Paper Menu >>
Journal Menu >>
 Vol.2, No.10, 1184-1190 (2010) Health doi:10.4236/health.2010.210174 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ Association of CYP2C19 genotype with type 2 diabetes Carlos Hoyo-Vadillo1*, Jaime Garcia-Mena2, Adán Valladares3, Caterina R. Venturelli1, Niels Wacher-Rodarte4, Jesús Kumate5, Miguel Cruz3 1Department of Pharmacology, Cinvestav-IPN Zacatenco, México City, Mexico; *Corresponding Author: citocromo@cinvestav.mx 2Departamento de Genética y Biología Molecular, Cinvestav-IPN Zacatenco, México City, Mexico 3Unidad de Investigación Médica en Bioquímica, Hospital de Especialidades, Centro Médico Nacional Siglo XXI del IMSS, México City, Mexico 4Unidad de Investigación Médica en Epidemiología, Hospital de Especialidades, Centro Médico Nacional Siglo XXI de IMSS, México City, Mexico 5Fundación IMSS, México City, Mexico Received 4 July 2010; revised 13 July 2010; accepted 20 July 2010. ABSTRACT Background: CYP2C19 is a major isoform of cytochrome P450 that metabolizes a number of commonly prescribed drugs such as omepraz- ole, diazepam, tolbutamide and propranolol. Its expression is regulated by the constitutive and- rostane receptor (CAR), involved in glucocorti- coids synthesis. Since a number of crosslinks have been described for CYPs and some horm- ones, an association of CYP2C19 with type 2 diabetes is likely. Methods: Two groups were studied, 352 diagnosed with type 2 diabetes patients and 342 healthy volunteers form Mexi- co City. Both groups were tested for CYP2C19*2 and *3 alleles. We carried out an allelic discri- mination using TaqMan assay for *2, and used FRET sensor and anchor probes for *3. Results: Ninety one percent of the subjects had the wild type allele, 9% have the *2 allele; no subject presented the *3 allele. The CYP2C19*2 allele is associated with type 2 diabetes (p = 0.012). Ad- mixmap program was used to correct the adm- ixture of this population and get the correlation. This was further confirmed in a linear model with a 67% power and by the method of Strom and Wienker for association on subjects within the mean range of Amerindian ancestry only (60%). Conclusion: Type 2 diabetes patients have sign- ificatly more *2 allele than healthy volunteers, more evident for the patients with the homocyg- ous genotype. Keywords: Pharmacogenomics; CYP2C19; Type 2 Diabetes; Mexicans; Allele Frequency 1. INTRODUCTION The P450 Cytochrome gene super-family is involved in the synthesis of steroids, as well as in the metabolism of cholesterol [1]. Cytochrome genes exhibit a number of mutations that alter their activity [2]. In the case of CYP- 2C19 the most common mutations *2 and *3 show no enzymatic activity. For this isoform, CYP2C19, known for metabolizing both, mephenytoin and omeprazole it has been demonstrated to have the greatest genetic varia- bility among human populations [3]. Some of them, Asians for example [4], present a very high frequency of mutations, Latin-Americans on the other hand, have a small frequency of inactive alleles [5,6], while Eskimo Inuits do not seem to present any mutations at all [7]. CYP2C19 metabolizes an estimated of 8% of all the- rapeutic