Advances in Microbiology
Vol. 2 No. 4 (2012) , Article ID: 26214 , 12 pages DOI:10.4236/aim.2012.24079
Molecular Characterization of Listeria monocytogenes Based on the PFGE and RAPD in Korea
1Seoul Metropolitan Government Research Institute of Public Health and Environment, Seoul, Korea
2Animal Disease Diagonsis Division, National Veterinary Research and Quarantine Service, Anyang City, Korea
3Veterinary Pharmacology Division, National Veterinary Research and Quarantine Service, Anyang City, Korea
4College of Veterinary Medcine, Chungnam National University, Daejeon, Korea
Email: *sanghun93@seoul.go.kr
Received September 25, 2012; revised October 27, 2012; accepted November 5, 2012
Keywords: Listeria monocytogene; PFGE; RAPD
ABSTRACT
This study was performed to characterize 35 L. monocytogenes isolates from animals, foods, environmental samples collected between 1997 and 2007 with no apparent epidemiological relations, and five reference isolates using serotypic, genotypic and molecular typing methods to understand the pattern of strain distribution in Korea. For this study, we used serotyping and detected 6 different virulence-associated genes (inlA, inlB, plcA, plcB, hlyA, and actA) and 16s rRNA using multiplex-PCR. We also compared RAPD and PFGE to determine genetic characterization of L. monocytogenes strains to define the genetic diversity. Serotype patterns of the 30 L. monocytogenes strains was as follows; 9 isolates (30.0 %) belonged to serotype, 7 isolates (23.3%) belonged to serotype 4b, 4 isolates (13.3%) belonged to serotype 1/2b, 3 isolates (10.0%) belonged to serotype 1/2c, 2 (6.7%) isolates belonged to 4c, 2 (6.7%) isolates belonged to NT (Non Type), one isolate (3.2%) belonged to 3a and 3b, and 4a, respectively. Although, a limited number of isolates were analyzed in this study, molecular typing with RAPD and PFGE indicated that PFGE is more discriminatory for the subtyping L. monocytogenes than RAPD. Some L. monocytogenes isolates by RAPD and PFGE types are associated with specific sources. And, combining data obtained by these methods will increases the likelihood of strain discrimination.
1. Introduction
Listeriosis is a foodborne disease caused by L. monocytogenes, and a severe disease characterized by abortion, meningitis and septicemia with lethality around 20% - 30% of cases. L. monocytogenes is a ubiquitous and psychotropic micro-organism which can contaminate food at all steps of the food chain [1].
L. monocytogenes is a Gram-positive, non-spore forming, intracellular, facultative anaerobic rod, growing at refrigeration temperature that causes invasive, often fatal, disease in susceptible hosts [2].
Epidemiological investigations have revealed that L. monocytogenes is ubiquitously distributed throughout the environment and in foods [3]. Foodstuffs associated with listeriosis epidemics have included among other foods both milk and dairy products [4].
Phenotypic methods often yield a low power of discrimination in strains (e.g., serotyping), suffer from biological variability and may not be applicable to all strains [5]. The classical method of bacteriological identification for Listeria spp., including L. monocytogenes, is laborious and time consuming.
Recently, the use of multiple key virulence factors such as hlyA [6], internalin B (inlB) [7], actA, plcA [8], plcB [9], to detect L. monocytogenes have been described. Several molecular typing have been used to differentiate Listeria species that include RAPD (Random Amplification of Polymorphic DNA), AP-PCR (Arbitrarily Primed PCR), ERIC-PCR (Enterobacterial Repetitive Intergenic Consensus PCR), REP-PCR (Repetitive sequence-based PCR) and virulence gene sequencing have been used to characterize strains of L. monocytogenes [10-13]. PFGE (Pulsed Field Gel Electrophorosis) method is standardized, reliable and reproducible, and as such is useful when conducting comparative genetic analysis and for molecular subtyping of L. monocytogenes isolates [13].
The sources of infection in Korea are unknown to a great extent, and no food associated and environment cases have been reported. So, the purpose of the present study was to characterize epidemiologically unrelated environmental, animal and food L. monocytogenes isolates using serotypic, genotypic and molecular typing methods to understand the pattern of strain distribution in Korea.
