 J. Biomedical Science and Engineering, 2013, 6, 346-356 JBiSE http://dx.doi.org/10.4236/jbise.2013.63A044 Published Online March 2013 (http://www.scirp.org/journal/jbise/) A diffusion-weighted imaging based diagnostic system for early detection of prostate cancer Ahmad Firjani1,2, Ahmed Elnakib1, Fahmi Khalifa1, Georgy Gimel’farb3, Mohamed Abou El-Ghar4, Adel Elmaghraby2, Ayman El-Baz1* 1BioImaging Laboratory, Bioengineering Department, University of Louisville, Louisville, USA 2Department of Computer Engineering and Computer Science, University of Louisville, Louisville, USA 3Department of Computer Science, University of Auckland, Auckland, New Zealand 4Radiology Department, Urology and Nephrology Center, University of Mansoura, Mansoura, Egypt Email: *aselba01@exchange.louisville.edu Received 18 January 2013; revised 21 February 2013; accepted 1 March 2013 ABSTRACT A new framework for early diagnosis of prostate cancer using Diffusion-Weighted Imaging (DWI) is proposed. The proposed diagnostic approach consists of the following four steps to detect locations that are suspicious for prostate cancer: 1) In the first step, we isolate the prostate from the surrounding anatomical structures based on a Maximum A Posteriori (MAP) estimate of a new log-likelihood function that ac- counts for the shape priori, the spatial interaction, and the current appearance of prostate tissues and its background (surrounding anatomical structures); 2) In order to take into account any local deformation between the segmented prostates at different b-values that could occur during the scanning process due to local motion, a non-rigid registration algorithm is employed; 3) A KNN-based classifier is used to clas- sify the prostate into benign or malignant based on three appearance features extracted from registered images; and 4) The tumor boundaries are determined using a level set deformable model controlled by the diffusion information and the spatial interactions between the prostate voxels. Preliminary experiments on 28 patients (17 malignant and 11 benign) resulted in 100% correct classification, showing that the pro- posed method is a promising supplement to current technologies (biopsy-based diagnostic systems) for the early diagnosis of prostate cancer. Keywords: Prostate Cancer; 3D Markov-Gibbs Random Field; Nonrigid Registration; Diffusion-Weighted Imaging 1. INTRODUCTION Prostate cancer is a major health problem, and the most frequently diagnosed malignancy in the American male population [1]. Recent prostate cancer studies reported an estimated 241,740 new cases and a mortality rate of close to 28,170 in 2012 [2]. Fortunately, early diagnosis of prostate cancer increases the survival rate of the pa- tients [3]. 1.1. Current Imaging Modalities for Prostate Cancer Diagnosis Currently, there are different techniques that are used for early diagnosis of prostate cancer. However, the accuracy of these techniques are clearly unsatisfactory. For exam- ple, Prostate Specific Antigen (PSA) screening doesn’t offer accurate information about the location and extent of the lesion(s) [4]. In add ition, PSA is associated with a high risk of over di agnosis of p rostate can c er. On the other hand, imaging tests using different imag- ing modalities, such as Transrectal Ultrasound (TRUS) [5], Computed Tomography (CT) [6], MR Spectroscopy (MRS) [7], Dynamic-Contrast Enhanced Magnetic Reso- nance Imaging (DCE-MRI) [8], and Diffusion-Weighted Imaging (DWI) [9] are still critically needed. TRUS im- aging [10] is widely used for guided needle biopsy due to the real time nature of the imaging system, ease of use, and portability. However, TRUS images have low Sig- nal-to-Noise Ratio (SNR) making it difficult to detect malignant tissues [11]. Another traditional imaging mo- dality is CT. It is widely used for diagnosis and follow- up of prostate cancer [12]. However, it has poor soft- tissue contrast resolution that does not allow precise dis- tinction of the internal or external anatomy of the pros- tate and thus CT images have shown limited specificity for prostate dia gn osi s [1 3] . On the other hand, MR image-based modalities, such as T2-weighted MR, MRS, DCE-MRI, and DWI, have also been widely employed for early detection of prostate cancer [14]. Despite widely use of T2-weighted MR im- ga*Corresponding author. OPEN ACCESS  A. Firjani et al. / J. Biomedical Science and Engineering 6 (2013) 346-356 347 aging for prostate cancer, the technique is limited by un- satisfactory sensitivity and specificity for cancer detec- tion and localization [15]. To improve the diagnostic performance of MR imaging in evaluations for prostate cancer, various other techniques have been applied. MRS provides metabolic information about prostate tissue by demonstrating the relative concentration of chemical compounds. However, MRS has its own limitations, such as the need of an additional software and longer acquisi- tion time [16], which lead to increased costs and de- creased throughput. Furthermore, MRS suffers from lack of spatial resolution. In add ition, signal from periprostatic fat and seminal vesicles can distort spectral waveforms [14]. DCE-MRI has been recently suggested for im- proved visualization and localization of the prostate can- cer [17]. It provides valuable pathologic and anatomical information. However, DCE-MRI has the drawback of intravenous contrast agent (e.g., gadolinium) administra- tion which is harmful to the kidney [18] and requires a longer setup time. Recently, DWI has emerged as an imaging modality that has shown more capabilities in determining the size and the shape of the prostate gland and localizing the cancer foci [19]. DWI is non-contrast functional imaging technique, whereby the image contrast is determined by the random microscopic motion of water protons, i.e., the Brownian motion [19]. Moreover, DWI has the distinct advantage of being acquired very rapidly, without the use of any intravenous contrast material or specialized hard- ware, and this is the main motivation behind this work. 1.2. Clinical Studies for Prostate Cancer Diagnosis Using DWI In recent years, a growing number of clinical studies [19- 30] have