drugs in current use [8], knowing the variants which do not present enzymatic activity is of huge inter- est for physicians prescribing drugs such as tolbutamide, omeprazole, proguanil, fluoxetine, citalopram, diazepam, propanolol, diclofenac, indomethacine and others. Previous studies have identified the presence of CYP2C19*2 in Bolivians [5] and in Colombians [6]. Whereas in Inuits no mutations have been identified [7]. Asians have a high frequency of 2C19 mutations, actu- ally they are so far the only population with 2C19*3 mutations. For this reason it is somehow obvious to look for the presence of this allele in our Mexican subjects since Amerindian populations came from Asia 30,000 to 45,000 years ago [9]. In addition, characterizing our populations is an important advance towards a personal- ized medicine. Nuclear receptors are transcription factors that modu- late cellular responses to small lipophyllic molecules such as steroids and others. They also regulate the expres-  C. Hoyo-Vadillo et al. / Health 2 (2010) 1184-1190 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 118 1185 sion of CYPs. Three of this nuclear receptors have been described to regulate the expression of CYP2C19, CXR (chicken xenobiotic receptor), PXR (pregnane X recep- tor) and CAR [10]. All this elements are a linkage of CYP2C19 with its role on cholesterol elimination and on the physiology of other steroid hormones like cortisol. Both process are tightly involved in the development of obesity and type 2 diabetes. This regulation of the CYP2C19 is regulated by a complex crosslinking in- volving cholesterol levels, glucocorticoid activity and the action of other steroid hormones; therefore, the in- terest to look for a possible association of CYP2C19 with type 2 diabetes. 2. SUBJECTS, MATERIALS AND METHODS 2.1. Subjects We included 367 men and 330 women (age range: 18-76 years old; body mass index averages of 28 for males and 30 for females, Table 1) from the DNA library of the Biochemistry Research Unit of Medical Center 21st cen- tury, IMSS, located in Mexico City. No relatives were included in the groups. The corresponding ethics and research committees approved the study. All participants signed the informed consent. About half of the subjects participating in this study were patients with type 2 diab- etes, and because they were already part of other studies, we had previously characterized their population ad- mixture. 2.2. Autosomal Ancestry-Informative Markers (AIMs) Stratification in the subjects of this study was corrected using previously reported data [11], these markers have large frequency differences among populations of Native American (65%), European (30%), and West African (5%) ancestry. 2.3. Biochemical Profile Clinical evaluation was made after 12 h overnight fas- ting. Blood samples were taken and used to measure gl- ucose, triglycerides, total, high and low density choles- terol levels were assayed using an ILab 350 Clinical Ch- emistry System (Instrumentation laboratory, Mexico). All participants were interviewed by a physician who colle- cted data about their weight, height, systolic and diasto- lic blood pressure. Quantitative measurements of fasting plasma insulin were carried out using the chemilumines- cence’s assay according to the manufacturer’s instruct- tions (Immulite, France). 