Paper Menu >>
Journal Menu >>
 J. Biomedical Science and Engineering, 2010, 3, 1021-1028 JBiSE doi:10.4236/jbise.2010.310133 Published Online October 2010 (http://www.SciRP.org/journal/jbise/). Published Online October 20 10 in SciRes. http://www.scirp.org/journal/jbise Ensemble-based active learning for class imbalance problem Yanping Yang, Guangzhi Ma School of Computer Science and Technology; Huazhong University of Science and Technology, Wuhan, China. Email: maguangzhi.hust@gmail.com Received 8 September 2010; revised 20 September 2010; accepted 25 September 2010 ABSTRACT In medical diagnosis, the problem of class imbalance is popular. Though there are abundant unlabeled data, it is very difficult and expensive to get labeled ones. In this paper, an ensemble-based active learning algorithm is proposed to address the class imbal- ance problem. The artificial data are created ac- cording to the distribution of the training dataset to make the ensemble diverse, and the random sub- space re-sampling method is used to reduce the da- ta dimension. In selecting member classifiers based on misclassification cost estimation, the minority class is assigned with higher weights for misclassi- fication costs, while each testing sample has a vari- able penalty factor to induce the ensemble to cor- rect current error. In our experiments with UCI disease datasets, instead of classification accuracy, F-value and G-means are used as the evaluation rule. Compared with other ensemble methods, our method shows best performance, and needs less labeled samples. Keywords: Class Imbalance, Active learn ing, Ensemble, Random Subspace, Misclassification Cost 1. INTRODUCTION In the medical diagnosis, it is common that there is a huge disproportion in the number of cases belonging to different classes [1]. For example, the number of cancer cases is much smaller than that of the healthy. The tradi- tional classifiers, however, are incapable of countering such class imbalance problem, because they favor the majority class. Moreover, the minority class is much more important in real applications. In addition, in real world, there are abundant unlabeled data but labeled instances are difficult, time-consuming or expensive to obtain. It will in turn make the labeled minority class much fewer further, which often degrades the perform- ance of traditional classifiers greatly. As a result, active learning with unlabeled imbalanced data becomes an important issue in machine learning [3]. To address the class imbalance problem, the direct way is to reduce the imbalance by re-sampling original dataset. Some methods try to under-sampling majority class, like Tomek link [4], condensed nearest neighbor rule [5] and neighborhood cleaning rule [6][7]. In these methods, the majority samples in certain area are con- sidered as useless and can be removed from training dataset. But, there is a risk of missing representative samples. Other methods, like SMOTE [8], try to over- sampling the minority class. In SMOTE method, the artificial datasets are created according to the distribu- tion of the minority class. However, the enhancement will be little, if the created artificial datasets have the same properties as the labeled samples Finding proper classifier for minority class is another way to counter class imbalance problem. Joshi [9] once modified the Boosting algorithm by assigning the minority class with a weight different from that of the majority class. Akbni [10] adjusted the SVM’s decision-boundary by modifying the kernel function. But, the certain classifier is only efficient in countering specific class imbalance, and cannot be extended to other applications. Another trend is to use ensemble of classifiers, which often has better performance than single classifier. But, the per- formance depends on the diversity of the ensemble [11]. If classifiers in an ensemble have the same property, there will be less improvement of performance even with more classifiers. Active learning techniques are conventionally used to solve problems where there are abundant unlabeled data but rare labeled ones [3]. Recently, various approaches on active learning from imbalanced datasets have been proposed in literatures [12]. For instance, as a good