evaluated the utility of DWI, either in combina- tion with or in comparison with other MRI techniques, for the detection of prostate cancer. These studies have reported various sensitivities and specificities of cancer diagnosis. Earlier studies [19,20] have investigated the abilities of DWI for prostate cancer diagnosis using an endorectal coil. However, the reported results demonstrated low diagnostic sensitivity. To increase the sensitivity of di- agnosis, Shimofusa et al. [21] suggested the addition of strong magnetic field gradient pulses (b-values) to the pulse sequence instead of using endorectal coil. In their study [21], they detected the tumor in the central zone of the prostate in five of eight total patients using DWI with strong magnetic field gradient pulses. Alternatively, the compared diagnostic results with T2-weighted imaging, detected the tumor only in one of the eight patients. Since then, DWI was used for the detection of cancerous tissue in later studies [22-30]. For example, Tan et al. [30] compared the performance of T2-weighted MRI, DCE- MRI, and DWI for the detection of cancer within the prostate gland. In their study they reported that DWI alone showed better specificity than DCE-MRI alone. It is also showed better overall specificity than combined DWI and T2-weighted imaging. To the best of our knowledge, there are a very limited number of image-based approaches for automated com- puter-aided diagnosis of prostate cancer using DWI. These related works are discussed in the following sec- tion. 1.3. Image-Based Computer-Aided Diagnostic (CAD) Systems for Prostate Cancer Detection In literature, a limited number of CAD systems for pros- tate cancer diagnosis have been proposed. For example, Chan et al. [31] proposed an in-vivo CAD system using multimodal MRI to estimate malignancy likelihood in the peripheral zone. They constructed statistical maps from T2-weighted MRI, DWI, and Proton Density (PD) images. These statistic maps were combined with tex- tural and anatomical features of prostate cancer areas in order to detect the cancerous regions. However, this study doesn’t include benign regions. Huisman et al. [32] developed a CAD system for prostate lesion classifica- tion using a Hessian-based blob detection algorithm [33]. Results showed an accuracy of 92% in classification within the peripheral region and an accuracy of 83% in classification within transitional zones of the prostate. However, their study focused on the peripheral and tran- sitional zones of the prostate gland and excluded central zones in which up to 30% of prostate cancers can occur. Viswanath et al. [34] generated similar likelihood maps by combining information from multimodal MR images using mathematical descriptors. Their study showed, on a voxel basis, that the discrimination be- tween benign and malignant tissue is feasible with good performances. The unsupervised classification by k- means clustering achieved an accuracy of 77%. Unfor- tunately, th e corresponding slice still needs to be selected between different modality. A study by Langer et al. [35] focused on the peripheral zone of the prostate gland and excluded the central and transitional zones. However, detailed anatomic studies have suggested that 70% of cancers arise in the peripheral zone of the prostate, but up to 30% of prostate cancers occur between transition zones and the central zone of the prostate [36]. To increase the sensitivity of diagnosis, accurate de- lineation of the prostate region is mandatory. Basically, manual outlining of the prostate borders is the most ac- curate segmentation that en ables pr ecise determination o f the prostate volume. However, it is prohibitively time Copyright © 2013 SciRes. OPEN ACCESS  A. Firjani et al. / J. Biomedical Science and Engineering 6 (2013) 346-356 348 consuming and is prone to intra- and inter-observer vari- ability. Traditional edge detection methods (e.g., [39]) are unable to extract the correct boundaries of the pros- tate since the gray-level distributions of the prostate and the surrounding organs are hardly distinguishable. There- fore, other automated segmentation methods are desir- able. However, multiple challenges stemming from 1) the large variations of prostate shape within a specific time series as well as across subjects; 2) lack of strong edges and diffused prostate boundaries; and 3) the simi- lar signal-intensity profile of the prostate and surround- ing tissues, complicates the segmentation process. The most successful known approaches (e.g., [37-45]) have addressed the segmentation challenges of the pros- tate by modeling object appearance and shape. In par- ticular, Zhu et al. [40] used a combination of an Active Shape Model (ASM) and 3D statistical shape modeling to segment the prostate. Toth et al. [41] presented an al- gorithm for the automatic segmentation of the pros tate in multi-modal MRI. Their algorithm starts by isolating the Region-Of-Interest (ROI) from MRS data. Then, an ASM within the ROI is used to obtain the final segmentation. Klein et al. [42] presented an atlas-based segmentation approach to extract the prostate from MR images. The segmentation of the prostate is obtained as the average of the best-matched registered atlas set to the test image (image to be segmented). Recently, Vikal et al. [43] used a priori knowledge of prostate shape to detect the contour in each slice and then refined them to form a 3D prostate surface. Martin et al. [44] developed an atlas-based ap- proach for segmenting the prostate from 3D MR images by mapping probabilistic anatomical atlas to the test im- age. The resulting map is used to constrain a deformable model-based segmentation framework. 1.4. Current Limitations and Motivation for Our Proposal The above-mentioned CAD systems for analyzing DWI are not sufficiently accurate and reliable for several rea- sons: 1) The majority of CAD systems used multimodal MRI which is cost inefficient [45]. 2) The majority of these studies require user interac- tion to select a ROI (a small window) around the pro state. Unfortunately, such approaches not only prone to inter- observer variability, but also ROI selection biases the final decision by over- or under-estimating the problem in the entire graft, just as with biopsy. 