2.4. DNA Samples DNA was extracted from blood cells using the Qiagen columns according to the manufacturer’s recommenda- tions (Qiagen, Chatsworth, CA, USA). Purity was veri- fied by UV absorption at 260/280 nm and DNA integrity was checked by electrophoresis in 0.8% agarose gels, stained with ethidium bromide. Table 1. General characteristics of the group studied. Control T2D Variable units Males Females Males Females Number 259 93 107 235 Age years 41.6(8.2) 40.5(8.7) 57.8(9.9) 55.8(9.6) Height m 1.68(0.07) 1.56(0.06) 1.64(0.08) 1.54(0.07) Weight kg 77.9(10.5) 69.4(11.9) 76.1(11.3) 71.2(13.8) Body Mass Index (BMI) kg/m2 27(3) 29(5) 29(5) 30(5) Waist Hip Index 0.87(0.05) 0.84(0.07) 0.95(0.04) 0.89(0.07) SBP mmHg 118(11) 114(8) 122(12) 124(15) DBP mmHg 74(7) 73(8) 95(25) 92(24) Glucose mg/dL 85.7(9.5) 84(8.7) 163.5(73.3) 154.2(60.9) Insulin IU/ml 16(6.6) 15.6(7.6) 9.2(12.2) 8.4(11.5) Glycosilated haemoglobin (A1c) % 4.2(0.6) 4(0.5) 7.6(2.4) 7.9(2) Total cholesterol mg/dL 195(40) 189(36) 196(38) 206(46) High density lipoprotein (HDL-C) mg/dL 51(23) 59(23) 40(16) 42(19) Low density lipoprotein (LDL-C) mg/dL 106(46) 98(41) 119(39) 130(39) Triglycerides mg/dL 173(102) 128(62) 225(134) 207(127) Serum Creatinine mg/dL 1.00(0.16) 0.76(0.13) 0.93(0.98) 0.81(0.76) Values are Mean (Standard Deviation).  C. Hoyo-Vadillo et al. / Health 2 (2010) 1184-1190 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 1186 2.5. Allele-Specific TaqMan PCR for CYP2C19*2 DNA samples were analyzed for CYP2C19*2 using the TaqMan PCR assay to detect G > A polymorphism acc- ording to the manufacturer’s instructions. Amplification and detection were made using the ABI PRISM 7000 (Applied Biosystems, USA) system with the following profile: a denaturing cycle of 95ºC for 10 min and 40 cycles of 92ºC for 15 s, extension phase of 60ºC for 1 min. Samples were judged positive for *2 when the va- lue of the emitted fluorescence was greater than the th- reshold calculated by the instrument’s software. Display- ed as an allelic discrimination plot. Wild type allele CYP2C19*1 was labeled with VIC and *2 allele with FAM. VIC was for TTCCCGGGAACCCA and FAM for ATTTCCCAGGAACCCA (SNP showed in bold- face). 2.6. CYP2C19*3 Evaluation by FRET Technology This polymorphism was identified following the method described by Borlak and Thum [12] using fluorescence resonance energy transfer (FRET) to assay genomic G > A Sensor and anchor probes were labeled with fluoresce- ein and Red640, respectively. PCR was run with a 5 min denaturation step. Fifty cycles with 7 sec of annealing (48ºC) and 14 sec of extension (72ºC) in a Light Cycler (Model 1.2, Roche diagnostics, Germany). The melting curve was done from 40ºC (with 30 sec of previous stab- ilization) to 80ºC with a 0.1ºC/min slope. Tms were 61ºC for the wild CYP2C19*1 type and 67ºC for the CYP2 C19*3 genotype. Software used was Light Cycler (Ro- che, Version 4). Control probes for *1 and *3 were used as validation of fusion points. The assay was validated using standards provided by Roche. 