clas- sifier, support vector machine (SVM) was proposed in active learning for the imbalance problem [14]. To re- duce the co mputational co mplexity in dealing with large imbalanced datasets, this method was implemented in a random set of training populations, instead of the entire training dataset. In [16], bootstrap-based over- sampling was proposed to reduce the imbalance in the application  Y. P. Yang et al. / J. Biomedical Science and Engineering 3 (2010) 1021-1028 Copyright © 2010 SciRes. JBiSE 1022 of word sense disambiguation. Facing the class imbal- ance issue, however, both re-sampling and classifier strategy have their own advantage as well as disadvan- tage. The best way is to combine them together [17]. Bu t progress in this field is little. In this paper, an ensemble-based active learning with artificial samples is proposed to address class imbalanced problem by using unlabeled data. Different from random sampling, we try to use active selection strategy to label the sample with potential benefit to the ensemble’s di- versity. In add ition , we will create artificial datasets from the distribution as the training dataset. The conversely labeling of each artificial data will bring diversity to the ensemble. Both the training dataset and the artificial dataset will be re-sampled according to random subspace concept. It will release the difficulty of traditional sam- pling methods while facing with high-dimension data. Further, when choosing member classifiers according to misclassification cost, the minority class is assigned with a higher weight for misclassification cost, and each test- ing sample has a variable penalty factor to induce the ensemble to correct current error. In the experiments with UCI disease datasets, instead of accuracy, F-value and G-mean are used to evaluate the performance, since they are better for minority classification tasks. The rest of this paper is organized as follows. In Sec- tion 2, the proposed ensemble is described in detail, in- cluding the creation of artificial datasets, random sub- space re-sampling and misclassification cost estimation. Section 3 introduces how to implement active learning with our proposed ensemble method. In experiment part, the new evaluation rules are introduced. Based on ex- periments on the UCI datasets, our proposed method is compared with other state-of-art methods. 2. RANDOM SUBSPACE ENSEMBLE WITH AR TIFICIAL DATASETS In our ensemble-based active learning, the ensemble algorithm is the core. So, in this section, we will intro- duce our Random Subspace Ensemble with Artificial Data (RSEAD) in detail. 2.1. Overview Figure 1 is the algorithm of our Random Subspace En- semble with Artificial Data (RSEAD). Each member classifier in the ensemble is created via the iteration steps in Figure 1. At the beginning of the algorith m, the training dataset T will be mapped into another dataset T, in a m-dimension subspace. Then a classifier will be created based on T, and used to initiate the ensemble C*. Also, the misclassification cost of current ensemble will be calculated. Whereafter, the algorithm will enter follow- ing iteration: 1) According to th e distribution of the training dataset T, an artificial dataset will be generated. The size of the artificial dataset will be in a certain ratio, Rsize, to that of training dataset. They will be labeled with a class dif- ferent from what the ensemble predicts 2) In the m-dimension subspace, both T and R will be re-sampled to T, and R,. 3) A new classifier C, will be learned from both la- beled R, and T,. In order to guarantee the performance of the ensemble when pursuing the diversity, the misclassi- fication cost of the new ensemble with C, is calculated. Compared with the previous ensemble, if the new classi- fier brings more misclassification cost, it will be re- moved; otherwise, it will be kept in the ensemble; 4) The above steps will be iterated until algorithm re- turns the expected size of ensemble, or the number of iterations reaches the limited value. To predict the class of an unlabeled sample x, each member classifier Ci in ensemble C, will assign x with an membership probability, , ˆ() i Cy Px . Then the ensemble will calculate membership probability of each class y for sample x