American Journal of Molecular Biology
Vol. 2 No. 4 (2012) , Article ID: 23928 , 14 pages DOI:10.4236/ajmb.2012.24033
Early expression of the tbx22 gene in zebrafish influences positioning of pharyngeal arch cartilages
![]()
1Department of Biology, University of Massachusetts Dartmouth, Dartmouth, USA
2Department of Bioengineering, University of Massachusetts Dartmouth, Dartmouth, USA
3Department of Periodontology, Institute of Oral Health Research, University of Alabama at Birmingham, Birmingham, USA
Email: tferreira@umassd.edu
Received 17 August 2012; revised 20 September 2012; accepted 28 September 2012
Keywords: Zebrafish; tbx Signaling; Pharyngeal Pouch
ABSTRACT
Mutations in human tbx22 cause X-linked cleft palate with anklyoglossia syndrome. The two zebrafish tbx22 splice isoforms, tbx22-1 and tbx22-2, encode proteins of 444 and 400 amino acids, respectively. Zebrafish tbx22 mRNA expression mirrors mammalian tbx22 expression and is consistent with early patterning of the vertebrate face. In zebrafish, tbx22 mRNA is strongly expressed during early pharyngeal arch development in the ventral mesenchyme, and a later expression domain is found in ectomesenchymal cells underlying the stomodeum, a bilaminar epithelial structure demarcating the early forming mouth. Therefore, tbx22 is hypothesized to be involved in craniofacial development. The objective of this work is to characterize the role of tbx22 during craniofacial development in zebrafish. tbx22 knockdown revealed that defects in tbx22 signaling cause mild clefting, joint defects and dorsoventral patterning defects in cartilages. Quantitative PCR and in situ analysis revealed that knockdown of tbx22 also causes a dramatic decrease in expression of osr1 and gdf5. Craniofacial patterning is dependent on proper signals from endoderm, mesoderm and ectoderm. The early influence of tbx22 on signals within the ventral mesenchyme impacts the domains of several key pharynxgeal arch signals, thereby helping to regulate proper patterning of the developing jaw.
1. INTRODUCTION
Craniofacial development relies on signals emanating from the endoderm, ectoderm and the brain. Proper formation and positioning of craniofacial cartilages is driven by numerous secreted signals in each of these three cell layers. As the Cranial Neural Crest (CNC) cells migrate ventrally from the dorsal neural tube they are influenced by signals such as sonic hedgehog produced by the foregut endoderm [1], forebrain expression of FGF-R [2], and Bone Morphogenetic Protein (BMP) and Noggin influence skeletal formation [3,4]. Within each pharyngeal arch there are dorsal and ventral components which are specified by additional signals such as those found in the endothelin (edn) pathway [3]. The regulation of these secreted signals is accomplished through a variety of transcription factors including members of the MSX, DLX, OSR and tbx families [5-8]. It is therefore critical that in addition to understanding how secreted ligands and receptors regulate craniofacial development, that we also unravel the regulation of their transcription.
The T-box genes are an ancient family of transcription factors that are well conserved throughout all metazoans [9,10]. All T-box genes share a conserved homology domain that encodes a polypeptide region known as the T-box [9]. The T-box region of the protein possesses DNA binding activity and binds to a specific sequence of DNA called the T-site [9,11]. T-box proteins are thought to be important regulators of early mesoderm induction, specification, movement, patterning, as well as somite formation [10,11]. T-box genes are usually expressed in highly specific patterns and tbx genes expressed in the same area can function additively or antagonistically to directly regulate genes that control patterning and differentiation of cell types in the region where they are expressed [9-11].
Characterization of several tbx genes suggests a role for these genes in craniofacial development [12-14]. tbx22 is a member of the tbx1 subfamily, which consists of tbx1, 10, 15, 18, 20, 22 [9]. tbx1 has been shown to regulate oral epithelial adhesion and palatal development [15] and the van gogh/tbx1 zebrafish mutant results in severe craniofacial defects similar to the complex defects found in DiGeorge syndrome [16]. Mutations in the human tbx22 gene cause syndromic, X-linked cleft palate/ankyloglossia and also strongly contribute to nonsyndromic cleft lip and/or cleft palate and cleft palate alone [13,17-21]. Therefore, we asked whether or not zebrafish tbx22 plays a similar role in palate formation and what role it plays in craniofacial development.
Characterization of zebrafish tbx22 revealed two splice isoforms, tbx22-1 and tbx22-2 [13]. Tbx22-1 encodes a protein of 444 amino acids and resembles canonical Tbx22 orthologs, while tbx22-2 encodes a 400 amino acid protein that lacks conserved N-terminal sequences [13]. Experiments examining a combined profile of both isoforms revealed that zebrafish tbx22 is expressed as early as 6 hours post fertilization (hpf) as a low level transcript, and maintains a low level of expression into adulthood [13]. Discrete expression domains are first visible in the pharyngeal mesenchyme and segmental paraxial mesoderm tissue at 28 hpf and are no longer visible at 30 hpf. By 38 hpf a second discrete domain of tbx22 expression is also found in the perioral mesenchyme underlying the early mouth and in early presumptive jaw joints [13]; however, the function of tbx22 is still unknown. Zebrafish tbx22 expression overlaps with expression of bapx1, a homeobox transcription factor known to be essential for jaw joint formation [22]. The expression pattern of tbx22 in zebrafish positions it to be involved in jaw and/or pharyngeal arch development.
Here we focus on the role of tbx22-2 in the formation of the orofacial complex. We show that knockdown of tbx22-2 results in mild clefting, ventrally restricted craniofacial cartilages and specific joint defects. Our analysis indicates that the early expression domain of tbx22-2 is a positive regulator of osr1. Early in arch development, decreased osr 1 expression causes endodermal patterning changes that alter the domains of key dorsal/ventral arch signals such as edn1 and bapx1 in later development. Furthermore, loss of tbx22-2 expression may be impacting gdf5 through decreased osr1 expression. This work provides insight into the role of tbx22 in craniofacial development and indicates that tbx22-2 influences early ventral mesenchyme signals causing dorsoventral defects in positioning of arch elements. The loss of tbx22-2 also results in joint defects due to altered joint domain expression of bapx1 and decreased gdf5 expression.
2. MATERIALS AND METHODS
2.1. Morpholino Injections
Morpholinos were created by Gene Tools Inc. (Philomath, OR), based on the sequences of tbx22-1 and tbx22-2. The tbx22-2 translation blocking morpholino is 5’-GGAAATGCAGAGTTCAATGTAAACG-3’. The translation blocking mismatch (tbx22-2-MM) morpholino is 5’-CACGAGTGTTAAACTTTCGTCATAG-3’. The splice blocking morpholino sequences are as follows: Tbx22 exon5-MO 5’-TCCATGAATTGGCAAGTTACCTGTT-3’; tbx22 exon4-MO 5’-TACTGAAAAAGGGCACTCACATGGC -3’. The tbx22-exon-MM morpholino 5’-tttcaaatgt ctctccactctaccT-3’. Each morpholino was prepared at a concentration of 1.0mM. Various dilutions, 1:5 (6.7 ng), 1:7 (5.3 ng), 1:10 (3.0 ng) were used to inject approximately 1.5 nl into 1 - 4 cell stage embryos to determine a concentration that produced moderately severe phenotypes. Morpholinos were injected using a picoinjector PLI-100. The 1:5 diltion produced a moderately severe phenotype and was used for all remaining experiments. Missense morpholinos were also injected to confirm that any developmental defects were the result of tbx22 knockdown and not a side effect of physical injection. Missense morpholinos produced no phenotype and are referred to as “wild-type” in figures. The embryos were monitored daily and collected at various stages of development for study.
