-1084. doi:10.1128/AEM.67.3.1076-1084.2001
- A. S. S. Ibrahim, M. A. El-Tayeb, Y. B. Elbadawi and A. A. Al-Salamah, “Isolation and Characterization of Novel Potent Cr(VI) Reducing Alkaliphilic Amphibacillus sp. KSUCr3 from Hypersaline Soda Lakes,” Electronic Journal of Biotechnology, Vol. 14, No. 4, 2011.
- A. S. S. Ibrahim, M. A. El-Tayeb, Y. B. Elbadawi and A. A. Al-Salamah, “Bioreduction of Cr (VI) by Potent Novel Chromate Resistant Alkaliphilic Bacillus sp. Strain KSUCr5 Isolated from Hypersaline Soda Lakes,” African Journal of Biotechnology, Vol. 10, No. 16, 2012, pp. 7207-7218.
- R. Wani, K. M. Kodam, K. R. Gawai and P. K. Dhakephalkar, “Chromate Reduction by Burkholderia cepacia MCMB-821 Isolated from the Pristine Habitat of Alkaline Crater Lake,” Applied Microbiology and Biotechnology, Vol. 75, No. 3, 2007, pp. 627-632. doi:10.1007/s00253-007-0862-7
- D. J. Opperman and E. van Heerden, “Aerobic Cr(VI) Reduction by Thermus scotoductus Strain SA-01,” Journal of Applied Microbiology, Vol. 103, No. 5, 2007, pp. 1907-1913. doi:10.1111/j.1365-2672.2007.03429.x
- U. Thacker and D. Madamwar, “Reduction of Toxic Chromium and Partial Localization of Chromium Reductase Activity in Bacterial Isolate DM1,” World Journal of Microbiology and Biotechnology, Vol. 21, No. 6-7, 2005, pp. 891-899. doi:10.1007/s11274-004-6557-7
- U. Thacker, R. Parikh, Y. Shouche and D. Madamwar, “Hexavalent Chromium Reduction by Providencia sp.,” Process Biochemistry, Vol. 41, No. 6, 2006, pp. 1332-1337. doi:10.1016/j.procbio.2006.01.006