via following equation: Figure 1. Algorithm of RSEAD ensemble. Algorithm: The RSEAD ensemble Input: BaseLearn– Base Learner L - Training Set R - Artificial dataset m - Dimension of random subspace Csize - Target size of subspace Imax - Maximum number of iterations Rsize - Ratio between the size of dataset R and L (1) i = 1 ; (2) trials = 1 ; (3) Preprocessing the training set based on m-dimension subspace : '()TRSMsamplingT (3) (') i CBaseLearnT (4) *{} i CC (5) Calculate the ensemble error, ; i = i + 1 (6) While i < Csize and trials < Imax { (7) Create artificial dataset , the size will be R size T ; (8) Assign each artificial sample a lab el different from C*’s prediction. (9) Re-sample the training set and artificial set in m-dimension subspace: '()TRSMsamplingT '() R RSMsampling R (10) '''TTR (11) (') i CBaseLearnT ** {} i CC C (12)Calculate the misclassification cost of new ensemble, ' (13) If ' then {' , i = i + 1 } (14) else {** ' {}CC C ; trials = trials + 1}  Y. P. Yang et al. / J. Biomedical Science and Engineering 3 (2010) 1021-1028 Copyright © 2010 SciRes. JBiSE 1023 *, * ˆ() ˆ() i i Cy CC y Px Px C (1) Equation (1) reflects the probability of x belonging to class y. Therefore, the label with largest membership probability will be assigned to x: *ˆ ( )argmax( ) y yY Cx Px (2) 2.2. Creation and Labeling of Artificial Datasets The diversity is a critical factor for a successful ensem- ble [11]. An ensemble will have less diversity if its member classifiers have the same property. To bring more diversity, Bagging [19] divides the training set into several smaller one, while Boosting adjusts the distribu- tion of the training dataset according to the chosen clas- sifier [20]. Further, in Random Forest [21], both training dataset and feature space are divided into smaller ones to train different classifiers. However, all these methods depend on the training dataset to induce the diversity. Therefore, if the training dataset is not big enough, the diversity will be limited. In our active learning method, the RSEAD ensemble’s diversity will be guaranteed in three ways: 1) with active learning, the large pool of unlabeled data can be sampled to get good training datasets; 2) besides the training da- taset, the artificial data are also created for training clas- sifier; 3) both the original training and the artificial da- tasets will be re-sampled in subspace to enhance diver- sity. In this part, we will focus on the creation of artifi- cial dataset and their labeling. In our method, the artificial data are created by ran- domly picking data points from an approximation of the training dataset distribution. The numeric attributes are defined according to the mean and the standard deviation of the training dataset, and generated in Gaussian distri- bution. For a nominal attribute, its value is based on the probability of the occurrence of each distinct value in its domain. The Laplace smoothing is used if a certain no- minal attribute is absent in the training dataset. Further, to construct an artificial data, there is a simplifying as- sumption that the attributes are independent, because it will cost much time and labeled data to accurately esti- mate the joint probability distribution of these attributes. In each iteration shown in Figure 1, the ensemble will predict the class label for each artificial data x. Firstly, ensemble will give a membership probability of x be- longing to certain class y. The zero membership prob- ability will be replaced by a small non-zero value in case that it may act as a denominator. Then the artificial data will be labeled a class that is different from what the ensembles predict. Therefore, if current ensemble pre- dicts the probability of x belonging to y is ˆ() y P x, then, the choice of label for x will be based on ' ˆ() y Px: 'ˆ 1/() ˆ() ˆ 1/() y y y y Px Px Px (3) Let us show this labeling method with a two-class problem. For instance, for an artificial sample x, the en- semble estimates that it has 20% probability of being a positive sample and 80% probability of being a negative one. In other words, the ensemble believes that x is more likely a negative sample. In our method, to create a new classifier with more diversity, x will be assigned with a positive label, and th en used to train a new classifier. The ensemble often has higher accuracy than single member classifier if each member classifier is not related with others. Therefore, our method of labeling artificial data can reduce the relevancy between classifiers, which will in turn bring the ensembl e with higher accuracy and less generalization error. 