2. Materials and Methods
2.1. Isolation of L. monocytogenes
A total of 35 L. monocytogenes isolates were used for experiment. 16 L. monocytogenes isolates were obtained from Dr. Byeong Yeal Jung (National Veterinary Research & Quarantine Service, Anyang). In particular, L. monocytogenes obtained from different animal species (porcine and bovine), environment (poultry slaughtering plant and dairy plant waste water) and meat (imported beef and pork) and from different foods (milk and cheese) from 1997 to 2007, were used. And five additional strains were purchased from the ATCC. Data are summarized in Table 1.
Twenty-five mL of buffer including the swabs were inoculated in 225 mL of Fraser Broth (Becton, Dickinson and company sparks, USA), and blended. All samples were incubated at 30˚C for 48 h. A portion (10 μL) of the enrichment broth was streaked on Palcam agar plate (Merk, Germany). After 24 - 48 h incubation at 37˚C the plates were examined for typical L. monocytogenes colonies, which were streaked for purity on horse blood agar plates. Hemolytic colonies on horse blood agar were confirmed as L. monocytogenes by API Listeria kit (Biomerieux, Korea).
2.2. Serotyping
Serotyping was performed using commercial Listeria antisera according to the instructions given by the manufacturer (DENKA SEIKEN CO., LTD., Tokyo, Japan). This method differentiates L. monocytogenes into 13 different serotypes based on the association of somatic (O) and flagellar (H) antigens with a series of polyvalent and monovalent antisera.
2.3. Preparation of Genomic DNA
Genomic DNA was extracted using the AccuPrep® Genomic DNA Extraction Kit (Bioneer, Korea) according to the manufacturer’s instruction.
2.4. Identification by the Multiplex-PCR of Virulence-Associated Genes
All of the primers used for specific PCR amplifications of the entire coding sequences of virulence-associated genes are reported in Table 2. PCR was performed in a PCR 9600 thermal cycler (Perkin-Elmer Corporation). A 50-μL aliquot contained buffer (10 mM Tris-HCl, 50 mM KCl, 2.5 mM MgCl2 [pH 8.3]), the dNTP mixture (TaKaRa, Japan) 0.25 mM each, 10 pmols of primer, 25 ng of DNA, and 0.8 U of Taq DNA polymerase (TaKaRa, Japan). The cycling conditions were the template DNA was denatured at 94˚C for 3 min followed by 35 cycles of amplification (each cycle consisted of denaturation at 94˚C for 1 min, annealing at 60˚C for 2 min and elongation at 72˚C for 1 min). 5 microlitre of the amplified products was separated by electrophoresis in 1.5% agarose gel containing ethidium bromide, and visualized under UV.
2.5. RAPD Fingerprinting
For RAPD-PCR, the HLWL 74 (5’-ACG TAT CTG C-3’) was used. The PCR mixture consisted of buffer (10 mM Tris-HCl, 50 mM KCl, 2.5 mM MgCl2 [pH 8.3]), the dNTP mixture (TaKaRa, Japan) 0.25 mM each, 10 pmols of primer, 25 ng of DNA, and 0.8 U of Taq DNA polymerase (TaKaRa, Japan) in a total volume of 25 μL. Each sample was subjected to an initial denaturation step of 95˚C for 4 min, followed by 45 amplification cycles of 1 min of 95˚C, 2 min at 35˚C, and 2 min at 72˚C 1 min and followed by final extension of 72˚C for 10 min. All PCR amplifications were carried out in a TGRADIENT (Biometra, Germany) [14].
2.6. PFGE
PFGE was performed according to the PulseNet standardized protocol, with ApaI as restriction endonuclease (Roche, Germany). Bacteria were grown on BHI agar plates at 37˚C for 16 - 18 h. Cell were removed from the plate to plastic tubes (Falcon 2057, 14 mL 17 × 100 mm) containing 3 mL of TE buffer using a sterile cotton swab and the cell density adjusted. The electrophoretic parameters used were as follows; initial switch time, 4.0 s; final switch time, 40.0 s; sum time. 22 h; angle, 120˚; gradient, 6.0 V/cm; temperature, 14˚C; ramping factor, linear. After electrophoresis, the gels were stained for 15 - 20 min in 250 mL of deionized water containing 25 μL of ethidium bromide (10 mg/mL) and destained by three washes of 20 - 30 min each using 500 mL of deionized water [15].