Morpholino rescues were performed using synthetic capped mRNA synthesized from full length tbx22-1/2 clones using the Ambion Mmessage machine kit (Ambion, Austin Tx). Approximately 1 - 4 nl of a 250 ng/ul solution was injected into 1 - 4 cell stage embryos. An equal mixture of tbx22-1 and tbx22-2 mRNA was used for rescue of the splice blocking morpholinos.
2.2. Alcian Blue Cartilage Staining
Alcian Blue Staining was used to visualize cartilages in larvae [23]. Six day old missense MO injected (wild type), tbx22-2-MO and tbx22 exon-MO injected larvae were euthanized in tricaine and fixed in 4% paraformaldehyde overnight. Larvae were washed with PBS-Tween (PBT) to remove paraformaldehyde, bleached in 30% hydrogen peroxide to remove pigment, and then washed in PBT again. Larvae were incubated in 0.3 mg/ml alcian green/acetic acid for 4 hours, rehydrated in a staged ethanol series and treated with 25 mg/ml trypsin to dissolve brain tissue and allow visualization of pharyngeal arch cartilages. Larvae were observed in 75% glycerol. Morpholino injected larvae were catalogued for presence/absence and defects in each craniofacial element compared with wild type larvae. Craniofacial cartilages of wild type (5 larvae) and tbx22 exon4-MO (15 larvae) embryos were dissected out of the larvae in glycerol and were flat mounted on glass slides. Bright-field images were taken of each individual cartilage element using the Olympus 12.5 megapixels color digital camera.
2.3. RNA Extraction and Quantitative RT-PCR
Developmentally staged zebrafish were collected homogenized in trizol and stored at –80˚C. Total RNA was isolated using Trizol according to manufacturer’s instructions (Invitrogen, Carlsbad, CA), and 4.5 mg of each total RNA sample was used in RT reactions using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA). The resulting RT products (1 ml) were used as template in 50-ml PCR reactions, performed with Platinum PCR SuperMix High Fidelity (Invitrogen, Carlsbad, CA), using the following reaction conditions: denaturation at 95˚C for 5 minutes followed by 35 cycles of 95˚C for 40 seconds, 60˚C for 1 minute, and 72˚C for 2 minutes, followed by a final extension at 72˚C for 7 minutes. Negative control is total RNA with no reverse transcriptase added in cDNA reaction. Internal primers were designed to span introns to ensure that no gDNA contamination occurred.
Quantitative-PCR—Real-Time PCR was performed with SYBR-Green chemistry, using a Roche Lightcycler. β-actin served as an internal reference. The following primers were designed for quantitative RT-PCR: β-actin Q For 5’-CTTCTTGGGTATGGAATCTTGC-3’, rev 5’-GTACCACCAGACAATACAGTG-3’, tbx22aQFor5’-cacgcgacaagtggatcata-3’, rev-5’CTCAGATCCTGTGCGTCAAA -3’, tbx22-2QFor 5’-cacgcgacaagtggatcata-3’, rev 5’- CACTAACCCTGTGCGTCAAA -3’; mef2ca Qfor-5’- CGCACGAGAGTCGGACTAAT-3’, mef2ca Qrev-5’- GGTCGATGTCCTCGTTGATT-3’, edn1Q for 5’-CTGGAATACGGGACTTGCAT-3’, edn1 Qrev 5’-TGTCCAGGTGGCAAAAGTAG-3’; gdf5 Qfor 5’-AGCCTTCTTCGTGGTGTTTG-3’, gdf5 Qrev 5’-GCATCTCTGTTTGGGGTTCT-3’; hand2 Qfor 5’-CAGACGCCAAAGAAGAAAGG- 3’, hand2 Qrev 5’-GTTCAGATGGCCTCATTTCG -3’; osr1 Qfor 5’-CATGCTGAGGAAGACGAACA -3’, osr1 Qrev 5’-AAGAAGGGTGAAGAGGCACA- 3’; chd1 Qfor 5’-GGTCTGATGCACTGCGTTAT-3’, chd1 Qrev 5’-CATGATTTTGCAGCAGTGTCC-3’. For the time course experiment an arbitrary value of 1 was assigned to the average expression at the 1 hpf cell stage and expression level at other time points was normalized to this denominator. For analysis of various genes in MO-injected embryos the Wt sample was assigned a value of 1 and expression level at other time points was normalized to this denominator. Plots of quantitative RT-PCR are the average 3 individual biological replicates.
2.4. Whole Mount in Situ Hybridizations
Bacterial cultures (bapx1, dlx2a, edn1, nkx2.3, sox9a) were inoculated with transformed cells and incubated overnight at 37˚C. Plasmids were isolated using QIaprep miniprep protocol (Qiagen, Valencia, CA). Plasmids were linearized as follows: bapx1/BamHI, dlx2a/BamHI, edn1/EcoR1; foxa2l/Sac1, nkx2.3/HindIII. The restriction digest was cleaned using equal volume phenol: chloroform extraction and Manual Phase Lock Gel (5-Prime, Gaithersburg, MD) columns. The linear template was precipitated with 1/10 the volume 3 M sodium acetate and 3 times the volume of 95 percent ethanol. Riboprobe templates for hand2 and osr1 were synthesized by PCR using platinum PCR supermix. PCR conditions were as follows: 94˚, 5 minutes, 94˚, 40 seconds, 60˚, 30 seconds, 72˚, 1 minute, 72˚, 5 minutes, 35 cycles. Primer sequences: osr1 foward 5’-TGGATAACCGTATTACCGCC-3’ osr1 reverse 5’-CGCGCAATTAACCCTCACTAAAGCACTAGTCATACCAGGATC-3’ hand2 forward 5’-TGGATAACCGTATTACCGCC-3’, hand2 reverse 5’CGCGCAATTAACCCTCACTAAAGCACTAGTCATACCAGGATC-3’. (Underline indicates T3 polymerase binding site). The PCR product was purified using the Montage PCR centrifugal filter device protocol (Millipore, Woburn, MA).
Riboprobes were synthesized using the DIG RNA Labeling Kit (T3//T7) (Roche Applied Science, Indianapolis, IN). WISH was performed as previously described [24], using a modification of published protocols [25]. Embryos were cleared in glycerol and photographed. All images were captured on an Olympus SZX12 microscope and assembled using Adobe Photoshop.