2.3. Re-Sampling in Subspace Re-sampling is the popular way to deal with class im- balance problem. However, most of sampling methods, like SMOTE, often work in the whole feature space, which is not efficient in countering high-dimension da- tasets. In addition, they often try to consider the class imbalance and the properties of the dataset as a whole. The data, however, often exhibit characteristics and properties at a local level, rather than the global level. Hence, it is important to study the dataset in a reduced subspace. Although a certain feature subspace may only lead to a weak classifier, the ensembling of such weak ones can make a strong classifier [22], since it induces higher diversity, which is an important condition for a classifier with good performance. To this end, we proposed the Random-Subspace- Mapping Sampling (RSM-sampling) algorithm. Suppose we have a dataset L, which has a n-dimension space: |L| = l, 12 { ,,...,} n F FF F. Any data PL can be represented as 12 { ,,...,} n PPP P, where i P is the value of the related feature i F in the feature space F. If the dimension of each subspace is set to m, m<n, the number of the likely subspace will be max m n kC. When [/2]mn , kmax has its biggest value. For our algorithm, each feature subspace will bring a candidate classifier. We often choose Cszie< kmax classifiers to construct an ensemble, since not every candidate classifier will help enhance the ensemble,s performance. Before re-sampling, a subspace S should be randomly selected from the feature space F. F mn . 12 {, ,...,} m SSS SF . Then, in the feature subspace,  Y. P. Yang et al. / J. Biomedical Science and Engineering 3 (2010) 1021-1028 Copyright © 2010 SciRes. JBiSE 1024 each data PL will be mapped into 12 { ,,...,} Sss sm PPP P. In each iteration step of our algorithm, both the train- ing dataset L and the artificial dataset R are re-sampled in chosen sub-space. 2.4. Misclassification Cost Estimation When pursuing the diversity, the performance of ensem- ble should be guaranteed too. To address the problem of class imbalance, the misclassification cost is used to replace the traditional classification error. A new classi- fier will be kept in the ensemble if it helps decrease the misclassification cost; otherwise, it will be removed. In our algorithm, the minority class is assigned with a higher weight of misclassification cost than that of the majority class. Also, each test sample will be assigned with a penalty factor. If current ensemble makes wrong decision on it, its penalty factor will be increased; oth- erwise, its penalty factor will be decreased. In this way, the ensemble will choose the new classifier that helps to correct the error of current ensemble. Also, since the minority samples have more chance to be misclassified, this penalty factor will bring an ensemble proper for minority class. Suppose we have t samples to evaluate the ensemble based on misclassification cost. Firstly, each sample’s penalty factor will be initialized as: 11/ 1 i dtit (4) The misclassification cost of the ensemble gotten in k-th iteration can be represented as: * 1 cos (,()) tk kikii i tyC xd (5) In Equation (5), yi is the correct class label of testing sample xi, *() ki Cx is the predicted class o f the en semble for xi. k i d is xi,s penalty factor for the k-th iteration. * cos (,()) iki tyC x is the weight of misclassifying a sam- ple with label yi as class *() ki Cx. When*() iki y Cx, there is* cos (,())0 iki tyC x because classification is correct. If the misclassification cost in the k-th iteration is less that that in the (K–1)-th iteration, then newly created classifier will be kept in ensemble. There, we have the coefficient of perf o rmanc e enhancing: ln((1) /) / 2 kkk (6) Each testing sample’s penalty factor will be modified according to current ensemble’s prediction. If current ensemble makes correct classification on xi, its penalty factor 1k i d will be decreased to: 1exp( ) kk ii k dd a (7) Otherwise, it will be increased to: 1exp( ) kk ii k dd a (8) Please note, all samples, new penalty factors will be normalized as following: 1 11 tk ki i Z d 11 1 / kk iik ddZ . (9) 1k i d will be used in the (k + 1)-th iteration. The design of misclassification cost weigh * cos (,()) iki tyC xand penalty factor1k i d will help the ensemble to pick the classifiers that can better deal with minority class. 