2.7. Discrimination Index
The discriminatory power of the subtyping methods employed in this study was determined by calculating the discrimination index (DI) using the formula of Hunter and Gaston [16].
3. Results
The serotype distribution of the 30 L. monocytogenes
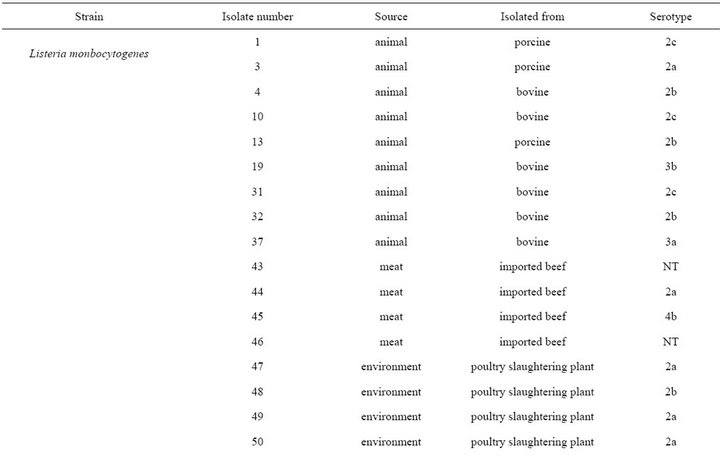

Table 1. List of the strains used in this study.
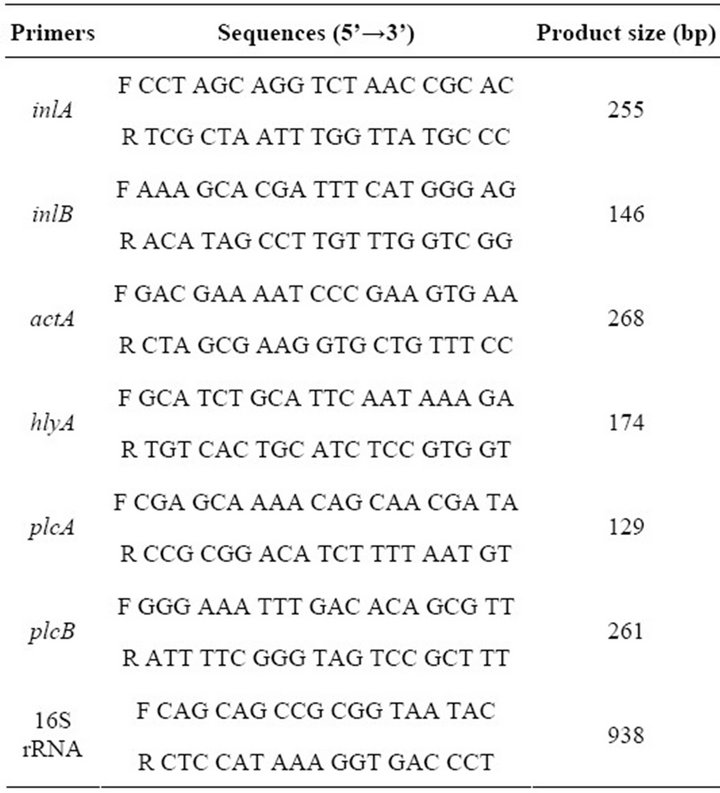
Table 2. Primer pairs used for amplification of virulence genes and 16s rRNA in Listeria isolates.
strains was as follows; 9 isolates (30.0 %) belonged to serotype 1/2a, 7 isolates (23.3%) belonged to serotype 4b, 4 isolates (13.3%) belonged to serotype 1/2b, 3 isolates (10.0%) belonged to serotype 1/2c, 2 (6.7%) isolates belonged to 4c, 2 (6.7%) isolates belonged to NT (Non typeable), one isolate (3.2%) belonged to 3a and 3b, and 4a, respectively (Table 1).