3) Automated prostate segmentation methods have one of the following limitations: Deformable model-based methods without adequate appearance and shape priors fail under excessive noise, poor resolution, diffused boundaries, or oc- cluded shapes in the images; Segmentation based only on the shape prior still re- sults in large errors caused by discontinuities in ob- ject boundaries, large image noise, and other inho- mogeneities; Parametric shape-based models are unsuitable for discontinuous prostate objects due to a very small number of distinct landmarks. 4) The majority of CADs assumes that the prostates (prostate contours) remain exactly the same from scan to scan. However, prostate contours may not always exactly match due to patient movement or breathing effects; therefore, image registration schemes should be applied first before ROI selection/segmentation. To overcome these limitations, we propose an auto- matic framework for analyzing DWI images building on our previous work in [46-48]. The proposed approach consists of the following steps as shown in Figur e 1: 1) Segmentation of the prostate from DWI (Section 2.1) based on a Maximum a Posteriori (MAP) estimate of a new likelihood function that accounts for both ap- pearance features of the prostate (Section 2.1.1) and th eir 3D spatial voxel interactions (Section 2.1.2), as well as a 3D shape prior (Section 2.1.3). 2) A non-rigid registration approach is employed to account for any local deformation that could occur in the prostate during the scanning process based on the solu- tion of the Laplace equation (Section 2.3). 3) KNN classifier to classify the prostate into benign or malignant based on three appearance features ex- tracted from registered images (Section 3.2). 2. MATERIALS AND METHODS In this paper we introduce a new, automated, and non- invasive framework for early diagnosis of prostate cancer from DWI. Figure 1 demonstrates the steps of the pro- posed CAD system. Below, we will illustrate each of these steps. 2.1. Segmentation of the Prostate Using a Joint MGRF Model The segmentation of the prostate is a challenge, since the gray-level distribution of the prostate and surrounding organs is not highly distinguishable and because of the anatomical complexity of prostate. This stage proposes a powerful framework for prostate segmentation based on a learned shape model and an identifiable joint Markov- Gibbs Random Field (MGRF) model of DWI and “ob- ject-background” region maps. The joint-MGRF model is fundamentally a model that relates the joint probability of an image and its object- background region map, to geometric structure and to the nergy of repeated patterns within the image. The basic e Copyright © 2013 SciRes. OPEN ACCESS  A. Firjani et al. / J. Biomedical Science and Engineering 6 (2013) 346-356 Copyright © 2013 SciRes. 349 Figure 1. Flowchart of the proposed CAD system for automatic detection of cancer from 3D DWI. theory behind such models is that they assume that the signals associated with each pixel depend on the signals of the neighboring pixels, and thus explicitly take into account their spatial interactions, and other features, such as the shape. Let , , and 0,1, ,1QQ ,ob bgL 0, 1U be a set of integer gray-level, a set of object (“ob”) and background (“bg”) labels, and a unit interval, respec- tively. Let a 3D arithmetic grid Q ,,:0,1, ,1; 0,1, ,1;0,1, ,1 xyz xX yYzZ R Figure 2. Aligning a 3-D joint Markov-Gibbs random field model with shape prior of DWI. support a grayscale DWI and their binary region maps , and probabilistic shape model . The shape model allows for registering (aligning) 3D prostate DWI. The DWI data :gR Q :mR L :sR U and their region maps are described with a joint probability model [49,50]: m shape model provides the voxel-wise object and back- ground probabilities be ing used, together with the condi- tional image intensity model P m, to build an initial region map. The final Bayesian segmentation is per- formed using the identified joint MGRF model of the DWI and region maps. ,h PPP m mm (1) where is a 2nd-order MGRF of region maps and h Pm P m is a conditionally independent random field of image intensities given the map. The map model 2.1.1. Con dition al Intensity M o d el The specific visual appearance of the prostate in each data set to be segmented is taken in to account by model- ing a marginal gray level distribution with a Linear Com- bination of Discrete Gaussians (LCDG) [50,51]. Close approximation with LCDG separates each factor of the joint empirical gray level distribution, hs PPPmm s Pm m has two parts: a shape prior prob- ability being a spatially varian t independen t random field of region labels , for a set of co-aligned training DWI data, and a 2nd-order MGRF model Pm of a spatially homogeneous evolving map. ,x y Pp g ,mix xy R, into two (object and back- ground) components, The Bayesian MAP estimate of the map, given the DWI data ,argmax,gmL gmm maximize the log- likelihoo d f unction: g ;,andq QLpq . The LCDG modeling restores transitions between these components more accurately than conventional mixtures of only positive Gaussians, thus yielding a better initial region map formed by voxel-wise classification of the image gray values, the similar intensity profile of the prostate and surrounding tissues. ,log log h LP P m mm In this work we focus on accu rate identification of the spatial interaction between the prostate voxels Pm, and the intensity distribution for the prostate tissues, P m, and the prior distribution of the pros- tate shape, as shown in Figure 2. s Pm2.1.2. Spatial Voxel In te raction M odel In order to overcome noise effect and to ensure the ho- mogeneity of the segmentation, spatially voxel interac- To perform the initial prostate segmentation, a given 3D DWI is aligned to one of the training 3D DWI. The OPEN ACCESS  A. Firjani et al. / J. Biomedical Science and Engineering 6 (2013) 346-356 350 tions between the region labels are also taken into ac- count using the popular Potts model, i.e., the MGRF with the nearest voxel 2 6-neighborhood (see Figure 3). A