3. ACTIVE LEARNING WITH THE ENSEMBLE RSEAD The ensemble with diversity will be used in active learning for selecting unlabeled data. Like the QBC [23], our proposed active learning method also chooses the unlabeled samples that have the biggest prediction dif- ference among the classifiers in the ense mble. Such pre- diction difference is often called uncertainty, which is calculated via margin measure in our algorithm. The margin is defined as the difference of membership probability between the samples most likely class and second most likely class. *12 ˆˆ (,) ()() yy M argin CxPxPx (10) where y1 and y2 are class labels of unlabeled sample x predicted by ensemble C*. y1 has the highest member- ship probability for x, while y2 is the second high est one. Then, the uncertainty can be represented as: *1 (,) (*, ) UncertaintyCxMargin Cx (11) where the is a small value in case margin is 0. The smaller margin is, the bigger the uncertainty is. For a two-class task, when12 ˆˆ () () yy Px Px,the margin will be 0, and x will have the biggest uncertainty, * (,)1/UncertaintyCx 4. EXPERIMENTS To evaluate our method,s effectiveness for medical di- agnosis, eight disease datasets from the UCI machine learning repository [24] are used in experiments. In this section, we will discuss the experiments in detail. 4.1. Evaluation Rule In a two-class task, a classifier will have four kinds of prediction results [25] for dataset with N samples, shown in Table 1. TP and FN responsively mean the number of correctly and wrongly classified positive samples, while 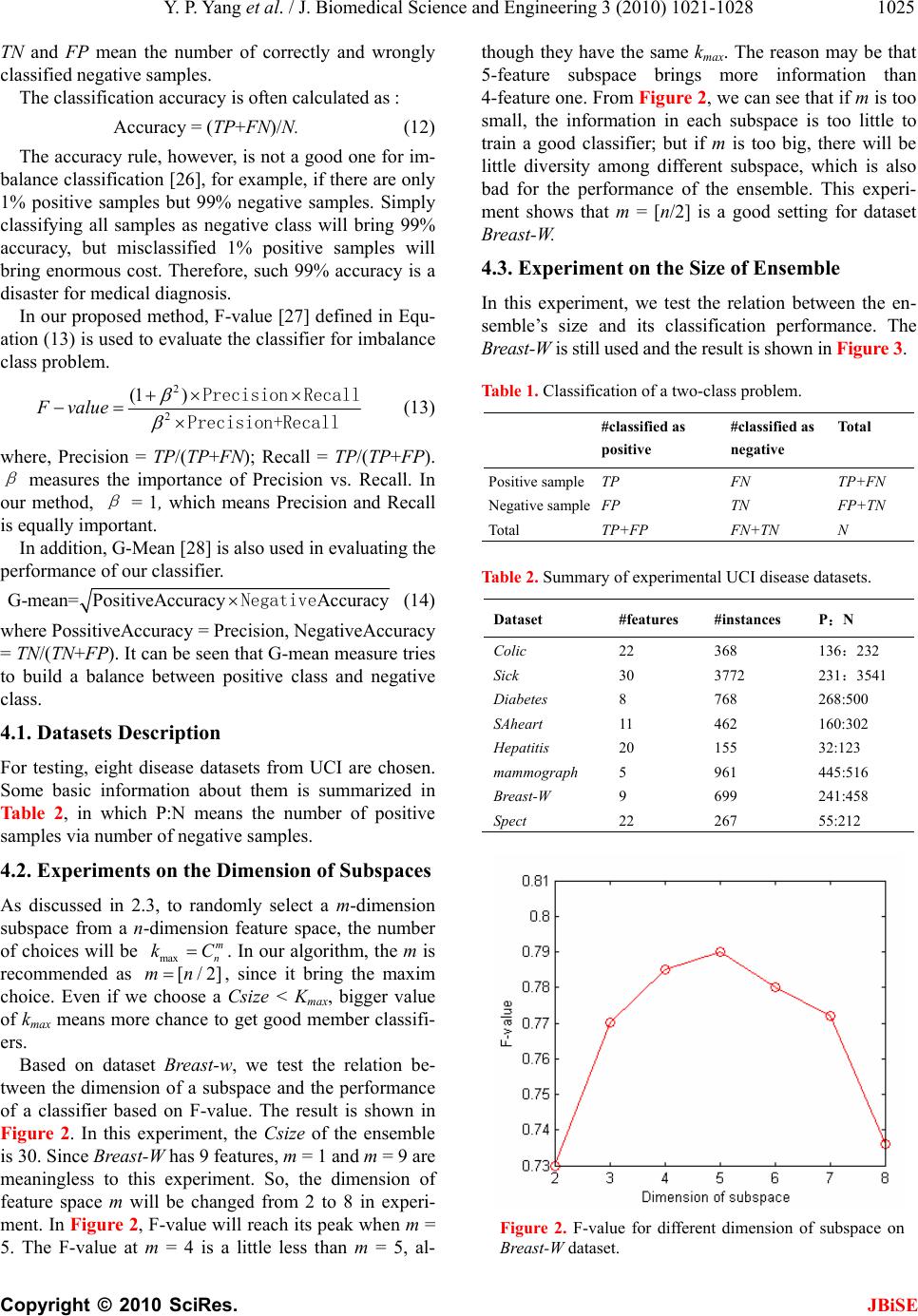 Y. P. Yang et al. / J. Biomedical Science and Engineering 3 (2010) 1021-1028 Copyright © 2010 SciRes. JBiSE 1025 TN and FP mean the number of correctly and wrongly classified negative samples. The classification accuracy is often calculated as : Accuracy = (TP+FN)/N. (12) The accuracy rule, however, is not a good one for im- balance classification [26], for example, if there are only 1% positive samples but 99% negative samples. Simply classifying all samples as negative class will bring 99% accuracy, but misclassified 1% positive samples will bring enormous cost. Therefore, such 99% accuracy is a disaster for medical diagnosis. In our proposed method, F-value [27] defined in Equ- ation (13) is used to evaluate the classifier for imbalance class problem. 2 2 (1 ) F value Precision Recall Precision+Recall (13) where, Precision = TP/(TP+FN); Recall = TP/(TP+FP). β measures the importance of Precision vs. Recall. In our method, β = 1, which means Precision and Recall is equally important. In addition, G-Mean [28] is also used in evaluating the performance of our classifier. G-mean= PositiveAccuracyAccuracyNegative (14) where PossitiveAccuracy = Precision, NegativeAccuracy = TN/(TN+FP). It can be seen that G-mean measure tries to build a balance between positive class and negative class. 4.1. Datasets Description For testing, eight disease datasets from UCI are chosen. Some basic information about them is summarized in Table 2, in which P:N means the number of positive samples via number of negative samples. 4.2. Experiments on the Dimension of Subspaces As discussed in 2.3, to randomly select a m-dimension subspace from a n-dimension feature space, the number of choices will be max m n kC. In our algorithm, the m is recommended as [/2]mn, since it bring the maxim choice. Even if we choose a Csize < Kmax, bigger value of kmax means more chance to get good member classifi- ers. Based on dataset Breast-w, we test the relation be- tween the dimension of a subspace and the performance of a classifier based on F-value. The result is shown in Figure 2. In this experiment, the Csize of the ensemble is 30. Since Breast-W has 9 features, m = 1 a nd m = 9 ar e