3. RESULTS AND DISCUSSION
3.1. Differential Expression of the tbx22-1 and tbx22-2 Transcripts throughout Larval Development
Initial analysis of the tbx22-1 and tbx22-2 transcripts using standard reverse transcriptase PCR (RT-PCR) revealed expression of both transcripts consistently throughout adulthood [13]. The initial studies indicated that tbx22-1 was the only maternal transcript, and the other stages examined revealed relatively equal levels of transcription of both isoforms using end-point PCR [13]. Since the tbx22-2 transcript is a mere 113 bp larger than the tbx22-1 transcript, assaying for differential expression of the two transcripts was not successful using whole mount in situ hybridization. We used quantitative PCR (Q-PCR) to determine if there was any difference in expression between the two transcripts. Q-PCR is more sensitive than standard RT-PCR, often revealing differences in expression not resolved by standard gel electrophoresis. Q-PCR revealed that tbx22-2 expression is higher than tbx22-1 as early as 11 somites(som). Tbx22-2 continues to be the dominant transcript from 11som through 28 hpf (Figure 1). The two transcripts become more equally expressed by 38 hpf. This time period is interesting as it is prior to the discrete expression in the stomodeum region we see by 38 hpf. By the 10 somite
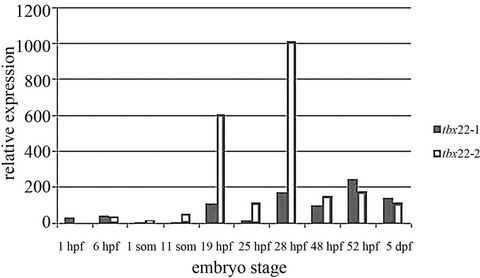
Figure 1. Developmental Q-PCR profile of tbx22-1/2 mRNAs. Quantitative PCR reveals dramatic increases in tbx22-2 transcript over tbx22-1 transcripts between 11 somite stage and 28 hours post fertilization (hpf). Q-PCR analysis reveals that at 19 hpf and 28 hpf tbx22-2 is expressed nearly 5× greater than tbx22-2.
stage neural crest cells begin to migrate from the dorsal neural keel, and are positioned in pharyngeal pouches by 28 hpf. As noted, the only other visible expression domain of tbx22 was the ventral pharyngeal mesenchyme and segmental paraxial mesoderm at 28 hpf [13], we will refer to this expression as the early tbx22-2 expression domain, while the bilateral expression after 38 hpf is the late expression domain.
While early tbx22 expression is likely to impact various tissues, our interest lies specifically in mouth formation. We next examined tbx22 expression in head vs. tail tissue. Reverse transcriptase PCR revealed that at 48 hpf the dominant transcript is tbx22-2 in both the head and tail, with minimal tbx22-1 expression in the head at this time (Figure 2). These results in conjunction with the peaks in expression led us to focus in on the role of tbx22-2 in development of the craniofacial region.
3.2. tbx22 Knockdown Results in Defects in Positioning of Pharyngeal Arches 1 and 2 Ventral Elements and Defects in Pharyngeal Arch Joint Formation
A morpholino (MO)-mediated gene knockdown strategy was used to examine the role of tbx22-2 on craniofacial cartilage development. Wild type (mismatch MO injected), tbx22-2-MO and splice blocking MO injected embryos were stained at 6 dpf with alcian blue, which stains acid glycosaminoglycans present in cartilage [25]. Each injected larvae was analyzed for the presence/absence of individual pharyngeal cartilage elements as well as any deviations from normal patterning when compared with normal embryos.
Tbx22-2-MO and tbx22 exon4/5-MO larvae displayed nearly identical phenotypes with clear defects in the ventral positioning of the first and second branchial arches, and incompletely fused trabeculae with a more severe

Figure 2. Analysis of Head and Tail expression of tbx22-1/2 mRNAs. Reverse Transcriptase PCR was used to examine transcript levels in severed heads and tails at 48 hpf. Head cDNA (A, B), and Tail cDNA (C, D). Tbx22-1 and tbx22-2 fragments are labeled 1 and 1 respectively (B,D). Beta-actin was used as a control (A,C). Results show that tbx22-2 is the predominant transcript in the head, while the tail has a higher level of tbx22-1 expression than the head but tbx22-2 is still the more abundant transcript.
defect in the tbx22 exon4/5-MO larvae (indicated by Figure 3). Translation blocking morpholinos target the ATG start site specifically of tbx22-2. We also used splice blocking morpholino’s to confirm that loss of tbx22 was causing the observed phenotype. Splice blocking morpholinos target an intron-exon boundary which causes an altered gene product vs preventing translation. This splice blocking approach can utilize RT-PCR to assay altered mRNA and hence overall decrease in functional protein. In our case, the splice blocking morpholino also blocks both isoforms of tbx22 making it a more severe defect. All tbx22-2-MO fish (n = 55) possessed the Meckel’s cartilage; however 54% of larvae had a ventrally restricted Meckel’s (Figures 3(B) and (B’)). After examining the dissected Meckel’s cartilage, it was clear that tbx22-2-MO injected larvae exhibited defects in the retroarticular process of the Meckel’s cartilage (Figures 4(a), Panel D). The ceratohyal cartilage was present in all tbx22-2-MO injected larvae but was ventrally restricted in 67% of larvae (Figures 3(B) and (B’)). The patterning defects in the Meckel’s cartilage and the ceratohyal cartilage were the most common defects seen in lower jaw elements in the injected larvae. Both the Meckel’s and ceratohyal cartilages develop from ventral condensations of cranial neural crest cells, suggesting that tbx22-2 plays a role in patterning ventral cartilages. The dorsal arch elements, palatoquadrate and hyosymplectic cartilages were smaller but normally positioned. However, tbx22-2-MO injected larvae had visible breaks in the hyosymplectic cartilage (53%) which corresponded to a noticeable loss of the interhyal, second arch joint

Figure 3. Cartilage phenotypes of tbx22 knockdown embryos. Alcian Blue cartilages staining of 6 dpf wild type, tbx22-2-MO, tbx22exon4-MO and tbx22exon5-MO larvae. A, A’) lateral and ventral views of mis-sense control (wt) larvae. B, B’) tbx22-2-MO injected embryo. C, C’) tbx22-2 mRNA rescued larvae D, D’) tbx22exon4-MO injected embryo E, E’) tbx22-1/2 mRNA rescued tbx22exon-MO larvae—both the Meckel’s cartilage and the ceratohyal are ventrally restricted and smaller than wild type larvae. Dissected trabeculae from each embryo represented with area of cleft indicated by red asterisk. Abbreviations: Ch-ceratohyal, M-Meckel’s.