generic MGRF of region maps accounts only for pairwise interaction between each region label and its characteristic neighbors. Generally, the interaction struc- ture and the Gibbs potentials can be arbitrary and are identified from the training data. By symmetry considerations, we assume that the po- tentials are independent of relative orientation of each voxel pair and depend only on intra- or inter-region posi- tion (i.e. whether the labels are equal or not). Under these restrictions, it is the 3D extension of the conventional auto-binomial, or Potts model differing only in that the potentials are estimated analyt ically. The 26-neighborhood has three types of symmetric pairwise interaction s specified by the absolute distan ce a between two voxels in the same and adjacent MRI slices (, 1a2, and 3, respectively): 1) the closest pairs with the inter-voxel co- ordinate offsets; 2) the diagonal pairs with the offsets 11,0,0, 0,1,0 , 0,0,1N 0,1,1,1, 0N , 1,1, 1,0 2; and 3) the farthest diagonal pairs with the offsets 3. The Gibbs potentials of each type are bi-valued because only label coincidence is accounted for: where if l 1, 1, 1 ; aaeq VVV N ,, aeqa VVll l ,,ane and ll ,, ane a VV if ll; 1,2, 3aA. Then the MGRF model of region maps is as follows [52,53]: ,, , ,, ,, 1exp , a haxy xyzak PVm z RA N m zxyzk m (2) where Z is the normalizing factor (partition function). To id en tif y th e MGR F in Eq.1, approximate analytical maximum likelihood estimate of th e 3D Gibbs potentials, and are derived in line with [52]: ,aeq V,ane V Figure 3. Pairwise voxel interac- tion for 26 neighborhood system in a 3D GGMRF. The reference voxel is shown in red. ,,, 1 22 aeqaneaeq VV fm (3) where, ,aeq m denotes the relative frequency of the equal labels in the equivalent voxel pairs ,, ,,,: ,,; ,,;,, a xyz xyzxyz xyz R RN of a region map of a given DWI aligned in accord with the prior shape model. m 2.1.3. Probabilistic Shape Model To enhance the segmentation accuracy, the expected shape of the goal object is constrained with a soft prob- abilistic 3D prostate shape model. Initially, a training database collected from different subjects are co-aligned by rigid, affine 3-D transformations. The shape prior is a spatially variant independent random field of region la- bels: ,, ,, mx y z xyz Ps R m where is the empirical probability that the voxel (x, y, z) belongs to the prostate (L = “ob”) or the backgro und ( L = “bg”) given the map. To enhance the segmentation of the current prostate volume, the prior probabilistic shape model is updated by adding the previous segmented 3D prostate data to the prior calculated shape model. The proposed prostate segmentation process can be summa- rize as follows: Perform an affine alignment of a given 3D MRI to an arbitrary prot ot ype prost ate f rom t he traini ng set usi ng mutual information [54] as a similarity measure to obtain the learned probabilistic shape model s Pm; Estimate the conditional intensity model Pgm by identifying the bimodal LCDG; Use the intensity model found and the learned prob- abilistic shape model to perform an initial segmenta- tion of the pros t a t e , i.e., to form an initial region map; Use the initial region map to estimate the potential for the Potts model and to identify the MGRF model Pm of region maps; Improve the region map using voxel-wise stochastic relaxation (Iterative Conditional Mode (ICM) [55]) through successive iterations to maximize the log likelihood function of Eq.1 until the log likelihood remains almost the same for two successive iterations; Output: The 3D prostate segmentation is the final estimate m. 2.2. Performance Evaluation of the Proposed Segmentation Algorithm The proposed segmentation is evaluated based on char- Copyright © 2013 SciRes. OPEN ACCESS 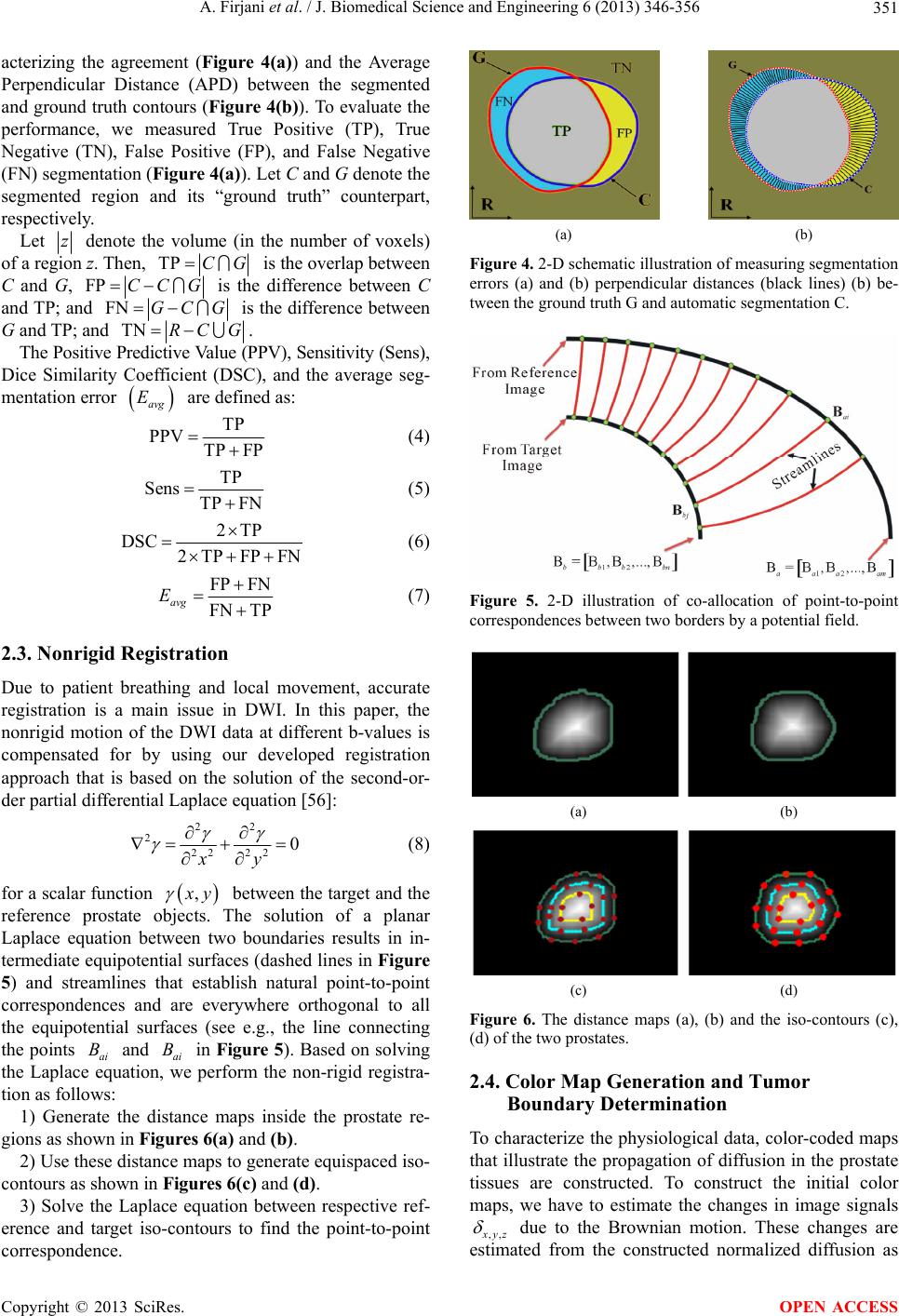 A. Firjani et al. / J. Biomedical Science and Engineering 6 (2013) 346-356 351 acterizing the agreement (Figure 4(a)) and the Average Perpendicular Distance (APD) between the segmented and ground truth contours (Figure 4(b)). To evaluate the performance, we measured True Positive (TP), True Negative (TN), False Positive (FP), and False Negative (FN) segmentation (Figure 4(a)). Let C and G denote the segmented region and its “ground truth” counterpart, respectively. Let z denote the volume (in the number of voxels) of a region z. Then, TPCG is the overlap between C and G, FP CCG is the difference between C and TP; and FNGC G is the differe nce between G and TP; and TNRC G. The Positive Predictiv e Value (PPV), Sensitivity (Sens), Dice Similarity Coefficient (DSC), and the average seg- mentation error are defined as: avg E TP PPV TP FP (4) TP Sens TP FN (5) 2TP DSC 2TPFPFN (6) FP FN FN TP avg E (7) 2.3. Nonrigid Registration Due to patient breathing and local movement, accurate registration is a main issue in DWI. In this paper, the nonrigid motion of the DWI data at different b-values is compensated for by using our developed registration approach that is based on the solution of the second-or- der partial differential Laplace equation [56]: 22 222220 xy (8) for a scalar function , y B between the target and the reference prostate objects. The solution of a planar Laplace equation between two boundaries results in in- termediate equipotential surfaces (dashed lines in Figure 5) and streamlines that establish natural point-to-point correspondences and are everywhere orthogonal to all the equipotential surfaces (see e.g., the line connecting the points ai and ai in Figure 5). Based on solving the Laplace equation, we perform the non-rigid registra- tion as follows: B 1) Generate the distance maps inside the prostate re- gions as shown in Figures 6(a) and (b). 2) Use these distance maps to generate equispaced iso- contours as shown in Figures 6(c) and (d). 3) Solve the Laplace equation between respective ref- erence and target iso-contours to find the point-to-point correspondence. (a) (b) Figure 4. 2-D schematic illustration of measuring segmentation errors (a) and (b) perpendicular distances (black lines) (b) be- tween the ground truth G and automatic segmentation C. Figure 5. 