2.7. Statistical Analysis In order to correct the stratification of this population, ADMIXMAP program was employed to look for case- control (shown in Table 2) associations using logistic regression and to measure the Hardy-Weinberg equilib- rium. Iterations were tested to 4000, burning was sele- cted to 200, “every” parameter to 7. The rest of parame- ters were defaults. Co-variables were log of age and ge- nder. Hardy-Weinberg equilibrium was also tested after the Sasieni method implemented by Strom and Wienker [13] (available at: http://ihg2.helmholtz-muenchen.de/ cgi-bin/hw/hwa1.pl) [14]. Proportions of genotypes were compared by the Fisher exact test. Odds ratios and their corresponding 95% confidence intervals were also esti- mated. All p values < 0.05 were considered statistically significant. The association between total cholesterol, HDL-C, LDL-C levels with the CYP2C19*2 allele was analyzed by the type III linear model with and without co-variables, including the stratification of the population, health, age, gender, height, body mass, waist-hip index, and BMI. Linear models were also tested using model I for interaction, with an Amerindian index as co-variable as well as log of age and gender. In all tests significance was considered for values less than 0.05. 3. RESULTS 3.1. Characteristics of the Sample and Stratification of the Population At total of 697 individuals were studied (Table 1). This sample of Mexico City population had been previously characterized for its ancestry informative makers by ad- mixture mapping analysis, showing an estimated propor- tion of 65% component of Amerindian, 30% European Table 2. Linear model for CYP2C19 associtation with type 2 diabetes. Source Type I Sum of Squares df Mean Square F Sig. Noncent. Parameter Observed Power(a) Corrected Model 48.1(b) 4 12,042 87,292 0.000 349,169 1,000 Intercept 94,616 1 94,616 685,863 0.000 685,863 1,000 Homocygotic 0.8 1 0.800 5800 0.016 5,800 0.671 Amerindian index 0.186 1 0.186 1,346 0.247 1,346 0.212 Sex 16,589 1 16,589 120,254 0.000 120,254 1,000 In (Age) 30,593 1 30,593 221,769 0.000 221,769 1,000 Error 58,216 422 0.138 Total 201,000 427 Corrected Total 106,384 426 a: Computed using alpha = 0.05; b: r Squared = 0.453 (Adjusted R Squared = 0.448). Homocygotic referes to 2,2 genotype for CYP2C19. Amerindian index was obteinded from admixmap and equals the integer of ameriandian component multiplied by 10 and divided by 4. 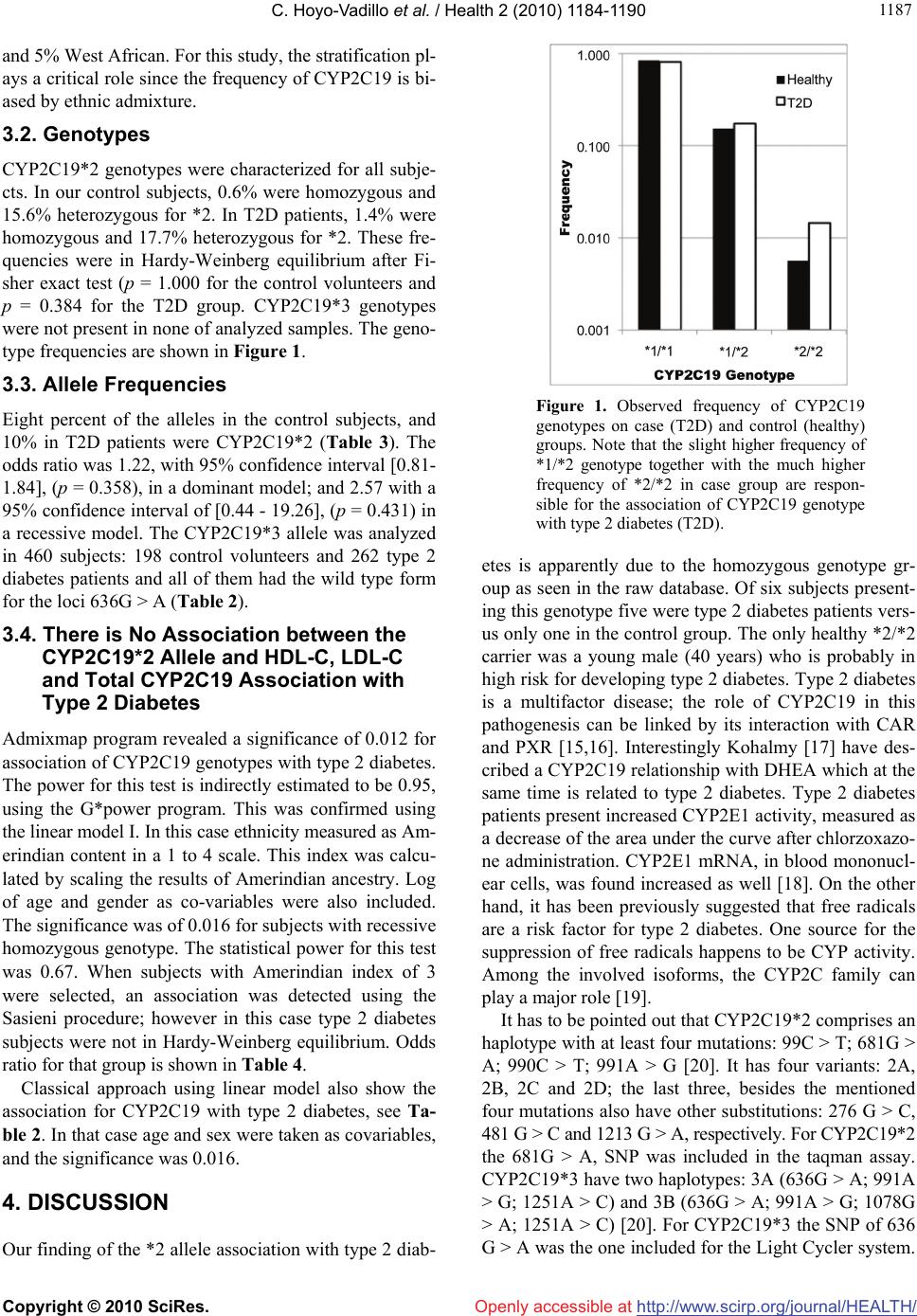 C. Hoyo-Vadillo et al. / Health 2 (2010) 1184-1190 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 118 1187 and 5% West African. For this study, the stratification pl- ays a critical role since the frequency of CYP2C19 is bi- ased by ethnic admixture. 3.2. Genotypes CYP2C19*2 genotypes were characterized for all subje- cts. In our control subjects, 0.6% were homozygous and 15.6% heterozygous for *2. In T2D patients, 1.4% were homozygous and 17.7% heterozygous for *2. These fre- quencies were in Hardy-Weinberg equilibrium after Fi- sher exact test (p = 1.000 for the control volunteers and p = 0.384 for the T2D group. CYP2C19*3 genotypes were not present in none of analyzed samples. The geno- type frequencies are shown in Figure 1. 