meaningless to this experiment. So, the dimension of feature space m will be changed from 2 to 8 in experi- ment. In Figure 2, F-value will reach its peak when m = 5. The F-value at m = 4 is a little less than m = 5, al- though they have the same kmax. The reason may be that 5-feature subspace brings more information than 4-feature one. From Figure 2, we can see that if m is too small, the information in each subspace is too little to train a good classifier; but if m is too big, there will be little diversity among different subspace, which is also bad for the performance of the ensemble. This experi- ment shows that m = [n/2] is a good setting for dataset Breast-W. 4.3. Experiment on the Size of Ensemble In this experiment, we test the relation between the en- semble’s size and its classification performance. The Breast-W is still used and t he result is shown in Figure 3. Table 1. Classification of a two-class problem. #classified as positive #classified as negative Total Positive sampleTP FN TP+FN Negative sampleFP TN FP+TN Total TP+FP FN+TN N Table 2. Summary of experimental UCI disease datasets. Dataset #features #instances P:N Colic 22 368 136:232 Sick 30 3772 231:3541 Diabetes 8 768 268:500 SAheart 11 462 160:302 Hepatitis 20 155 32:123 mammograph 5 961 445:516 Breast-W 9 699 241:458 Spect 22 267 55:212 Figure 2. F-value for different dimension of subspace on Breast-W dataset.  Y. P. Yang et al. / J. Biomedical Science and Engineering 3 (2010) 1021-1028 Copyright © 2010 SciRes. JBiSE 1026 Figure 3. F-Value for different size of an ensemble on Breast-W dataset. In the experiment, the dimension of subspace is fixed as [/2][9/2] 5mn. Therefore, there will be 5 9126Cchoices of subspaces to train Csize classifiers for the ensemble. In Figure 3, the F-value increases quickly when Csize grows from 10 to 30, but the en- hancement is not big when Csize is changed from 30 to 120. It shows that for dataset Breast-W, 30 subspaces with 5 dimension s are enough to build a good ensemble. The additional subspace will contribute little to the di- versity of ensemble, and there will be no much en- hancement in performance, though the computation cost grows much. Therefore, 30 is a trade-off between per- formance and computation cost for dataset Breast-W. 4.3. Experiment Result In this experiment, we firstly test the performance of our proposed RSEAD ensemble algorithm. For comparison, two state-of-art classification algorithms, Bagging and Adaboost, are chosen. For fair comparison, C4.5 is used as base learner, and is configured with the default setting in Weka [29]. In the evaluation of performance, F-value and G-mean are used in experiments with 10-fold cross validation. For RSEAD algorithm, it has a setting with [/2]mn,k = 30, and Imax = 50. Shown in Table 3 is the F-value for the minority class in each dataset, while Ta b le 4 is the G-value for every whole dataset. For each dataset, the highest value is marked in bold. For convenience of comparison, the base learner C4.5 is also used as the reference. In the tables, Ada represents the Adaboost algorithm. In Ta bl e 3 and 4., all 3 ensembles have good F-value and G-mean than C4.5 on eight datasets. Compared with Bagging and Adaboost, our RSEAD has higher F-value and G-mean on most of dataset. From Tab le 3, it can be concluded that RSEAD has the best performance for minority class on 6 datasets. On dataset mammo g raph , the difference between ensembles is not significant. The reason may be that ratio between the minority and ma- jority classes is near 4:5, which has a very small class imbalance. Also, dataset mammograph is defined only by 5 features, which leaves little room for our random subspace re-sampling method to enhance the ensemble's performance. In the evaluation based on G-mean, our RESEAD wins for all 8 datasets. From Tables 3 and 4, it can be seen that our ensemble RESEAD has better per- formance than Bagging and ADABOOST in countering problem of imbalance class. This advantage comes from the unique way of creating each member classifier as well as the misclassification cost based decision in se- lecting proper classifiers. Compared with Bagging, Adaboost has better performance, because Adaboost introduces different cost weight for different