2-D illustration of co-allocation of point-to-point correspondences between two borders by a potential field. (a) (b) (c) (d) Figure 6. The distance maps (a), (b) and the iso-contours (c), (d) of the two prostates. 2.4. Color Map Generation and Tumor Boundary Determination To characterize the physiological data, color-coded maps that illustrate the propagation of diffusion in the prostate tissues are constructed. To construct the initial color maps, we have to estimate the changes in image signals ,, yz due to the Brownian motion. These changes are estimated from the constructed normalized diffusion as Copyright © 2013 SciRes. OPEN ACCESS  A. Firjani et al. / J. Biomedical Science and Engineering 6 (2013) 346-356 352 the difference between the signals of image sequences at two different b-values. DWI is performed with at least two b values, including a b value of 0 sec/mm2 and a higher b value of 500 - 1000 s/mm2 depending on the body region or organ being imaged [57]. At b = 0 s/mm2, there is no diffusion sensitizing gradient with free water molecules have high signal intensity. We used b = 800 s/mm2 because imaging of solid organs requires high b value specially in prostate and using h igh b values allows differentiation of areas of restricted from the normal high signal at the peripheral zone. During our trials we found the b = 800 s/mm2 allows lesions differentiation with least degradation of image quality as the image quality decrease with the high b values. To preserve continuity (remove inconsistencies), the initial estimated ,, yz values are considered as samples from a Generalized Gauss-Markov Random Field (GGMRF) image model [58] of measurements with the 26-voxel neighborhood (Figure 3). Continuity of the constructed 3-D volu me (Figure 7) is amplified by using their MAP estimates [51]: ,, ,, ,,,, ,, ,,, , ,,,, , ,, ˆargmin xyz xyz xyzxyzxyz xyzxy z xyzx y z xyz where ,, yz and ,, yz denote the original values and their expected estimates, ,, yz v is the 26-neighborhood voxel set (Figure 7), ,,,, , yzx y z is the GGMRF po- tential, and and are scaling factors. The parame- ter 1.01,2 2.0 controls the level of smoothing (e.g., smooth, , vs. relatively abrupt edges, 1.01 ). The parameter determines the Gaussian, 1, 2 2 , or Laplace, 2 , prior distribution of the esti- mator. Then, the color maps are generated based on the final estimated (see Figure 8). Finally, to allocate the boundary of the detected tu- mors, which is important to determine the cancer stag e in case of malignancy, we used a level set-based deform- able model controlled by a stochastic speed function [59]. The latter accounts for the perfusion information and 3-D GGMRF For Continuity Preservation Final Estimation Initial Estimation Figure 7. Enhanced perfusion estimation and continuity analy- sis using the 3-D GGMRF image model. Color Scale Before 3-D GGMRFAfter 3- D GGMRF Figure 8. Color-coded maps for three of the test subjects (col- umnwise) before and after the 3-D GGMRF smoothing with ρ = 1, λ = 5, β = 1:01, α = 2, and ,, ,, ,2 xyzx y z and their re- spective color-coded maps. The red and blue ends of the color scale relate to the maximum and minimum changes, respec- tively. spatial interactions between the prostate voxels. 3. EXPERIMENTAL RESULTS The performance of the proposed framework has been evaluated by applying it on DWI prostate images col- lected from 28 patients. These patients had biopsy- proven prostate cancer and underwent DWI at 1.5-T (SIGNA Horizon, General Electric Medical Systems, Milwaukee, WIS). DWI were then obtained using mono- directional gradients and a multi-section Fast Spin Echo type (FSE) echo-plan ar sequence in the axial plan e using a body coil with the following imaging parameters: TE: 84:6 ms; TR: 8.000 ms; Band Width 142 kHz; FOV 34 cm; slice thickness 3 mm; inter-slice gap 0 mm; seven excitations, water excitatio n with b value of 0 s/mm2 and 800 s/mm2. Fifty four slices were obtained in 120 secon d. to cover the prostate in each patient. Note all the subjects were diagnosed using a biopsy (ground truth). 3.1. Segmentation Results The proposed segmentation approach has been tested on 28 independent data sets of DWI images. Figure 9 shows sample examples of prostate segmentation from different data sets, with respect to the ground truth segmentation. The ground truths were obtained by manual delineation of the prostate borders by an MR imaging expert. To highlight the advantages of our segmentation technique, we compare it to the shape-b ased segmentation approach proposed by Tsai et al. [60]. We re-implemented the method described in [60] and tested it on our locally- acquired data. Figure 9 compares qualitatively the accuracy of our approach and the shape-based approach [60] with respect to the ground truth. The segmentation accuracy for all data sets has been evaluated using the average segmenta- tion error, given by Eq.7. Differences between the mean Copyright © 2013 SciRes. OPEN ACCESS  A. Firjani et al. / J. Biomedical Science and Engineering 6 (2013) 346-356 353 (a) (b) (c) Figure 9. 3-D prostate segmentation projected onto 2-D. (a) Our segmentation (red) in comparison with the ground truth (white); (b) The segmentation with the algorithm in [60] (red) comparison with the ground truth; and (c) 2-D visualization for our segmented prostates for three of the test subjects. errors for our segmentation and the shape-based ap- proach [60] in Ta ble 1 are statistically significant by the unpaired t-test and thus highlight the advantages of the proposed integration of the shape prior, prostate/back- ground marginal intensity distributions, and spatial in- teraction characteristics into MAP-based seg mentation. Moreover, the accuracy of our segmentation approach has been evaluated, with respect to the expert tracing, using the PPV, Sens, DSC [61], and the APD between the borders of ground truth G and automatic segmentation C (see Figure 10). Table 2 compares the segmentation over all the test data sets with the ground truth obtained by manual tracing by an imaging exp e rt. 3.2. Diagnostic Results The ultimate goal of the proposed framework is to dis- tinguish between benign and malignant detected tumors. The malignant tissues show higher signal intensity with a b-value