 (a)
(a)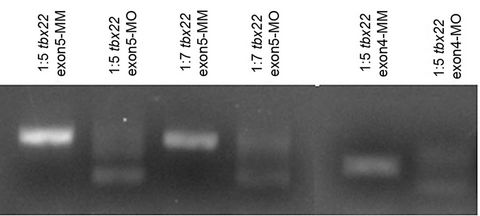 (b)
(b)
Figure 4. Dissected tbx22 knockdown cartilages. (a) Dissected and flat-mounted cartilage elements from wild type (A, C, E, G), tbx22exon4-MO injected (B, D, F, H) 6 dpf larvae. Arch 1 (mandibular) dorsal (d) and ventral (v) components are the palatoquadrate (pq) and the Meckel’s (m), respectively. Arch 2 (hyoid) arch dorsal and ventral components are the hyosymplectic (h) and the ceratohyal (ch), respectively. (b) Reverse Transcriptase (RT) PCR analysis of altered splicing of exon5 or exon4 upon morpholino injection. Loss of exon 5 results in a product 168 bp smaller, while loss of exon 4 results in a loss of 175 bp.
region, which articulates at the location of the break (Figures 4(a), panel F). Each defect was rescued by injection of tbx22-2 mRNA (Figures 3(C) and (C’)).
Splice blocking morpholinos (tbx22 exon4-Mo, tbx22 exon5-MO) were designed to test the specificity of the effect of the translation blocking morpholino. The Meckel’s cartilage was ventrally restricted in 79% - 81% (Table 1) of the splice blocking-MO injected larvae (Figures 3(D) and (D’)) and this was accompanied by defects in the joint region of the Meckel’s cartilage (Table 1) similar to the defects seen in the tbx22-2-MO injected larvae. Similarly, the ceratohyal elements were ventrally restricted in 82% - 85% of the splice blocking-MO injected larvae (Table 1). The palatoquadrate cartilage was smaller than controls but normally shaped, and the hyosymplectic possessed similar defects and loss of the second arch joint in 69% - 75% of cases (Table 1).
Loss of tbx22 signals consistently results in clefting of the zebrafish palatal element, the trabeculae. Similar to the other elements, the splice-blocking morpholino causes a more severe cleft than the tbx22-2-MO. Analysis of cDNA from splice blocking-MO injected embryos reveals a dose dependent decrease in wild type splicing which correlated with the observed phenotype (Figure 4(b)). The highest concentration of morpholino injected (6.7 ng) revealed consistent patterning defects and higher concentration of morpholino, both splice blocking and tbx 22-2-MO, resulted in severe developmental defects that prevented analysis of craniofacial cartilages. While the overall patterning defects were the same with each morpholino tested, there was a higher percent of defects in the splice blocking-MO injected embryos. This is likely due to the fact that any splice blocking morpholino will cause defects in both tbx22-1 and tbx22-2 transcripts since these only differ by a small region.
3.3. Gene Expression Analysis
Previous studies revealed two discrete expression domains for tbx22 [13]. While tbx22 is expressed as early as 6 hpf, the first domain of expression is visible in the ventral pharyngeal mesenchyme by 28 hpf. A later domain of expression is found from 38 hpf through 60 hpf as discrete, bilateral domains at either corner of the forming mouth. This later oral ectoderm domain has overlapping expression with bapx1 [13].
To determine the role of the early tbx22-2 expression, we examined the patterning of the pharyngeal arches as well as pharyngeal endoderm in the tbx22-MO injected larvae. By 28 hpf the pharyngeal pouch system is well developed with NCC’s streaming into endodermal pouches which lay the foundation for cartilage differentiation. We asked if tbx22 expression was required for patterning of the pharyngeal pouches. Analysis of the NCC’s as well as the endoderm lining the pouches reveals mis-patterned pouches that do not appear to be normally angled towards the anterior of the embryo. Instead, they appear to migrate more ventrally. This ventral position can be seen in the pattern of streaming NCC’s, dlx2a expression (Figure 5(A’)), as well as in the positioning of the endoderm, nkx2.3 staining (Figure 5(C’)). The pouch structure is perturbed as early as 24 hpf as indicated in the sox10:eGFP line (Figure 5(B’)), Levels of foxa2 expression do not seem dramatically affected by tbx22 knockdown, however, there is a patterning defect in the endoderm. Tbx22 morphants have a narrowed pharyngeal endoderm but foxa2 expression is normal (Figure 5(D’)).

Table 1. Effect of tbx22 knockdown by translation-blocking and splice-blocking morpholinos on ventral position of meckels and ceratohyal cartilages, and defects in the first and second arch joint elements.
Surprisingly, osr1, which is also expressed in the endoderm, anterior ventral mesenchyme (avm) as well as in the branchial arches (Figure 5(E’)) is severely reduced in the tbx22 morphants. Osr1 is a gene that belongs to the odd-skipped gene family, and has been shown to be expressed in the mouse intermediate mesoderm, branchial arches, and ventral mesenchyme [27,28]. In zebrafish osr1 is expressed robustly in the ventral head mesenchyme and branchial arches but its role in craniofacial development has not been examined. Osr1 has been shown to be essential for kidney formation [29], and recently has been shown to interact with tbx5a during pectoral fin development [30]. Here we show that osr1 is severely disrupted by loss of tbx22-2 expression. Quantitative PCR analysis revealed a 60% - 70% decrease in Osr1 expression between 36 - 48 hpf (Figures 7(c) and (d)).
The second, later expression domain of tbx22, in the developing mouth region overlaps with bapx1 [13]. Bapx1 is a member of the endothelin (edn1) signaling pathway and is involved in joint formation [22]; furthermore, all tbx22-MO injected embryos have ventral positioning defects. Since the edn1 pathway is the major ventral specification pathway, we examined the effect of shape of the developing pharyngeal pouches.
Edn signaling is a tightly regulated pathway that has many intermediates. Tbx22 is a mesodermally expressed transcription factor and is likely to be exerting its effect on molecules within the mesoderm. Alterations in mesodermal patterning could cause arch positioning defects, so we next asked what molecules influence edn1 levels that are positioned well to be influenced by tbx22. Mef2ca is a transcription factor that plays a critical role in mesoderm development and has been shown to be required to effect edn1 signaling in the cranial neural crest [31]. Loss of mef2ca signaling results in less severe craniofacial defects than edn1 mutants, however, loss of mef2ca has striking similarities to the phenotype observed in tbx22 knockdowns [31]. Analysis of spatial expression patterns of several major edn1 pathway components by whole mount in situ analysis reveals slight patterning changes in tbx22-MO knockdowns. The bapx1 expression domain in tbx22-2-MO injected embryos reveals a posterior shift and cells that are positioned more laterally (Figures 6(A) and (B)), similar to changes the endothelin expression domains (Figure 6(D), asterisks). The cluster of bapx1 expressing cells also appears to be more ventrally positioned. Bapx1 is downstream of edn1 and examination of edn1 in tbx22 morphants reveals a gap in the edn1 expression domain (Figure 6(D) asterisks). The edn1 domain appears narrowed in the A-P plane but extended laterally (Figure 6(D), open arrow) mirroring the narrowed ventrally restricted bapx1 expression domain. Hand2 expression levels and patterning appear unaffected by tbx22-2 knockdown (Figures 6(E) and (F)). The observed changes in ventral positioning of these cell populations may result from changes in patterning of the pharyngeal endoderm and shape of the developing pharyngeal pouches.
Alterations in mesodermal patterning could cause arch positioning defects, so we next asked what molecules influence edn1 levels that are positioned well to be influenced by tbx22. Mef2ca is a transcription factor that plays a critical role in mesoderm development and has been shown to be required to effect edn1 signaling in the cranial neural crest [31].