misclassi- fication. It also indirectly proves the correctness of our misclassification cost estimation. To further test the performance of our active learning method with RSEAD ensemble, the Bagging and Ada- boost are also merged into the active learning architec- ture for comparison. Single RSEAD is tested further as reference. Tab le 5 shows how many samples each algo- rithm needs to get certain F-value on each dataset. Compared with RSEAD, the active learning methods need fewer samples to get the same F-value. Among the three active learning methods, our Active-RSEAD has significant advantage, which benefits from the design of RSEAD ensemble. Table 3. F-vaule for minority class in each dataset. Dataset C4.5 RSEAD Bagging Ada Colic 76.54 80.97 79.71 80.03 Sick 87.65 93.23 90.44 91.43 Diabetes 61.4 71.8 67.9 69.8 SAheart 55.3 75.2 67.4 73.1 Hepatitis 52.8 68.4 67.2 68.5 mammograph 79.5 81.2 82.1 83.2 Breast-W 89.7 95.6 92.3 94.0 Spect 73.1 79.76 76.6 77.5 Table 4. G-mean for each dataset. Dataset C4.5 RSEAD Bagging Ada Colic 81.5 85.5 83.4 84.51 Sick 91.2 95.8 95.6 95.2 Diabetes 64.3 76.4 71.4 74.3 SAheart 60.4 77.8 72.3 77.5 Hepatitis 58.4 76.3 74.3 73.4 mammograph 88.4 89.4 89.1 89.3 Breast-W 94.3 96.5 95.3 95.4 Spect 82.3 85.6 82.4 83.4  Y. P. Yang et al. / J. Biomedical Science and Engineering 3 (2010) 1021-1028 Copyright © 2010 SciRes. JBiSE 1027 Table 5. Number of sampling for target F-vlalue. Dataset RSEAD Active-RSEAD Active-Bagging Active Adaboost Target F-value Colic 41 23 35 37 85% Sick 321 134 178 165 93% Diabetes 245 101 114 106 75% SAheart 280 123 157 167 60% Hepatitis 117 45 56 54 95% mammograph 100 24 35 30 80% Breast-W 32 36 45 75 95% Spect 53 38 43 39 75% 5. CONCLUSIONS To address the problem of imbalance class in medical diagnosis, an ensemble-based active learning method is proposed. Our ensemble algorithm, RSEAD, introduces the subspace sampling method to reduce the complexity of computation and bring more diversity together with the creation of artificial datasets. Further, in evaluating the quality of each classifier candidate based on misclas- sification cost, the minority class is assigned with a higher weight for misclassification costs, while each testing sample has a variable penalty factor to induce the ensemble to correct current classification error. In above experiments, eight UCI disease datasets are chosen. The F-value and G-mean are used instead of classification accuracy to evaluate the performance of classifiers. The result shows that our proposed ensemble method has better performance than others. Moreover, in active learning experiment, having the same perform- ance with F-value rule, our method needs fewer samples. These experiments show that our ensemble-based active learning method has significant advantage than tradi- tional methods. Ensemble-based active learning is a promising method to counter the problem of class imbalance in medical diagnosis. But, there are still many issues for further studying. For example, our method only deals with two-class tasks, while the real world has many mul- ti-class tasks. In addition, the noise in a dataset is not considered in current study. Also, the weighting method in our method needs further improvement from both theory and implementation. Therefore, we will focus on these issues to improve our active-RSEAD method in following research work. REFERENCES [1] Japkowicz, N. and Stephen, S. (2002) The class imbal- ance problem: A systematic study. Intelligent Data Analysis, 6(5), 203-231. [2] Gustavo, E.A., Batista, P.A., Ronaldo, C., et a1. (2004) A study of the behavior of several methods for balancing machine learning training data. SIGKDD Explorations, 6(1), 20-29. [3] Settles, B. (2009) Active Learning Literature Survey. Computer Sciences Technical Report 1648, University of Wisconsin-Madison. [4] Tomek, I. (1976) Two modifications of CNN. IEEE Transaction on Systems Man and Communications, 6, 769-772. [5] Hart, P.E. (1968) The condensed nearest neighbor rule. IEEE Transaction on Information Theory, 14(3), 515- 516. [6] Laurikkala, J. (2001) Improving identification of difficult small classes by balancing class distribution. Proceedings of the 8th Conference on AI in Medicine, Cascais, Portu- gal, Europe: Artificial