of 800 s/mm2, and a lower Apparent Diffusion Coefficient (ADC) compared with benign and normal tissue due to the replacement of normal tissue. To dis- tinguish between the benign and malignant cases, we used a KNN classifier learning statistical characteristics Table 1. A comparative segmentation accuracy over all test data sets for our approach and [60]. Note that “STD” stands for standard deviation. Eavg% Algorithm Our [60] Min. Error % 0 0 Max. Error % 1.6005 2.7724 Mean Error % 0.5500 1.46 15 STD. % 0.3085 0.7687 P-value 0.0001 Table 2. Error statistics over all test data sets. Note that “STD” stands for standard deviation and “APD” values are in mm. Performance measures PPV Sens DSC APD Min. 0.857 0.882 0.841 0.00 Max 0.991 0.851 0.930 3.1 Mean 0.952 0.816 0.991 0.60 STD. 0.004 0.004 0.004 0.80 Figure 10. Prostate image with ground truth (blue) and automatic segmenta- tion (green) contours, and their associ- ated streamlines (red) obtained by the solution of the Laplace equation yield- ing the estimation of the APD. of the DWI. The characteristics are obtained from the training sets containing both benign and malignant cases. After training, three features namely are the mean inten- sity value of the DWI at 0 s/mm2, the mean intensity value of the DWI at 800 s/mm2, and the mean value of ADC maps [62], were chosen to classify the test cases. To build the KNN classifier that characterizes the prostate tissue, we used 13 subjects for training, and the other 15 subjects for testing. The diagnostic accuracy based on the combined three features resulted in correct classifications of all 28 data sets (i.e., 100% accuracy). Copyright © 2013 SciRes. OPEN ACCESS  A. Firjani et al. / J. Biomedical Science and Engineering 6 (2013) 346-356 354 (B) (M) Figure 11. Tumor’s contour determination (green) using the level set approach for multiple image sections for benign (B) and malignant (M) subjects. For regional display we explore pixel-by-pixel maps of the registered diffusion data. The diffusion was com- puted for each pixel and superimposed on an image slice to form a parametric image. Also, for visual assessment of the prostate tumor the tumor con tours are determined. Figure 11 shows the tumor contours determination for selected image sections for two subjects involved in our study. 4. CONCLUSIONS AND DISCUSSION In this paper, we present a novel fully automatic frame- work for detecting prostate cancer using DWI. The framework includes prostate segmentation, nonrigid reg- istration, and KNN-based classification. For prostate segmentation, we introduced a new 3D approach that is based on a MAP estimate of a new log-likelihood func- tion that accounts for the sh ape priori, the sp atial interac- tion, and the current appearance of the prostate and its background which increases the accuracy of automatic segmentation, evidenced by the error and the DSC analysis (Tables 1 and 2). Following segmentation, we used a nonrigid registration approach that deforms the prostate object on iso-contours instead of a square lattice, which provides higher degrees of freedom to obtain ac- curate deformation. In the classification step, the seg- mented prostate regions are classified into malignant or benign using the KNN classifier. Applications of the proposed framework can assist the radiologist in detect- ing all prostate cancer locations and could replace the use of current technologies to determine the type of prostate cancer. Although we have obtained promising results in this initial study using DWI data in 28 patients, potential widespread adoption would require confirmation by other groups, and investigation in a larger number of subjects. Our future work will focus on comparing the diagnostic accuracy of prostate cancer detection using other imaging modalities, such as DCE-MRI. REFERENCES [1] Siegel, R., DeSantis, C., Virgo, K., Stein, K., Mariotto, A., Smith, T., et al. (2012) Cancer treatment and survivorship statistics. A Cancer Journal for Clinicians, 62, 220-241. [2] American Cancer Society (2012) Cancer treatment and survivorship facts & figures 2012-2013. http://www.cancer.org/acs [3] Deem, S., DeFade, B., Lohri, J., Ti erney, J., M odak, A. and Emmett, M. (2010) Prostate cancer screening: A primary care survey. Health, 2, 1179-1183. doi:10.4236/health.2010.210173 [4] Stenman, U.H., Leinonen, J., Zhang, W.M. and Finne, P. (1999) Prostate-specific antigen. Seminars in Cancer Bio- logy, 9, 83-93. doi:10.1006/scbi.1998.0086 [5] Halpern, E.J. (2006) Contrast-enhanced ultrasound imag- ing of prostate cancer. Urology, 8, 29-37. [6] Ives, E.P., Burke, M.A., Edmonds, P.R., Gomella, L.G. and Halpern, E.J. (2005) Quantitative computed tomo- graphy perfusion of prostate cancer: Correlation with whole-mount pat hology. Clinical Prostate Cancer, 4, 109- 112. doi:10.3816/CGC.2005.n.018 [7] Shah, N., Sattar, A., Benanti, M., Hollander, S. and Cheuck, L. (2006) Magnetic resonance spectroscopy as an imag- ing tool for cancer: A review of the literature. Journal of the American Osteopathic Association, 106, 23-27. [8] Noworolski, S.M., Henry , R.G., Vigneron, D.B. and Kur- hanewicz, J. (2005) Dynamic contrast-enhanced MRI in normal and abnormal prostate tissues as defined by b io p sy , MRI, and 3D MRSI. Magnetic Resonance in Medicine, 53, 249-255. doi:10.1002/mrm.20374 [9] Kajihara, H., Hayashida, Y., Murakami, R., Katahira, K., Nishimura, R., Hamada, Y., et al. (2009) Usefulness of diffusion-weighted imaging in the localization of prostate cancer. International Journal of Radiation Oncology Bio- logy Physics, 74, 399-403. doi:10.1016/j.ijrobp.2008.08.017 [10] Ellis, W.J. and Brawer, M.K. (1995) Repeat prostate nee- dle biopsy: Who needs it? Journal of Urology, 153, 1496-1498. doi:10.1016/S0022-5347(01)67444-5 [11] Applewhite, J.C., Matlaga, B.R., McCullough, D.L. and Hall, M.C. (2001) Transrectal ultrasound and biopsy in the early diagnosis of prostate cancer. Cancer Control Journal, 8, 141-150. [12] Gossner, J. (2012) Computed tomography of the prostate- A review. The Internet Journal of Radiology, 14, 1. [13] Bouchelouche, K., Turkbey, B., Choyke, P. and Capala, J. (2010) Imaging prostate cancer: An update on positron emission