PCR products of the 6 different virulence-associated genes and 16s rRNA (Table 2) were obtained DNA from all Listeria strains considered in this study, except for strain No.37, 62, 80, 82, 83, 84, and s3 which was negative for the hlyA gene. All of the amplification products were of the expected size, except for the actA gene PCR products from 10 strains of serotypes 1/2b, 4b and 7 which had molecular weights lower than expected. Twenty-one isolates (70%) showed the expected actA product size of 268 bp, while nine isolates (30%) showed a higher product size of 385 bp. Most of the reference strains (s1, 2, 4, and s5) showed 385 bp, while s3 had 268 bp product. In this study, the isolates each had 16S rRNA and six virulence-associated genes (hlyA, plcA, plcB, inlA, inlB, and actA), suggesting that they are potentially pathogenic (Table 3).
RAPD (HLWL 74) types of all isolated strains are presented in Figutr 1 and Table 4. It demonstrates examples of all RAPD types indicated in our studies. Among the 35 strains of L. monocytogenes examined (30 isolates and 5 reference/type strains) it was possible to identify 19 different banding types at a relative genetic similarity of 80%. RAPD (HLWL74) of genomic DNA from L. monocytogenes isolates generated multiple DNA fragments in sizes ranging between 100 and 2000 bp. Among them, five types (D, O, P, Q, and S) had two more isolates and D and S types had same cluster having more than two different strains. RAPD (HLWL 74) type S was the dominant type (10 strains) and was isolated all animal (porcine and bovine) isolates, some imported beef (No.43), and cheese isolates (s3 and s4). The most predominant type was S1. Interestingly, an identical RAPD profile (S1) was observed in all porcine samples. Three types found in bovine belonged to E1, K1, and S1. Especially, profile S1 was detected in isolates from the bovine (66.7%, 4/6). Four types had shown in imported beef and poultry slaughtering plant, respectively. Bovine and raw milk had three types. S1 cluster represented 66.7% (n = 4) in bovine and D cluster accounted for 50% (n = 2) in raw milk. Dairy plant waste water had Q1 (75%, n = 3) and R1 (25%, n = 1) and pork isolates was identified in O1 (40%, n = 2) and P1 (60%, n = 3). Five reference strains belonged to four types (K1, M1, N1 and S1), and profile S1 represented of two reference strains (s3 and s4).
PFGE (ApaI) types of all isolated strains are presented in Figure 2 and Table 5. It demonstrates examples of all PFGE types indicated in our studies. Among the 35 strains of examined (30 isolates and 5 reference/type strains), dendrogram analyses of the PFGE profiles showed that the 35 L. monocytogenes isolates from 21 different PFGE profiles with discriminatory indexes 0.937 at 80% relative genetic similarity. The most predominant type was R. Interestingly, an identical PFGE profile (R3) was observed in all porcine samples. Also profile R3 was detected in isolates from the bovine (33.3%, n = 2). Dairy plant waste water and pork were clustered in (B1, F, and G1) and (I1 and L1), respectively. Five reference strains belonged to four types (E1, H1, and R2), and profile S represented of two reference strains (s1 and s4).
Among the 30 L. monocytogenes and 5 reference strains examined, results calculated for RAPD (HLWL74) and PFGE (ApaI) typing at 70%, 80%, 90%, and 95% relative genetic similarity were shown in Table 6.