Loss of mef2ca signaling results in less severe craniofacial defects than edn1 mutants, however, loss of mef2ca has striking similarities to the phenotype observed in tbx22 knockdowns [31]. Mef2ca mutants have open mouths, ventrally restricted jaws and joint loss [31], similar to the defects we see in tbx22-2 knockdown embryos. We assayed the level of expression of mef2ca as well as several edn1/bapx1 related genes. Previous work had demonstrated that edn1 signaling is required for bapx1 and hand2 expression [22]. Gene clusters can be grouped by those with direct interaction with edn1 or genes that act downstream of edn1. We examined two
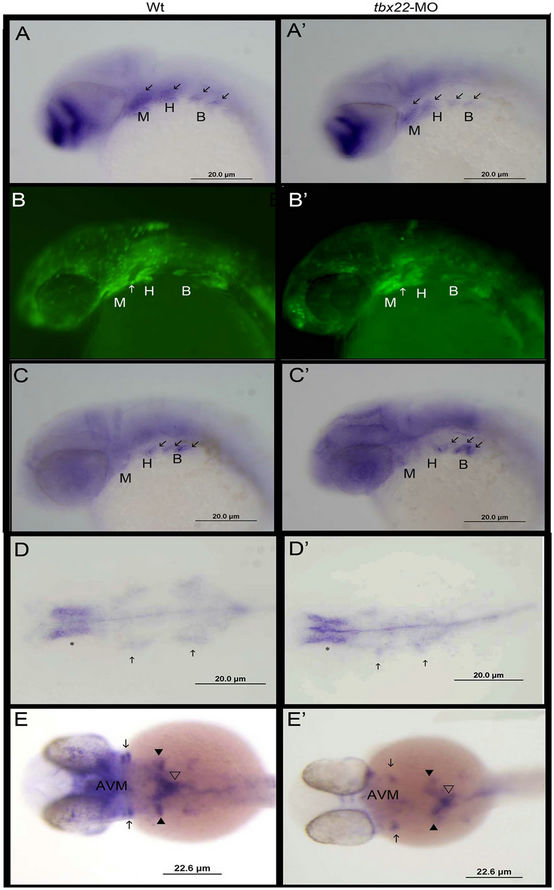
Figure 5. Analysis of pharyngeal arch/pouch patterning in tbx22 knockdowns. Lateral views of dlx2a expression in 32 hpf wild type (A) and tbx22exon4- MO (A’) injected embryos. dlx2a is expressed in neural crest cells in the pharyngeal pouches as well as the diencephalon of the brain. Fluorescent sox10 expression in 24 hpf wild type (B) and tbx22exon4- MO(B’) injected zebrafish embryos. Sox10 is expressed in neural crest cells in 24 hpf embryos. By 24 hpf cranial neural crest cells have migrated into the pharyngeal arches. As with the dlx2a staining the arches seem more ventrally oriented compared to the anterior streaming position of the normal arches. Nkx2.3 is expressed in the endodermal cells of the pouches at 35 hpf in wild type (C) and tbx22exon4-MO (C’) injected embryos. Specifically, the domain between the Meckels (M) and Hyoid (H) arch (white arrow, C-C’) is oriented more ventrally compared the pouch in the normal embryos. Similar ventral restriction is observed. Overall endoderm patterning was examined using foxa2 at 28 hpf. Foxa2 is expressed in the pharyngeal endoderm (arrows) and appears normal in tbx22 morphants (D’) compared to wild type (D), although the endoderm is mediolaterally narrowed in the tbx22 morphants. At 32 hpf osr1 expression (E,E’) in the anterior ventral mesenchyme (avm) is severely reduced as well as in the anterior branchial arches (arrows), but appears normal in the more posterior ceratobranchial arches (black arrow heads).
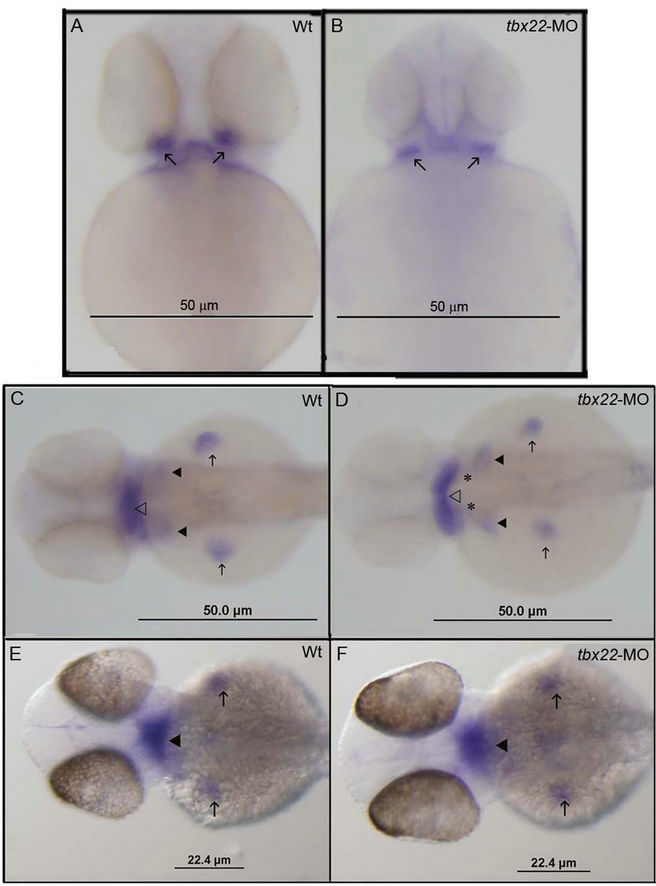
Figure 6. Spatial expression defects in edn signals in tbx22 knockdown embryos. Bapx1 expression in 50 hpf wild type embryos (A) and tbx22-2-MO injected embryos (B). tbx22-2-MO injected embryos appear to exhibit an altered domain of bapx1 expression in the intermediate joint region compared with wild type embryos. Arrows indicate bapx1 expression in the early jaw joint. Ventral view of edn1 expression in 48 hpf zebrafish in wild type (C) and tbx22-2-MO injected (D) embryos. Edn1 is expressed in the pharyngeal arches, heart and mesoderm (open arrowhead), the early ceratobranchial arches (arrowheads) and the pectoral fins (arrows). The edn1 expression domain appears to be laterally extended in tbx22- 2-MO injected embryos, especially in the anterior mesoderm (open arrowhead). Ventral views of hand2 expression at 48 hpf in wild type embryos (E) and tbx22-2-MO embryos (F). Hand2 expression in the pharyngeal arches (arrowheads) and in the pectoral fins (arrows) does not appear to be affected.
