American Journal of Molecular Biology
Vol.4 No.1(2014), Article ID:41627,4 pages DOI:10.4236/ajmb.2014.41003
A method to synthesize cDNA constructs by homology based recombination cloning
Department of Genetics, University of Delhi, South Campus, New Delhi, India
Email: *pburma@south.du.ac.in
Received 11 November 2013; revised 5 December 2013; accepted 30 December 2013
ABSTRACT
We introduce a homology-based recombination approach for generating a cDNA construct. This method depends on amplifying several exon fragments and their fusions by the homology-based recombination. This method provides a way to generate the cDNA sequence of any gene without any need for its mRNA. The paper describes the strategy by assembling cDNA of the MYB1 and MYB2 genes of Arabidopsis thaliana.
Keywords:Homology Based Recombination Cloning; cDNA Assembly
1. INTRODUCTION
In several instances, the cDNA of a gene is required for its expression in heterologous or transgenic systems. Traditionally cDNA is synthesized by using mRNA as a template. In some instances, availability of mRNA is limited due to a narrow time window of expression, tissue specificity or lack of information. In such instances, the cDNA sequence can be assembled by chemical synthesis of DNA or by recursive PCR, followed by its cloning in appropriate vectors. In recent years, Homology Based Recombination (HBR) [1] has been used to assemble recombinant molecules. In the present work, we have used HBR cloning to assemble cDNA fragments. This involves amplification of exon fragments with overlapping homology. Reverse primers used in amplification of exons, are designed to have at least 15 base pair (bp) homology sequence with the adjacent exon fragment. By HBR cloning, the amplified exons of the target gene are linked to each other via the regions of overlap to generate the cDNA sequence which then can be cloned in a suitable plasmid vector using the same strategy.
2. MATERIALS AND METHODS
2.1. Polymerase Chain Reactions
All PCRs were carried out in a total volume of 50 µl with Phusion high fidelity DNA polymerase (Thermo Fisher Scientific, USA) amplification kit. 100 ng of genomic DNA isolated from A. thaliana was used for amplification of the exons.After an initial denaturation at 98˚C for 5 minutes, 30 cycles of denaturation at 98˚C for 30 s, annealing at temperatures according to Tm of primer pairs for 30 s and extension at 72˚C for 1 min was carried out. This was followed by a final extension for 8 min at 72˚C. The PCR product was then size-fractionated on a 1% Agarose gel in TAE buffer. The corresponding band was excised and DNA purified for further assembly. Sequences of primers used in amplification of MYB1 and MYB2 genes are summarized in Tables 1 and 2, respectively.
2.2. Homology Based Recombination Cloning Reaction
After amplification of exons, In-Fusion reactions were performed using In-Fusion® HD Cloning Kit (Clonetech Laboratories, Inc.). The recommended protocol was modified for assembling two exon fragments in a vector (Three-way assembly) and for three exon fragments ina vector (Four-way assembly). In both types of assembly, each exon fragment was used in ten-fold molar excess to the vector. 70 to 100 ng of vector was used in a 20 µl reaction volume consisting of exon fragments, digested vector DNA and 4 µl of Infusion HD Enzyme Premix as supplied in the kit against a recommended use of 2 µl of
Table 2. MYB2 primers.
the premix per reaction. The mix was incubated at 37˚C for 15 min followed by incubationat 50˚C for 15 min. 5 µl of the reaction mixture was used for transformation of E. coli.
3. RESULTS AND DISCUSSION
We present examples of developing cDNA constructs for two genes of Arabidopsis thaliana.
3.1. cDNA Assembly of MYB1 Gene from A. thaliana
The transcribed region of the MYB1 gene (Acc. no. NC_003074), is 1.9 Kb long and carries a single intron of 494 bp. The aim was to assemble the cDNA and clone it downstream to a CaMV35S promoter available in the vector pRT101 [2]. In order to amplify the exons, two sets of primers were designed. Primer sets 1, 2 and 3, 4 (Table 1) were designed to amplify exons 1 and 2 respectively. Further 5’ ends of primer 1 and 4 had 15bp overhangs which were homologous to the ends of XhoI - KpnI digested vector molecule, where the cDNA sequence had to be inserted. Similarly primer 2 had a 15 bp overhang homologous to 5’ end of exon 2 as shown in Figure 1. While exon 2 could be easily amplified (Figure 2(A)), exon 1 could not be amplified using primer 1 and 2 in spite of several modifications in the PCR conditions. We felt that the 15 bp overhang of primer 2 complementary to the 5’ end of exon 2 was leading to unproductive annealing (Figure 1). A similar situation was also observed in our second example (Figure 3), which is described later.