3.3. Allele Frequencies Eight percent of the alleles in the control subjects, and 10% in T2D patients were CYP2C19*2 (Table 3). The odds ratio was 1.22, with 95% confidence interval [0.81- 1.84], (p = 0.358), in a dominant model; and 2.57 with a 95% confidence interval of [0.44 - 19.26], (p = 0.431) in a recessive model. The CYP2C19*3 allele was analyzed in 460 subjects: 198 control volunteers and 262 type 2 diabetes patients and all of them had the wild type form for the loci 636G > A (Table 2). 3.4. There is No Association between the CYP2C19*2 Allele and HDL-C, LDL-C and Total CYP2C19 Association with Type 2 Diabetes Admixmap program revealed a significance of 0.012 for association of CYP2C19 genotypes with type 2 diabetes. The power for this test is indirectly estimated to be 0.95, using the G*power program. This was confirmed using the linear model I. In this case ethnicity measured as Am- erindian content in a 1 to 4 scale. This index was calcu- lated by scaling the results of Amerindian ancestry. Log of age and gender as co-variables were also included. The significance was of 0.016 for subjects with recessive homozygous genotype. The statistical power for this test was 0.67. When subjects with Amerindian index of 3 were selected, an association was detected using the Sasieni procedure; however in this case type 2 diabetes subjects were not in Hardy-Weinberg equilibrium. Odds ratio for that group is shown in Table 4. Classical approach using linear model also show the association for CYP2C19 with type 2 diabetes, see Ta- ble 2. In that case age and sex were taken as covariables, and the significance was 0.016. 4. DISCUSSION Our finding of the *2 allele association with type 2 diab- Figure 1. Observed frequency of CYP2C19 genotypes on case (T2D) and control (healthy) groups. Note that the slight higher frequency of *1/*2 genotype together with the much higher frequency of *2/*2 in case group are respon- sible for the association of CYP2C19 genotype with type 2 diabetes (T2D). etes is apparently due to the homozygous genotype gr- oup as seen in the raw database. Of six subjects present- ing this genotype five were type 2 diabetes patients vers- us only one in the control group. The only healthy *2/*2 carrier was a young male (40 years) who is probably in high risk for developing type 2 diabetes. Type 2 diabetes is a multifactor disease; the role of CYP2C19 in this pathogenesis can be linked by its interaction with CAR and PXR [15,16]. Interestingly Kohalmy [17] have des- cribed a CYP2C19 relationship with DHEA which at the same time is related to type 2 diabetes. Type 2 diabetes patients present increased CYP2E1 activity, measured as a decrease of the area under the curve after chlorzoxazo- ne administration. CYP2E1 mRNA, in blood mononucl- ear cells, was found increased as well [18]. On the other hand, it has been previously suggested that free radicals are a risk factor for type 2 diabetes. One source for the suppression of free radicals happens to be CYP activity. Among the involved isoforms, the CYP2C family