time points to assay early arch events and later arch events, 29 hpf and 48 hpf respectively (Figures 7(a) and (b)). At 29 hpf early mef2ca expression is decreased in tbx22-MO injected embryos and no significant change in edn1 or hand2 is observed (Figures 7(a) and (b)). By 48 hpf mef2ca levels appear to return to a more normal level of expression, and this is confirmed with quantitative PCR analysis at 50 hpf (Figure 7(a)). Since tbx22 has a very discrete bilateral expression domain surrounding the mouth, we also asked whether or not there is a local effect on bapx1 or its signaling partners. Endpoint RTPCR revealed a decrease in gdf5 but not chd at 50 hpf
 (a)
(a)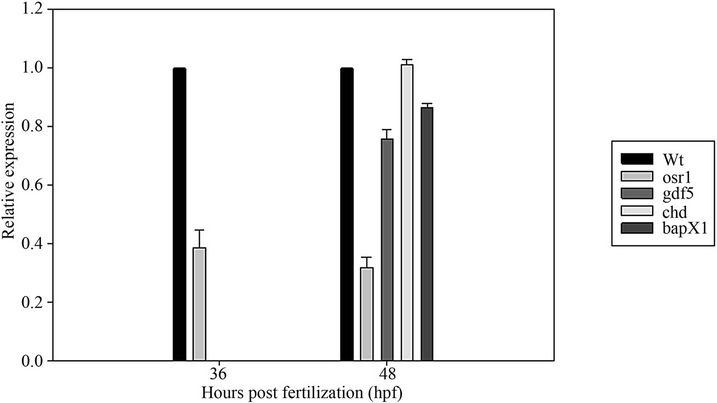 (b)
(b)
Figure 7. Analysis of endothelin signals in tbx22 knockdowns. Quantitative Reverse Transcriptase PCR (RT-PCR) analysis of various endothelin signaling partners in tbx22exon4-MO injected embryos. (a) RT-PCR analysis of mef2ca, edn1, hand2 with the b-actin control for relative expression level. Results reveal a significant (p < 0.001) decrease in mef2ca expression at 29 hpf and 50 hpf; (b) RT-PCR analysis of chordin (chd), gdf5, bapx1, osr1 at 50 hpf reveals a significant (p < 0.001) decrease in osr1 at both 36 and 48 hpf, and gdf5 and bapx1 at 48 hpf. Expression was normalized to beta-actin in each sample which represented 1× expression level and tbx22 MO knockdowns are presented as a percentage of normal expression.
(Figure 7(b)), Q-PCR analysis of bapx1 and its signaling partners gdf5 and chd reveal a 15% decrease in bapx1 expression and a 25% decrease in gdf5 expression in tbx22 morphants (Figure 7(b)).
4. CONCLUSIONS
In the present study we have examined the role of zebrafish tbx22 in craniofacial development. This work has demonstrated that loss of tbx22 signals causes ventral arch positioning defects as well as first and second arch joint defects, and a mild clefting of the zebrafish palatal element. This palatal clefting is similar to mammalian cleft palate which has been linked to defects in Tbx22 signals. These preliminary studies suggest that the zebrafish will be a useful model to investigate the role of tbx22 in palate development. Dissecting the requirement of any gene during development must take into consideration the timing of knockdown and whether or not defects are due to developmental delays or specific expression of key signals. For this reason we have described what we believe to be the role of early tbx22-2 expression in the developing zebrafish embryo. The first discrete tbx22 expression domain is the ventral pharyngeal mesenchyme and somitic mesoderm. We have shown that loss of tbx22-2 signals at this early time, 28 hpf, resulted in defects in the shape of the developing endodermal pouches (Figure 5).
This early mis-patterning has consequences, such as mis-positioned arch elements and an overall size reduction in the craniofacial complex of tb22-morphants. We have also revealed another role for zebrafish osr1 in development. Loss of tbx22 signals dramatically decreases osr1 expression as seen in whole mount stains and quantitative PCR.
Osr genes have been well characterized for their involvement in kidney formation, as well as pectoral fin development. Vertebrate Osr genes are expressed in many tissues and have been shown to be required for proper formation and/or patterning of the heart, endoderm, teeth, palate, the bones and synovial joints in the limbs [28,32-36]. While no studies have defined the specific requirement of osr1 in zebrafish craniofacial development, recent work has shown that osr1 and tbx5a interact to drive proper formation of the pectoral fins [30].
The second later domain of tbx22 expression poses a challenge in regards to analyzing its role in mouth formation. Clearly, the morpholino gene knockdown technique causes a loss in the early expression of tbx22-2 which impacts overall endodermal patterning preventing analysis of the discrete mouth domains by 38 hpf. However, the later expression of tbx22 which overlaps with bapx1 expression positions it well to interact with endothelin 1 signals.
Edn is known to act as a morphogen affecting two developmental fates, joint formation and ventral cartilage formation [22]. Edn1 acts through bapx1 in specifying joint formation while hand2 is an intermediate for ventral cartilage formation [22]. Mef2ca, which is expressed in postmigratory cranial neural crest within pharyngeal arch primordia, has recently been shown to be an effector in the edn1 pathway [31]. Mef2ca function is required for expression of many edn1-dependent target genes including bapx1 [31]. Members of the MEF2 family have been shown to bind Tbx transcription factors, specifically MEF2C binds tbx5 in heart formation [37]. Furthermore, previous work has shown that tbx1 is required for pharyngeal arch expression of edn1 [38], and tbx22 is grouped within the TBX1 subfamily [39]. Knowing that evidence exists for a substantial role for tbx-binding proteins in regulating endothelin signaling, and with zebrafish tbx22 being expressed with bapx1, we examined the effect of tbx22-2 knockdown on components of the edn1 pathway.
Analysis of the endothelin-1 signaling pathway reveals that decreased tbx22 results in decreased expression of bapx1 and altered expression domains of bapx1 and edn-1. There is, however, no decrease in hand2 expression. We believe that knockdown of tbx22-2 causes perturbations in tissue patterning resulting in spatial defects in edn signals. These results do not support a direct role for tbx22 in the edn pathway. The decrease in mef2ca expression at 28 hpf is likely due to changes in heart expression of mef2ca and not a specific change in neural crest expression. Our results reveal that the role of tbx22 on mef2ca expression is timing specific, and this change in mef2ca expression may be causing observed edema and heart abnormalities. Analysis of a detailed time series of tbx22 morphants as well as other mef2ca signaling intermediates, dlx5a, dlx6a and dlx3b, will provide more detail into the specific requirements for tbx22 throughout the early stages of arch formation and joint patterning.
While examining the signals downstream of bapxI, we did uncover a decrease in gdf5 expression in the tbx22-2 knockdown embryos. This decrease in gdf5 could be the result of a decrease in bapxI expression, but this is not supported by the chd expression which is unaltered. It is more likely that the gdf5 levels are being influenced by another factor independent of bapx1. Osr1 has been shown to be required for maintenance of expression of signaling molecules critical for joint formation, including Gdf5 [30]. This work has also revealed a significant loss of osr1 signaling in tbx22 morphants which positions osr1 as a candidate for mediating the effect of loss of early tbx22 signals in craniofacial development. Normal expression of osr1 positions it well to influence craniofacial development, and osr1 is expressed in the ventral pharyngeal arch mesenchyme, similar to the pharyngeal mesenchyme expression domain of tbx22 (Figure 8) [13,40]. The zebrafish osr1 gene does have a putative t-binding domain located –419 bp upstream of the transcriptional start site. We predict that tbx22 binds and influences osr1 transcription which in turn impacts patterning of the endoderm and is likely to impact gdf5 levels and joint formation. Furthermore, conditional Osr1 knockouts in mice have demonstrated a requirement for Osr1 in heart formation [33] which overlaps with our observed heart defects and decreased mef2ca expression during heart formation stages. Current studies are examining the interaction of osr1 and tbx22.