This problem was circumvented by first amplifying the complete MYB1 gene using primers 1 and 4, followed by its cloning into pRT101 vector by HBR cloning reaction. By using the restriction enzyme XmnI, exon 2 was deleted from the construct and exon 1 was then amplified using primers 1 and 2 (Figure 2(B)). A proper amplicon was observed in this reaction. This showed that primer 2 was properly designed and its homology with exon 2 led to unproductive annealing in the earlier instance.
The amplified exon 1, exon 2 and the digested vector were finally assembled by HBR cloning reaction. cDNA synthesis of MYB1 gene was confirmed by restriction digestion of plasmids isolated from several colonies. The size of the products observed on 1% agarose gel electrophoresis was in agreement with the predicted sizes (Figure 2(C)). Further the assembled cDNA from several clones was sequenced and found to carry no errors.
3.2. cDNA Assembly of MYB2 Gene from A. thaliana
In a second example, the cDNA of MYB2 (Acc no. NC_003071.7) gene of A. thaliana was assembled. This gene is 1.3 Kb long and carries two introns of 92 bp and 102 bp. Three sets of primers 1, 2a; 3, 4a and 5, 6 (Table 2) were designed for amplification of three exons of MYB2 gene similar to the initial strategy of example 1. While exon 3 could be amplified using genomic DNA as a template, with primer 5 and 6, exons 1 and 2 could not be amplified with their respective primers (Figure 3). This showed that the 15 bp overhang in the primer 2 and
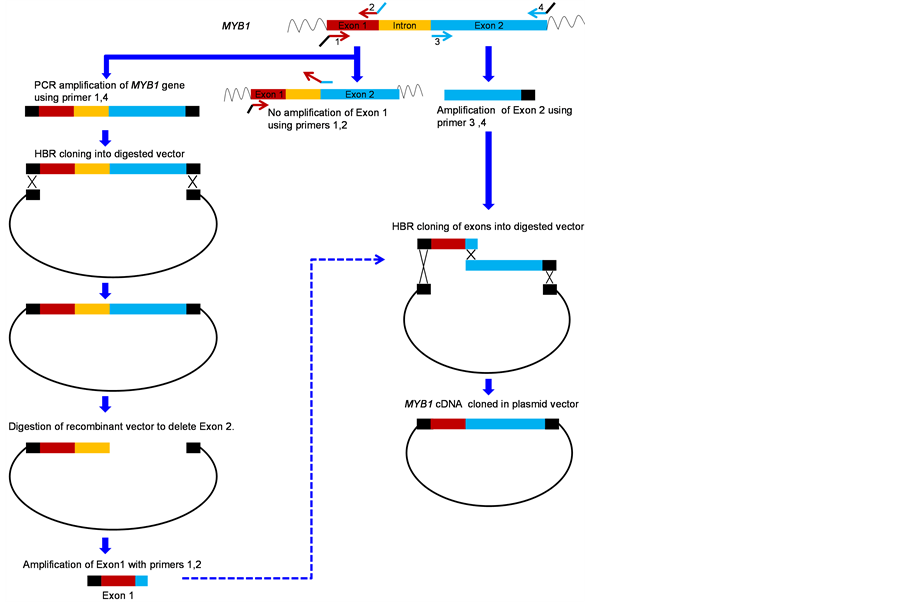
Figure 1. Outline of the strategy to assemble and clone the cDNA sequence of the MYB1 gene from A. thaliana.

Figure 2. (A) Amplification results of exon 2 and exon 1 of the MYB1 gene with primer set 3, 4 and 1, 2, respectively using genomic DNA as a template; (B) Amplification of exon1 with primer set 1, 2 using pRT101:MYB1 with deleted exon 2; (C) Analysis of clones with MYB1 cDNA in the vector pRT101 (lanes 1 - 4) and its comparison with a MYB1 genomic DNA clone (lane 5). Lanes M1 and M2 represent 100 bp and 1 Kb size markers, respectively.