can play a major role [19]. It has to be pointed out that CYP2C19*2 comprises an haplotype with at least four mutations: 99C > T; 681G > A; 990C > T; 991A > G [20]. It has four variants: 2A, 2B, 2C and 2D; the last three, besides the mentioned four mutations also have other substitutions: 276 G > C, 481 G > C and 1213 G > A, respectively. For CYP2C19*2 the 681G > A, SNP was included in the taqman assay. CYP2C19*3 have two haplotypes: 3A (636G > A; 991A > G; 1251A > C) and 3B (636G > A; 991A > G; 1078G > A; 1251A > C) [20]. For CYP2C19*3 the SNP of 636 G > A was the one included for the Light Cycler system.  C. Hoyo-Vadillo et al. / Health 2 (2010) 1184-1190 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 1188 Table 3. Observed frequencies of CYP2C19*2 and *3. Healthy T2D Healthy T2D Healthy T2D Genotype Number of subjects Observed Frequency H-W Expected freq. *1/*1 295 279 83.8% 80.9% 83.9% 80.5% *1/*2 55 61 15.6% 17.7% 15.4% 18.5% *2/*2 2 5 0.6% 1.4% 0.7% 1.1% *1/*3 0 0 0% 0% 0% 0% *2/*3 0 0 0% 0% 0% 0% *3/*3 0 0 0% 0% 0% 0% Allele *1 91.6% 89.7% Allele *2 8.4% 10.3% Allele *3 0% 0% Table 4. Odds ratio for genotypes in healthy males (control) versus diabetic males. Risk allele 2 [1]<->[2] [11]<->[12] [11+]<->[22] [11]<->[12+22] Common odds ratio Odds_ratio = 1.992 Odds_ratio = 1.724Odds_ratio = 5.663 Odds_ratio = 1.907Odds_ratio = 2.741 C.I. = [1.084-3.659] C.I. = [0.904-3.289] C.I. = [0.310-103.496] C.I. = [1.006-3.617] chi2 = 5.09 chi2 = 2.78 chi2 = 2.54 chi2 = 4.00 chi2 = 4.88 p = 0.02404 (Pearson) p = 0.09571 p = 0.11067 p = 0.04543 p = 0.02711 Risk allele 1 [2]<->[1] [22]<->[12] [22]<->[11] [11+12]<->[22] Common odds ratio Odds_ratio = 0.502 Odds_ratio = 0.298Odds_ratio = 0.177 Odds_ratio=0.193 Odds_ratio=0.543 C.I. = [0.273-0.922] C.I. = [0.016-5.714] C.I. = [0.010-3.227] C.I. = [0.011-3.522] chi2 = 5.09 chi2 = 1.46 chi2 = 2.54 chi2 = 2.33 chi2 = 4.88 p = 0.02404 (Pearson) p = 0.22749 p = 0.11067 p = 0.12663 p = 0.02711 For both alleles just one SNP was included as the stan- dard technique proceeds, because SNPs always come together for a given haplotype, in other words, one SNP, the one shared for all variants, is enough to characterize the haplotype. Complex networks of nuclear receptors, where PXR and CAR are outstanding representatives, regulate the ex- pression of CYP3A4 and CYP2C19. The latter is modu- lated by glucocorticoids which at the same time interact with insulin for energy balance altogether with the part- icipation of the neuropeptide Y. Since it has been demonstrated that CAR also regu- lates the expression of CYP2C19 [21] the CYP2C family can be considered part of the xenosensing mechanism in charge of the regulation of cholesterol, bile acids and in- directly the uptake of dietary cholesterol as Handschind [16] describes it. Furthermore, the crosstalk between all the orphan nuclear receptors has been pointed out by several authors [22,23]. After all CYPS are not very substrate specific and the interesting conclusion of the Meyer group that the organism manages xenobiotics and drugs as toxic bile salts [24]. These changes suggest a crosstalk between the factors deriving in type 2 diabetes