Intelligence Medicine, 63-66. [7] Wilson, D.L. (1972) Asymptotic properties of nearest neighbor rules using edited data.IEEE Transaction on Systems, Man and Communications, 2(3), 408-421. [8] Chawlan, V., Bowyer, K.W. and Hall, L.O. (2002) SMOTE: Synthetie minority over-sampling technique. Journal of Aflificial Intelligence Research, 16(1), 321- 357. [9] Joshi, M., Kumar, V. and Agarwal, R. (2001) Evaluating boosting algorithms to classify rare classes: Comparison and improvements. Proceedings of the 1st IEEE Interna- tional Conference on Data Mining. Washington DC: IEEE Computer Society, 257-264. [10] Akbani, R., Kwek, S. and Japkowicz, N. (2004) Applying support vector machines to imbalanced datasets. Pro- ceedings of the 15th European Conference on Machines Learning, Pisa, Italy, 39-50. [11] Krogh, A. and Vedelsby, J. (1995) Neural network en- sembles, cross validation and active learning. Advances in Neural Information Processing Systems, 7, 231-238. [12] Provost, F. (2000) Machine learning from imbalanced data sets 101. Invited paper for the AAAI, Workshop on Imbalanced Data Sets, Menlo Park, CA. [13] Abe, N. (2003) Invited talk: Sampling approaches to learning from imbalanced datasets: Active learning, cost sensitive learning and beyond. ICML-KDD Workshop: Learning from Imbalanced Data Sets. [14] Ertekin, S., Huang, J. and Giles, C.L. (2007) Active learning for class imbalance problem. Proceedings of Annual International ACM SIGIR Conference Research and development in information retrieval, Amsterdam, Netherlands, 823-824. [15] Ertekin, S., Huang, J., Bottou, L. and Giles, C.L. (2007) Learning on the border: Active learning in imbalanced  Y. P. Yang et al. / J. Biomedical Science and Engineering 3 (2010) 1021-1028 Copyright © 2010 SciRes. JBiSE 1028 data classification. Proceedings of the sixteenth ACM conference on Conference on information and knowledge management, November 6-8, Lisboa, Portugal, 127-136. [16] Zhu, J. and Hovy, E. (2007). Active Learning for Word Sense Disambiguation with Methods for Addressing the Class Imbalance Problem. In Proc. Joint Conf. Empirical Methods in Natural Language Processing and Computa- tional Natural Language Learning, Prague, 783-790. [17] Chawla, N.V., Lazarevic, A. and Hall, O. (2003) SMOTE- Boost: improving prediction of the minority class in boosting: knowledge discovery in databases. Proceeding of the 7th European Conference on Principles and Prac- tice of Knowledge Discovery in Databases. Cavtat Du- brovnik, 107-119. [18] Veropoulos, K., Campbell, C. and Cristianini, N. (2009) Con- trolling the sensitivity of support vector machines. Proc of Intemational Joint Confbrence on AI, 55-60. [19] Breiman, L. (1996) Bagging predictors. Machine Learn -ing, 24(2), 123-140. [20] Abe, N. and Mamitsuka, H. (1998) Query learning strate- gies using boosting and bagging. Proceedings of the In- ternational Conference on Machine Learning (ICML), Morgan Kaufmann, 1-9. [21] Breiman, L. (2001) Random forests. Machine Learning, 2001, 45(1), 5-32. [22] Kleinberg, E.M. (1990) Stochastic discrimination. Annals of Mathematics and Artificial Intelligence, 1(1-4), 207- 239. [23] Seung, H.S., Opper, M. and Sompolinsky, H. (1992) Query by committee. In Proceedings of the ACMWork- shop on Computational Learning Theory, 287-294. [24] Blake, C., Keogh, E., and Merz, C.J. UCI repository of machine lea r n i ng database s. http://www.ics.uci.edu [25] Su, C.T., Chen, L.S. (2006) Knowledge acquisition through information granulation for imbalanced data. Expert Systems with applications, 31, 531-541. [26] Joshi, M. (2002) On evaluating performance of classi- fiers for rare classes. Proceeding of the 2nd IEEE In- ternational Conference on Data Mining, Maebishi, Japan, 641-644. [27] Kotsiantis, S., Kanellopoulos, D., Pintelas, P. (2006) Handling imbalanced datasets: A review. GESTS Interna- tional Transactions on Computer Science and Engineer- ing, 30(1), 25-36. [28] Guo, H., Viktor, H. (2004) Learning from imbalanced data sets with boosting and data generation: the Data- Boost-IM approach. Sigkdd Explorations, 6(1), 30-39. [29] Witten, I. H., Frank, E. (2005) Data mining-pracitcal machine learning tools and techniques with JAVA im- plementations. 2nd Edition, Morgan Kaufmann Publishers. |