These studies cannot directly address the function of the later time point of tbx22 expression in the bilateral domains surrounding the developing mouth. Future studies will require either site specific gene knockdown or generation of a tbx22 knockout that is rescued through the early stages of development to assay specifically mouth formation. TALEN technology is advancing allowing for the potential to generate a tbx22 mutant

Figure 8. Schematic representation of early tbx22 expression domain overlap with osr1. Both tbx22 and osr1 are expressed in the ventral mesenchyme during the early stages of pharyngeal arch formation. Decreased expression of tbx22-2 (red dots) results in dramatic loss of osr1 (green dots) in the mesenchyme causing decreased endodermal tissue. The narrowed endoderm results in pharyngeal pouches that do not curve towards the anterior of the embryo (arrows).
[41,42]. However, in regards to the interaction of osr1 and tbx22 at this later time point, recently the signals involved in zebrafish palatogenesis have been eloquently displayed by Swartz et al., 2011 [40]. This work reveals neighboring expression domains of tbx22 and osr1 surrounding the oral ectoderm at 44 hpf. Therefore, tbx22’s influence on osr1 expression may also be relevant specifically in mouth formation.
Future work is required to determine the difference in function of the tbx22-1and tbx22-2 isoforms. The splice target morpholino’s result in similar defects as the tbx22- 2 translation blocking morpholino. Tbx22-1 may provide some redundant function to compensate for tbx22-2 loss, which is why the splice morpholino does have a higher % of joint defects and ventral displacement at similar concentrations. However, it is likely that tbx22-1 may also have another role in development. Efforts are underway to parse out unique expression domains of tbx22-1 and tbx22-2. Promoter analysis may also provide clarity as to the role for two tbx22 isoforms. Detailed analysis of these regulatory regions will provide much needed clarity into the regulation of tbx22 signals. Furthermore, promoter analysis may uncover a mouth specific regulatory element that would be helpful in designing experiments to drive mouth specific knockdown of signals that are redundant throughout development, making it very difficult to assay these later developmental events.
5. ACKNOWLEDGEMENTS
We wish to thank, Andrea Moreira and Lane Wilson for their technical assistance and The Molecular Genetics Core Facility of Children’s Hospital Boston. We would also like to thank Dr. Thomas Schilling for sharing the sox10: eGFP transgenic line. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the University of Massachusetts Institutional Animal Care and Use Committee (IACUC), Assurance Number: A4491-01. This research was supported in part by National Institutes of Health (NIH)/National Institute of Dental and Craniofacial Research (NIDCR) grant DE14683 (TLF), and a UMass Dartmouth Healey endowment grant.
REFERENCES
- Benouaiche, L., Gitton, Y., Vincent, C., Couly, G. and Levi, G. (2008) Sonic hedgehog signaling from foregut endoderm patterns the avian nasal capsule. Development, 135, 2221-2225. doi:10.1242/dev.020123
- Hu, D. and Marcucio, R.S. (2009) A SHH-responsive signaling center in the forebrain regulates craniofacial morphogenesis via the facial ectoderm. Development, 136, 1707-1758. doi:10.1242/dev.026583
- Foppiano, S., Hu, D. and Marcucio, R.S. (2007) Signaling by bone morphogenetic protein directs formation of an ectodermal signaling center that regulates craniofacial development. Developmental Biology, 312, 103-114. doi:10.1016/j.ydbio.2007.09.016
- Alexander, C., Zuniga, E., Blitz, I.L., Wada, N., Le, P, Javidan, P., Zhang, Y., Cho, T., Crump, J.G. and Schilling T.F. (2011) Combinatorial roles for BMPs and Endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development, 138, 5135-5146. doi:10.1242/dev.067801
- Brown, J.M., Wedden, S.E., Millburn, G.H., Robson, L.G., Hill, R.E, et al. (1993) Experimental analysis of the control of expression of the homeobox-gene Msx-1 in the developing limb and face. Development, 119, 41-48.
- Gibson-Brown, J.J., Agulnik, S.I., Silver, L.M. and Papaioannou, V.E. (1998) Expression of T-box genes tbx2-tbx5 during chick organogenesis. Mechanisms of Development, 74, 165-169. doi:10.1016/S0925-4773(98)00056-2
- Barlow, A.J. Bogardi, J.P. Ladher, R. and Francis-West, P.H. (1999) Expression of chick Barx-1 and its differential regulation by FGF-8 and BMP signaling in the maxilla primordia. Developmental Dynamics, 214, 291-302. doi:10.1002/(SICI)1097-0177(199904)214:4<291::AID-AJA2>3.0.CO;2-E
- Lan, Y. Liu, H., Ovitt, C.E., Wang, Q., Maltby, K.M. and Jiang, R. (2009) Distinct and synergistic roles of Osr1 and Osr2 in craniofacial development. Developmental Biology, 331, 490. doi:10.1016/j.ydbio.2009.05.390
- Papaioannou, V.E. and Silver, L.M. (1998) The T-box gene family. BioEssays, 20, 9-19. doi:10.1002/(SICI)1521-1878(199801)20:1<9::AID-BIES4>3.0.CO;2-Q
- Wardle, F.C. and Papaioannou, V.E. (2008) Teasing out T-box targets in early mesoderm. Genes & Development, 18, 418-425.
- Kiefer, J.C. (2004) The tbx files: The truth is out there. Developmental Dynamics, 231, 232-236. doi:10.1002/dvdy.20122
- Begemann, G., Gibert, Y., Meyer, A. and Ingham, P.W. (2002) Cloning of zebrafish T-box genes tbx15 and tbx18 and their expression during embryonic development. Developmental Dynamics, 114, 137-141. doi:10.1016/S0925-4773(02)00040-0
- Jezewski, P.A. Fang, P.K. Payne-Ferreira, T.L. and Yelick, P.C. (2009) Alternative splicing, phylogenetic analysis, and craniofacial expression of zebrafish tbx22. Developmental Dynamics, 238, 1605-1612. doi:10.1002/dvdy.21962
- Kochilas, L.K., Potluri, V., Gitler, A., Balasubramanian, K. and Chin, A.J. (2003) Cloning and characterization of zebrafish tbx1. Gene Expression Patterns, 3, 645-651.