and the regulation of cytochromes P450. At the moment the regulation of CYP2C19 has been demonstrated but the inverse can be only assumed as part of a feedback system. In conclusion our work shows that the mexican popu-  C. Hoyo-Vadillo et al. / Health 2 (2010) 1184-1190 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 118 1189 Figure 2. Frequencies of main alleles of CYP2C19 on different populations. Colombians, Bolivians and Mexicans have the lo- wer frequencies for *2 allele. But Inuits have no presence of them. While Asians have the greatest frequencies for the *2 and only they present *3. lation of Mexico City has the same frequencies for CY- P2C19 wild and *2 alleles than the populations reported in Bolivia and Colombia (Figure 2). The *3 allele was not detected in our population just as in other Latin- Americans. 5. ACKNOWLEDGEMENTS The collaboration of Dr. Esteban Parra is fully acknowledged for get- ting the ancestry informative proportions of our population and with the use of ADMIXMAP. Financial support from Fundación IMSS and from Cinvestav are greatly appreciated REFERENCES [1] Chan, J., Donalson, L.M., Kushwaha, R.S., Ferdinan- dusse, S., VandeBerg, J.F. and VandeBerg, J.L. (2008) Differential expression of hepatic genes involved in cho- lesterol homeostasis in high- and low-responding strains of laboratory opossums. Metabolism, 57(5), 718-724. [2] Bertilsson, L., Dahl, M.L., Dalen, P. and Al-Shurbaji, A. (2002) Molecular genetics of CYP2D6: Clinical rele- vance with focus on psychotropic drugs. British Journal of Clinical Pharmacology, 53(2), 111-122. [3] Wedlund, P.J. (2000) The CYP2C19 enzyme polymer- phism. Pharmacology, 61(3), 174-183. [4] Hamdy, S.I., Hiratsuka, M., Narahara, K., El-Enany, M., Moursi, N., Ahmed, M.S., et al. (2002) Allele and geno- type frequencies of polymorphic cytochromes P450 (CYP2C9, CYP2C19, CYP2E1) and dihydropyrimidine dehydrogenase (DPYD) in the Egyptian population. British Journal of Clinical Pharmacology, 53(6), 596- 603. [5] Bravo-Villalta, H.V., Yamamoto, K., Nakamura, K., Baya, A., Okada, Y. and Horiuchi, R. (2005) Genetic poly- morphism of CYP2C9 and CYP2C19 in a Bolivian population: An investigative and comparative study. European Journal of Clinical Pharmacology, 61(3), 179- 184. [6] Isaza, C., Henao, J., Martinez, J.H., Arias, J.C. and Beltran, L. (2007) Phenotype-genotype analysis of CYP2C19 in Colombian mestizo individuals. BMC Cli- nical Pharmacology, 7(1), 6. [7] Jurima-Romet, M., Goldstein, J.A., LeBelle, M., Aubin, R.A., Foster, B.C., Walop, W., et al. (1996) CYP2C19 genotyping and associated mephenytoin hydroxylation polymorphism in a Canadian Inuit population. Pharma- cogenetics, 6(4), 329-339. [8] Lewis, D.F. (2003) Human cytochromes P450 associated with the phase 1 metabolism of drugs and other xeno- biotics: A compilation of substrates and inhibitors of the CYP1, CYP2 and CYP3 families. Current Medicinal Chemistry, 10(19), 1955-1972. [9] Cavalli-Sforza, L.L., Menozzi, P. and Piazza, A. (1994) The history and geography of human genes. Princeton University Press, Princeton. [10] Handschin, C., Podvinec, M. and Meyer, U.A. (2000) CXR, a chicken xenobiotic-sensing