4, complementary to 5’ end of exon 2 and 3 were leading to unproductive annealing.
In this example, we circumvented the problem by designing two reverse primers 2b and 4b (Table 2) for exon 1 and 2 respectively. These primers were similar to 2a and 4a except that they did not carry the 15 bp overhang complementary to 5’ end of the adjacent exon. Exon 1 and exon 2 were first amplified using primers 1, 2b and 3, 4b respectively. The amplicons thus obtained were again amplified with primers 1, 2a and 3, 4a, respectively to introduce the region of homology in exons 1 and 2 (Figure 3).
The amplified exon 1, exon 2, exon 3 and the vector were finally assembled by the homology-based recombination reaction. The cDNA synthesis of MYB2 gene was confirmed by restriction digestion of plasmids isolated from several colonies. Further the assembled cDNA from several clones was sequenced and found to carry no errors.
In summary, this is the first report of a method to synthesize cDNA sequence of any intron containing gene, as well as its cloning in a suitable plasmid vector molecule by the homology-based recombination cloning. It is a simple, efficient and error free method.
ACKNOWLEDGEMENTS
This work was supported by a grant from University of Delhi. NV was supported by a research fellowship from Council of Scientific and
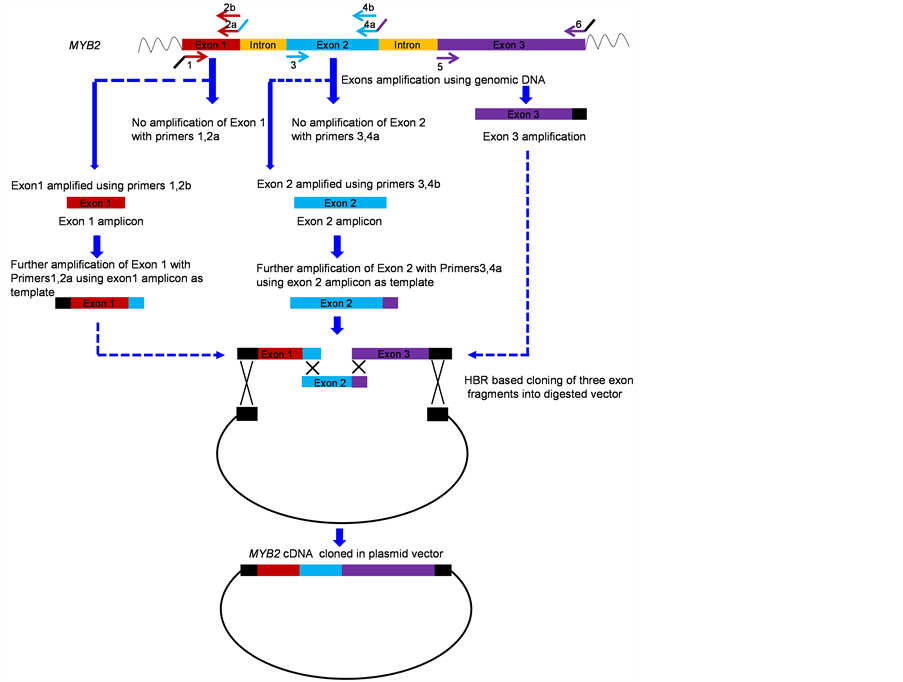
Figure 3. Outline of the strategy to assemble and clone the cDNA sequence of the MYB2 gene from A. thaliana.
Industrial Research (CSIR), Delhi, India.
REFERENCES
- Sleight, S.C., Bartley, B.A., Lieviant, J.A. and Sauro, H.M. (2010) In-fusion biobrick assembly and re-engineering. Nucleic Acids Research, 38, 2624-2636. http://dx.doi.org/10.1093/nar/gkq179
- Topfer, R., Matzeit, V., Bruno, G., Schell, J. and Steinbiss, H.H. (1987) A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Research, 15, 5890.
NOTES
*Corresponding author.