tomography and magnetic resonance imaging. Current Urology Reports, 11, 180-190. doi:10.1007/s11934-010-0105-9 [14] Fuchsjager, M., Shukla-Dave, A., Akin, O., Barentsz, J. and Hricak, H. (2008) Prostate cancer imaging. Acta Ra- diologica, 49, 107-120. doi:10.1080/02841850701545821 [15] Choi, Y.J., Kim, J.K., Kim, N., Kim, K.W., Choi, E.K. Copyright © 2013 SciRes. OPEN ACCESS  A. Firjani et al. / J. Biomedical Science and Engineering 6 (2013) 346-356 355 and Cho, K.S. (2007) Functional MR imaging of prostate cancer. RadioGraphics, 27, 63-75. doi:10.1148/rg.271065078 [16] Zakian, K.L., Eberhardt, S., Hricak, H., Shukla-Dave, A., Kleinman, S., Muruganandham, M., et al. (2003) Tran- sition zone prostate cancer: Metabolic characteristics at 1H MR spectroscopic imaging-initial results. Radiology, 229, 241-247. doi:10.1148/radiol.2291021383 [17] Punwani, S., Emberton, M., Walkden, M., Sohaib, A., Freeman, A., Ahmed, H., et al. (2012) Prostatic cancer surveillance following whole-gland high-intensity focused ultrasound: Comparison of MRI and prostate-specific an- tigen for detection of residual or recurrent disease. British Journal of Radiology, 85, 720-728. doi:10.1259/bjr/61380797 [18] Bhave, G., Lewis, Julia, B. and Chang, S. (2008) Asso- ciation of gadolinium based magnetic resonance imaging contrast agents and nephrogenic systemic fibrosis. Jour- nal of Urology, 180, 830-835. doi:10.1016/j.juro.2008.05.005 [19] Yu, K.K. and Hricak, H. (2000) Imaging prostate cancer. Radiologic Clinics of North America, 38, 59-85. doi:10.1016/S0033-8389(05)70150-0 [20] Ikonen, S., Karkkainen, P., Kivisaari, L., Salo, J.O., Taari, K., Vehmas, T., et al. (2001) Endorectal magnetic re-so- nance imaging of prostatic cancer: Comparison between fat-suppressed T2-weighted fast spin echo and three-di- mensional dual-echo, steady-state sequences. European Radiology, 11, 236-241. doi:10.1007/s003300000598 [21] Shimofusa, R., Fujimoto, H., Akamata, H., Motoori, K., Yamamoto, S. , Ueda, T ., et al. (2005) Diffusion-weighted imaging of prostate cancer. Journal of Computer Assisted Tomography, 29, 149-153. doi:10.1097/01.rct.0000156396.13522.f2 [22] Hacklander, T., Scharwachter, C., Golz, R. and Mertens, H. (2006) Value of diffusion-weighted imaging for diag- nosing vertebral metastases due to prostate cancer in comparison to other primary tumors. Rofo, 178, 416-424. [23] Yoshimitsu, K., Kiyoshima, K., Irie, H., Tajima, T., Asayama, Y., Hirakawa, M., et al. (2008) Usefulness of apparent diffusion coefficient map in diagnosing prostate carcinoma: Correlation with stepwise histopathology. Jour- nal of Magnetic Resonance Imaging, 27, 132-139. doi:10.1002/jmri.21181 [24] Lim, H.K., Kim, J.K., Kim, K.A. and Cho, K.S. (2009) Prostate cancer: Apparent diffusion coefficient map with T2-weighted images for detection a multireader study. Radiology, 250, 145-151. doi:10.1148/radiol.2501080207 [25] Haider, M.A., van der Kwast, T.H., Tanguay, J., Evans, A.J., Hashmi, A.T., Lockwood, G., et al. (2007) Com- bined T2-weighted and diffusion-weighted MRI for loca- lization of prostate cancer. American Journal of Roent- genology, 189, 323-328. doi:10.2214/AJR.07.2211 [26] Padhani, A.R., Liu, G., Koh, D.M., Chenevert, T.L., Thoeny, H.C., Takahara, T., et al. (2009) Diffusion- weighted magnetic resonance imaging as a cancer bio- marker: Consensus and recommendations. Neoplasia, 11, 102-125. [27] Kitajima, K., Kaji, Y., Fukabori, Y., Yoshida, K., Suga- numa, N. and Sugimura, K. (2010) Prostate cancer detec- tion with 3T MRI: Comparison of diffusion-weighted imaging and dynamic contrast-enhanced MRI in com- bination with T2-weighted imaging. Journal of Magnetic Resonance Imaging, 31, 625-631. doi:10.1002/jmri.22075 [28] Iwazawa, J., Mitani, T., Sassa, S. and Ohue, S. (2011) Prostate cancer detection with MRI: Is dynamic contrast- enhanced imaging necessary in addition to diffusion- weighted imaging? Diagnostic and Interventional Radiology, 17, 243-248. [29] Kim, C.K., Park, B.K. and Lee, H.M. (2009) Prediction of locally recurrent prostate cancer after radiation therapy: Incremental value of 3T diffusion-weighted MRI. Journal of Magnetic Resonance Imaging, 29, 391-397. doi:10.1002/jmri.21645 [30] Tan, C.H., Wang, J. and Kundra, V. (2011) Diffusion weighted imaging in prostate cancer. European Radiology, 21, 593-603. doi:10.1007/s00330-010-1960-y [31] Chan, I., Wells, III, W., Mulkern, R.V., Haker, S., Zhang, J., Zou, K.H., et al. (2003) Detection of prostate cancer by integration of line-scan diffusion, T2-mapping and T2-weighted magnetic resonance imaging; a multichannel statistical classifier. Journal of Medical Physics, 30, 2390- 2398. doi:10.1118/1.1593633 [32] Huisman, H., Vos, P., Litjens, G., Hambrock, T. and Barentsz, J. (2010) Computer aided detection of prostate cancer using t2w, DWI and DCE-MRI: Methods and clinic al ap plica tio ns. Proceedings of International Workshop, Held in Conjunction with MICCAI 2010, Beijing, 20-24 September 2010, 4-14. [33] Frangi, A.F., Niessen, W.J. , Vincken, K.L. and Viergever, M.A. (1998) Multiscale vessel enhancement filtering. Proceedings of the 1st Annual International Conference of the MICCA, Cambridge, 11-13 October 1998, 130-137. [34] Viswanath, S., Bloch, B.N., Genega, E., Rofsky, N., Len- kinski, R., Chappelow, J., et al. (2008) A comprehensive segmentation, registration, and cancer detection scheme on 3 Tesla in vivo prostate DCE-MRI. Medical Image Computing and Computer-Assisted Intervention, 11, 662- 669. [35] Langer, Deanna, L., van der Kwast Theodorus, H., Evans, Andrew, J., et al. (2009) Prostate cancer detection with multi-parametric MRI: Logistic regression analysis of quantitative T2, diffusion-weighted imaging, and dynamic contrast-enhanced MRI. Journal of Magnetic Resonance Imaging, 30, 327-334. doi:10.1002/jmri.21824 [36] Villeirs, G.M., L Verstraete, K., De Neve, W.J. and De Meerleer, G.O. (2005) Magnetic resonance imaging ana- tomy of the prostate and periprostatic area: A guide for radiotherapists. Radiotherapy and Oncology, 76, 99-106. doi:10.1016/j.radonc.2005.06.015 [37] Zhan, Y. and Shen, D. (2003) Automated segmentation of 3D US prostate images using statistical texture-based matching method. Proceedings of the 6th Annual Interna- tional Conference of the MICCA, Montreal, 15-18 Nove- mber 2003, 2878, 688-696. [38] Lixin, G., Pathak, S.D., Haynor, D.R., Cho, P.S. and Kim, Y. (2004) Parametric shape modeling using deformable superellipses for prostate segmentation. IEEE Transac- Copyright © 2013 SciRes. OPEN ACCESS  A. Firjani et al. / J. Biomedical Science and Engineering 6 (2013) 346-356 Copyright © 2013 SciRes. 