- Funato, N., Nakamura, M., Richardson, J.A., Srivastava, D. and Yanagisawa, H. (2012) Tbx1 regulates oral epithelial adhesion and palatal development. Human Molecular Genetics, 21, 2524-2537. doi:10.1093/hmg/dds071
- Piotrowski, T., Ahn, D., Schilling, T.F., Nair, S., Ruvinsky, I., Geisler, R., Rauch, G., Haffter, P., Zon, L.I., Zhou, Y., Foott, H., Dawid, I.B. and Ho, R.K. (2003) The zebrafish van gogh mutation disrupts tbx1, which is involved in the DiGeorge deletion syndrome in humans. Development, 130, 5043-5052. doi:10.1242/dev.00704
- Braybrook, C., Doudney, K., Marcano, A.C.B., Arnason, A., Bjornsson, A., Patton, N.A., et al. (2001) The T-box transcription factor gene tbx22 is mutated in X-linked cleft palate and ankyloglossia. Nature Genetics, 29, 179- 183. doi:10.1038/ng730
- Haenig, B., Schmidt, C., Kraus, F., Pfordt, M. and Kispert, A. (2002) Cloning and expression analysis of the chick ortholog of tbx22, the gene mutated in X-linked cleft palate and ankyloglossia. Mechanisms of Development, 117, 321-325. doi:10.1016/S0925-4773(02)00196-X
- Marcano, A.C.B., Doudney, K., Braybrook, C., Squires, R., Patton, M.A., Lees, M., et al. (2004) TBX22 mutations are a frequent cause of cleft palate. Journal of Medical Genetics, 41, 68-74. doi:10.1136/jmg.2003.010868
- Herr, A., Meunier, D., Muller, I., Rump, A., Fundele, R., Hilger-Ropers, H., et al. (2003) Expression of mouse tbx22 supports its role in palatogenesis and glossogenesis. Developmental Dynamics, 226, 579-586. doi:10.1002/dvdy.10260
- Bush, J.O., Lan, Y., Maltby, K.M. and Jiang, R. (2002) Isolation and developmental expression analysis of tbx22, the mouse homolog of the human X-linked cleft palate gene. Developmental Dynamics, 225, 322-326. doi:10.1002/dvdy.10154
- Miller, C.T., Yelon, D., Stainer, D.Y.R. and Kimmel, C.B. (2003) Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and jaw joint. Development, 130, 1353-1365. doi:10.1242/dev.00339
- Whiteman, P. (1973) The quantitative measurement of alcian blue-glycosaminogycan complexes. Biochemical Journal, 131, 343-350.
- Thisse, C., Thisse, B., Schilling, T.F. and Postlethwait, J.H. (1993) Structure of the zebrafish snail1 gene and its expression in wild-type spadetail and no tail mutant embryos. Development, 119, 1203-1215.
- Payne-Ferreira, T.L. and Yelick, P.C. (2003) Alk8 is required for neural crest cell formation and development of pharyngeal arch cartilages. Developmental Dynamics, 228, 683-696. doi:10.1002/dvdy.10417
- Carney, T.J., Dutton, K.A., Greenhill, E., Delfino-Machin, M., Dufourcq, P., Blader, P., et al. (2006) A direct role for Sox10 in specification of neural crest derived sensory neurons. Development, 133, 4619-4630. doi:10.1242/dev.02668
- Chai, Y. and Maxson R.E. Jr. (2006) Recent advances in craniofacial morphogenesis. Developmental Dynamics, 235, 2353-2375. doi:10.1002/dvdy.20833
- Tena, J.J., Neto, A., de la Calle-Mustiennes, E., BrasPereira, C., Casares, F. and Gomez-Skarmeta, J.L. (2007) Odd-Skipped genes encode repressors that control kidney development. Development Biology, 301, 518-531. doi:10.1016/j.ydbio.2006.08.063
- Mudumana, S.P., Hentschel, D., Liu, Y., Vasilyev, A. and Drummond, I.A. (2008) Odd skipped related 1 reveals a novel role for endoderm in regulating kidney versus vascular cell fate. Development, 135, 3355-3367. doi:10.1242/dev.022830
- Neto, A., Mercader, N. and Gómez-Skarmeta, J.L. (2012) The Osr1 and Osr2 genes act in the pronephric anlage downstream of retinoic acid signaling and upstream of wnt2b to maintain pectoral fin development. Development, 139, 301-311. doi:10.1242/dev.074856
- Miller, C.T., Swartz, M.E., Khuu, P.A., Walker, M.B., Eberhart, J.K. and Kimmel, C.B. (2007) Mef2ca is required in cranial neural crest to affect Endothelin1 signaling in zebrafish. Development Biology, 308, 144-157. doi:10.1016/j.ydbio.2007.05.018
- Gao, Y., Lan, Y. and Jiang, R. (2011) The zinc finger transcription factors Osr1 and Osr2 control synovial joint formation. Development Biology, 352, 83-91. doi:10.1016/j.ydbio.2011.01.018
- Lan, Y., Liu, H., Ovitt, C.E. and Jiang, R. (2011) Generation of Osr1 conditional mutant mice. Genesis, 49, 419- 422. doi:10.1002/dvg.20734
- Kawai, S., Yamauchi, M., Wakisaka, S., Ooshima, T. and Amano, A. (2007). Zinc-finger transcription factor oddskipped related 2 is one of the regulators in osteoblast proliferation and bone formation. Journal of Bone and Mineral Research, 22, 1362-1372. doi:10.1359/jbmr.070602
- Wang, Q., Lan, Y., Cho, E.S., Maltby, K.M. and Jiang, R. (2005) Odd-skipped related 1 (Odd1) is an essential regulator of heart and urogenital development. Development Biology, 288, 582-594. doi:10.1016/j.ydbio.2005.09.024
- Zhang, Z., Lan, Y., Chai, Y. and Jiang, R. (2009). Antagonistic actions of Msx1and Osr2 pattern mammalian teeth into a single row. Science, 323, 1232-1234. doi:10.1126/science.1167418
- Ghosh, T.K., Song, F.F., Packham, E.A., Buxton, S., Robinson, T.E. and Ronksley, J. (2009) Physical interaction between TBX5 and MEF2C is required for early heart development. Molecular and Cellular Biology, 29, 2205- 2218. doi:10.1128/MCB.01923-08
- Piotrowski, T., Ahn, D.G., Schilling, T.F., Nair, S., Ruvinsky, I., Gisler, R., et al. (2003) The zebrafish van gogh mutation disrupts tbx1, which is involved in the DiGeorge deletion syndrome in human. Development, 130, 5043-5052. doi:10.1242/dev.00704
- Naiche, L.A., Harrelson, Z., Kelly, R.G. and Papaioannou, V.E. (2005) T-box genes in vertebrate development. Annual Review of Genetics, 39, 219-239. doi:10.1146/annurev.genet.39.073003.105925
- Swartz, M.E., Sheehan-Rooney, K., Dixon, M.J. and Eberhart, J.K. (2011) Examination of a palatogenic gene program in zebrafish. Developmental Dynamics, 240, 2204-2220. doi:10.1002/dvdy.22713
- Huang, P., Xiao, A., Zhou, M., Zhu, Z., Lin, S. and Zhang, B. (2011) Heritable gene targeting in zebrafish using customized TALENs. Nature Biotechnology, 29, 699-700. doi:10.1038/nbt.1939
- Moore, F.E., Reyon, D., Sander, J.D., Martinez, S.A., Blackburn, J.S., Khayter, C., et al. (2012) Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs). PLoS One, 7, e37877. doi:10.1371/journal.pone.0037877