4. Discussion
Studies have implicated contaminated foods such as cheese, milk and beef in the transmission of L. monocytogenes to human [3,5,17]. Similarly, animals can be infected through the consumption of contaminated feeds and environment also can be contaminated with L. monocytogenes. It is also important to investigate relation of contaminated foods and source of L. monocytogenes (e.g. animals, raw materials, and environment source). Thus, we used serotyping and PCR (6 different virulence-associated genes and 16s rRNA). We also compared RAPD and PFGE analyses to determine ge-
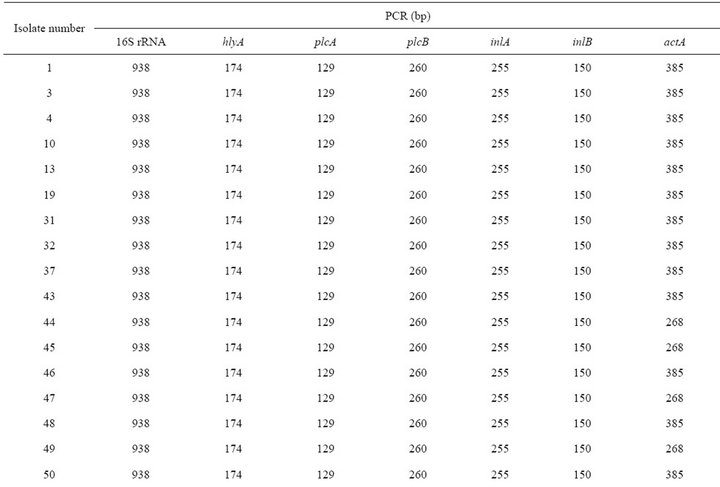
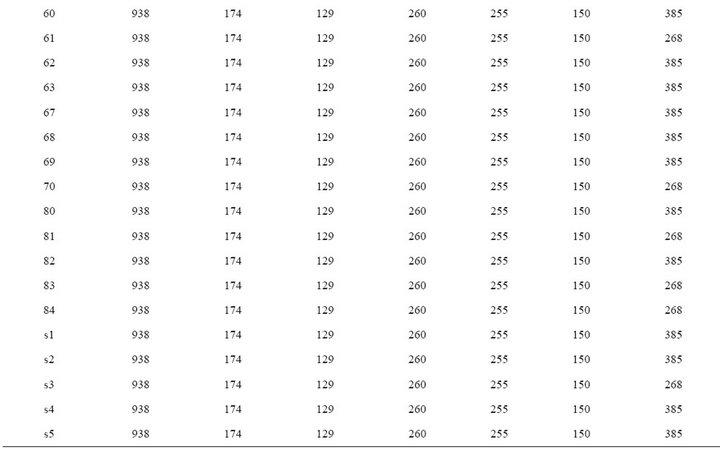
Table 3. Genotypic characterization of virulence genes in L. monocytogenes isolates.
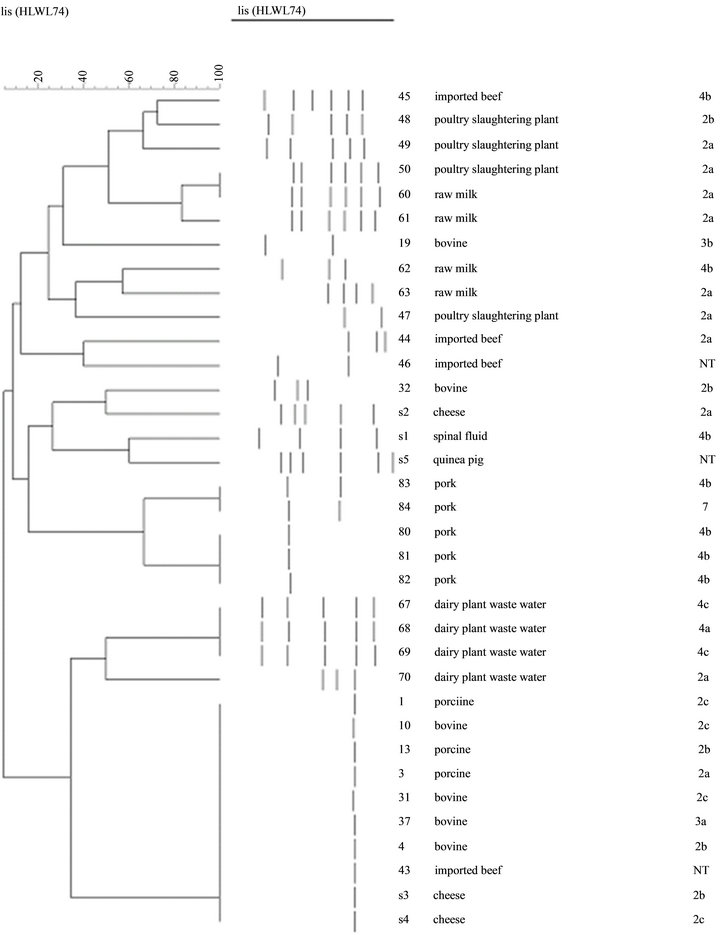
Figure 1. Dendrogram for 30 L. monocytogenes isolates and L. monocytogenes ATCC analyzed by RAPD (HLWL74). Percentages of similarity are shown above the dendrogram. Perpendicular line was stand for 80% relative genetic similarity. The origins of strains are shown in Table 1; 2a:1/2a, 2b:1/2b, 2c:1/2c.