356 OPEN ACCESS tions on Medical Imaging, 23, 340-349. doi:10.1109/TMI.2004.824237 [39] Zwiggelaar, R., Zhu, Y. and Williams, S. (2003) Semi- automatic segmentation of the prostate. Pattern Recog- nition and Image Analysis, 2652, 1108-1116. doi:10.1007/978-3-540-44871-6_128 [40] Zhu, Y., Williams, S. and Zwiggelaar, R. (2004) Segmen- tation of volumetric prostate MRI data using hybrid 2d+3d shape modeling. Medical Image Understanding and Analysis, 61-64. [41] Toth, R., Tiwari, P., Rosen, M., Kalyanpur, A., P ung avk ar, S. and Madabhushi, A. (2008) A multi-modal prostate segmentation scheme by combining spectral clustering and active shape models. Proceedings of the International Society of Optics and Photonic, San Jo se, 691 44 S- 6 9144S. [42] Klein, S., van der Heide, U.A., Raaymakers, B.W., Kot te, A.N.T.J., Staring, M. and Pluim, J.P.W. (2007) Segmen- tation of the prostate in MR images by atlas matching. Proceedings of the 4th Annual IEEE International Sym- posium on Biomedical Imaging: From Nano to Macro (ISBI), Washington DC, 12-15 April 2007, 1300-1303. [43] Vikal, S., Haker, S., Tempany, C. and Fichtinger, G. (2009) Prostate contouring in MRI guided biopsy. Proceedings of the International Society of Optics and Photonics, 7259, 72594A. [44] Martin, S., Troccaz, J. and Daanenc, V. (2010) Auto- mated segmentation of the prostate in 3D MR images us- ing a probabilistic atlas and a spatially constrained de- formable model. Journal of Medical Physics, 37, 1579- 1590. doi:10.1118/1.3315367 [45] Hambrock, T., Somford, D.M., Hoeks, C., Bouwense, S. A., Huisman, H., Yakar, D., et al. (2010) Magnetic re- sonance imaging guided prostate biopsy in men with re- peat negative biopsies and increased prostate specific an- tigen. Journal of Urology, 183, 520-527. doi:10.1016/j.juro.2009.10.022 [46] Firjani, A., Khalifa, F., Elnakib, A., Gimel’farb, G., El- Ghar, M.A., Elmaghraby, A. and El-Baz, A. (2011) 3D automatic approach for precise segmentation of the pr ost ate from diffusion-weighted magnetic resonance imaging. Proceedings of 18th Annual IEEE International Conference on Image Processing (ICIP), Brussels, 11-14 September 2011, 2285-2288. [47] Firjani, A., Khalifa, F., Elnakib, A., Gimel’farb, G., El- Ghar, M.A., Elmaghraby, A. and El-Baz, A. (2012) A novel image-based approach f or early detection of p ro sta te cancer. Proceedings of 19th Annual IEEE International Conference on Image Processing (ICIP), Orlando, 30 Se p- tember-3 October 2012, 2849-2852. [48] Firjani, A., Elnakib, A., Khalifa, F., Gimel’farb, G., El- Ghar, M.A., Elmaghraby, A. and El-Baz, A. (2011) A new 3D automatic segmentation framework for accurate extraction of prostate from diffusion imaging. Proceed- ings of 3rd Annual Biomedical Sciences and Engineering Conference (BSEC), Knoxville, 15-17 March 2011, 1-4. [49] El-Baz, A., Gimel’farb, G., Falk, R., Holland, T. and Shaffer, T. (2008) A new stochastic framework for accurate lung segmentation. Proceedings of the 11th Annual Inter- national Conference of the MICCA, New York, 6-10 Sep- tember 2008, 322-330. [50] El-Baz, A., Elnakib, A., Khalifa, F., Abou El-Ghar, M., McClure, P., Soliman, A. an d Gimel’farb, G. (2012) Pre- cise segmentation of 3-D magnetic resonance angio- graphy. IEEE Transactions on Biomedical Engineering, 7, 2019-2029. doi:10.1109/TBME.2012.2196434 [51] El-Baz, A. and Gimel’farb, G. (2007) EM based approxi- mation of empirical distributions with linear combinations of discrete Gaussians. Proceedings of 14th Annual IEEE International Conference on Image Processing (ICIP), San Antonio, 16-19 September 2007, 4, 373-376. [52] Farag, A.A., El-Baz, A. and Gimel’farb, G. (2006) P rec ise segmentation of multi-modal images. IEEE Transactions on Image Processing, 15, 952-968. doi:10.1109/TIP.2005.863949 [53] El-Baz, A. (2006) Novel stochastic models for medical image analysis. Ph.D. Thesis, University of Louisville, Louisville. [54] Viola, P. and Wells, W.M. (1997) Alignment by maxi- mization of mutual information. International Journal of Computer Vision, 24, 137-154. doi:10.1023/A:1007958904918 [55] Besag, J. (1986) On the statistical-analysis of dirty pic- tures. Journal of the Royal Statistical Society Series B- Methodological, 48, 259-302. [56] Khalifa, F., El-Baz, A., Gimel’farb, G. and Abu El-Ghar. M. (2010) Non-invasive image-based approach for early detection of acute renal rejection. Proceedings of the 13th Annual International Conference of the MICCA, Beijing, 20-24 September 2010, 1, 10-18. [57] Patterson, D.M., Padhani, A.R. and Collins, D.J. (2008) Technology insight: Water diffusion MRI—A potential new biomarker of response to cancer therapy. Nature Cli- nical Practice Oncology, 5, 220-233. doi:10.1038/ncponc1073 [58] Bouman, C. and Sauer, K. (1993) A generalized Gaussian image model for edge-preserving MAP estimation. IEEE Transactions on Image Processing, 2, 296-310. doi:10.1109/83.236536 [59] Khalifa, F., Beache, G.M., Gimelrfarb, G., Giridharan, G.A. and El-Baz, A. (2012) Accurate automatic analysis of cardiac cine images. IEEE Transactions on Biomedical Engineering, 59, 445-455. doi:10.1109/TBME.2011.2174235 [60] Tsai, A., Yezzi, Jr., A., Wells, W., Tempany, C., Tucker, D., Fan, A., et al. (2003) A shape-based approach to the segmentation of medical imagery using level sets. IEEE Transactions on Medical Imaging, 22, 137-154. doi:10.1109/TMI.2002.808355 [61] Dice, L.R. (1945) Measures of the amount of ecologic association between species. Ecology, 26, 297-302. doi:10.2307/1932409 [62] Walker-Samuel, S., Orton, M., Boult, J.K. and Robinson, S.P. (2011) Improving apparent diffusion coefficient esti- mates and elucidating tu mor heterogeneity using Baye sian adaptive smoothing. Magnetic Resonance in Medicine, 65, 438-447. doi:10.1002/mrm.22572
|