orphan nuclear re- ceptor, is related to both mammalian pregnane X receptor (PXR) and constitutive androstane receptor (CAR). Pro- ceedings of National Academic Sciences of USA, 97(20), 10769-10774. [11] Martinez-Marignac, V.L., Valladares, A., Cameron, E., Chan, A., Perera, A., Globus-Goldberg, R., et al. (2007) Admixture in Mexico City: implications for admixture mapping of type 2 diabetes genetic risk factors. Human Genetics, 120(6), 807-819. [12] Borlak, J. and Thum, T. (2002) Identification of major CYP2C9 and CYP2C19 polymorphisms by fluorescence resonance energy transfer analysis. Clinical Chemistry, 48(9), 1592-1594. [13] Strom, T.M.W. (2009) Case-control studies. http://ihg2. helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl [14] Sasieni, P.D. (1997) From genotypes to genes: Doubling the sample size. Biometrics, 53(4), 1253-1261. [15] Ourlin, J.C., Handschin, C., Kaufmann, M. and Meyer, U.A. (2002) A link between cholesterol levels and phe- nobarbital induction of cytochromes P450. Biochemistry and Biophysics Research Communication, 291(2), 378- 384. [16] Handschin, C., Podvinec, M., Amherd, R., Looser, R., Ourlin, J.C. and Meyer, U.A. (2002) Cholesterol and bile acids regulate xenosensor signaling in drug-mediated in- duction of cytochromes P450. Journal of Biological Chemistry, 277(33), 29561-29567. [17] Kohalmy, K., Tamasi, V., Kobori, L., Sarvary, E., Pas- cussi, J.M., Porrogi, P., et al. (2007) Dehydroepiandros- terone induces human CYP2B6 through the constitutive androstane receptor. Drug Metabolism and Disposition, 35(9), 1495-1501. [18] Z. Wang, S.D. Hall, J.F. Maya, L. Li, A. Asghar, J.C. Gorski, Diabetes mellitus increases the in vivo activity of cytochrome P450 2E1 in humans. British Journal of Clinical Pharmacology, 55(1), 77-85. [19] S. Elmi, N. A. Sallam, M. M. Rahman, X. Teng, A. L. Hunter, F. Moien-Afshari, et al. (2008) Sulfaphenazole treatment restores endothelium-dependent vasodilation in diabetic mice. Vascular Pharmacology, 48(1), 1-8.  C. Hoyo-Vadillo et al. / Health 2 (2010) 1184-1190 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 1190 [20] Sim, S.C. (2008) Home Page of the Human Cytochrome P450 (CYP) Allele Nomenclature Committee. http:// www.cypalleles.ki.se/ [21] Chen, Y., Ferguson, S.S., Negishi, M. and Goldstein, J.A. (2003) Identification of constitutive androstane receptor and glucocorticoid receptor binding sites in the CYP2- C19 promoter. Molecular Pharmacology, 64(2), 316-324. [22] Gerbal-Chaloin, S., Pascussi, J.M., Pichard-Garcia, L., Daujat, M., Waechter, F., Fabre, J.M., et al. (2001) In- duction of CYP2C genes in human hepatocytes in pri- mary culture. Drug Metabolism and Disposition, 29(3), 242-251. [23] Handschin, C. and Meyer, U.A. (2005) Regulatory net- work of lipid-sensing nuclear receptors: roles for CAR, PXR, LXR, and FXR. Archives of Biochemistry and Biophysics, 433(2), 387-396. [24] Handschin, C., Gnerre, C., Fraser, D.J., Martinez- Jimenez, C., Jover, R. and Meyer, U.A. (2005) Species- specific mechanisms for cholesterol 7alpha-hydroxylase (CYP7A1) regulation by drugs and bile acids. Archives of Biochemistry and Biophysics, 434(1), 75-85. |