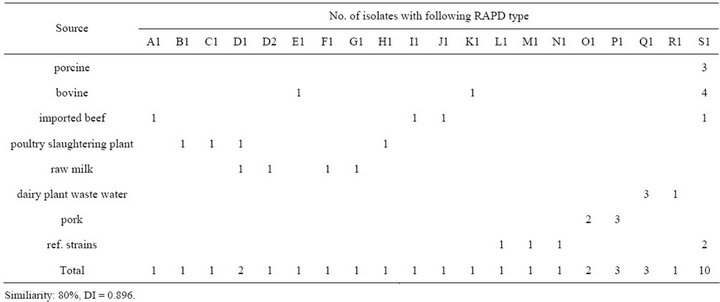
Table 4. Distribution of RAPD types determined with HLWL74 primer in L. monocytogenes isolates from different sources.
netic characterization of L. monocytogenes strains isolated from animals, raw materials, and environment to define the genetic diversity. Most of the pathogenic L. monocytogenes serotype is limited to 1/2a, 1/2b, and 4b. Especially, at least 95% of L. monocytogenes strains isolated from human listeriosis cases are of those three serotypes [18]. Out of 30 isolates, serovar 1/2a was predominant with 9 strains followed by serovars 4b and 1/2b with 7 and 4 strains, respectively. Figure 1 shows that serovars 1/2a, 1/2b, and 4b were predominant with 20 strains (66.7%) of 30 strains, in agreement with a recent report in Korea [19], showing that 90% of L. monocytogenes isolates were serotype 1 and only 4.1% were type 4. It was reported that these serotypes were widely distributed in food, animal, and environment of Korea. Chung HC et al. indicated that some serotype 1/2 isolates which showed that the H-antigenic factors could not be fully determined by traditional antiserum serotyping [20]. Because most human infections are reported to be associated with 1/2a, 1/2b, and 4b, our results were suggested that three major serotypes may be particularly important. Therefore, most of the organism isolated in this study that could lead to human infections and were then possibly pathogenic for human. 1/2c was shown in only animal. This finding may reflect a greater capacity of 1/2c strains to survive and multiply in the skin of animals. Also, it is interesting to note that serovar 4b, which causes 2/3 of the human infections, was not detected in animal. It may be assumed that serovar 4b does not play an important role in animal contamination. Out of 47 strains of L. monocytogenes, 16 belonged to serovar 1/2a, 30 to serovar 1/2c and only one to serovar 4b [21]. This study was similar to our results that most of the serovar was 1/2a, 1/2b, and 4b. Chung HC et al. reported that 54.4% (31/57) of human isolates belonged to serotype 4band 31.6% (18/57) belonged to serotypes 1/2a and 1/2b. These three serotypes were predominant among the L. monocytogenes isolated from human listeriosis cases and other types of isolates (food, environment, and animal) [19]. Many countries have seen a shift in the L. monocytogenes serotypes causing human infections from predominantly serotype 4b to 1/2a and 1/2b [22].
In general, PCR products of the virulence genes did not show polymorphism except for the actA gene [23]. Nine (30%) of these isolates showed the expected 268 bp actA gene products, whereas twenty one (70%) produced the 385 bp product. Among them, all isolates (100%) of animals (porcine and bovine) showed 385 bp products. Polymorphism of the actA gene for L. monocytogenes demonstrated by Wiedmann M et al. and can divided into two groups based on the actA gene sequence [24]. Many reports has been identified polymorphism of L. monocytogenes genes such as for hlyA [25], iap (murein hydrolase) [26] and inlA and inlB (internalin) [27]. However, in our study, we did not identify any polymorphism in the PCR products of these genes and other genes excepting actA. It might be that the primers used here have the target sequence located outside the areas of polymorphism, or the target regions may have been too small to detect any variations [23]. Jaradat ZW et al. also insisted that sequence analysis of these genes can possibly reveal a better picture of the relatedness of the isolates. Jacquet C et al. found that low molecular mass of ActA was due to a deletion in actA. As this deletion is detected in human strains, (i.e., pathogenic strains), it is probably not in the amino-terminal region, which is essential for F-actin assembly and movement [28]. In vitro motility assays showed that this deletion decreases the motility of the bacteria [29]. In mouse model, a mutant with a mutation in the proline-rich region was less virulent than the
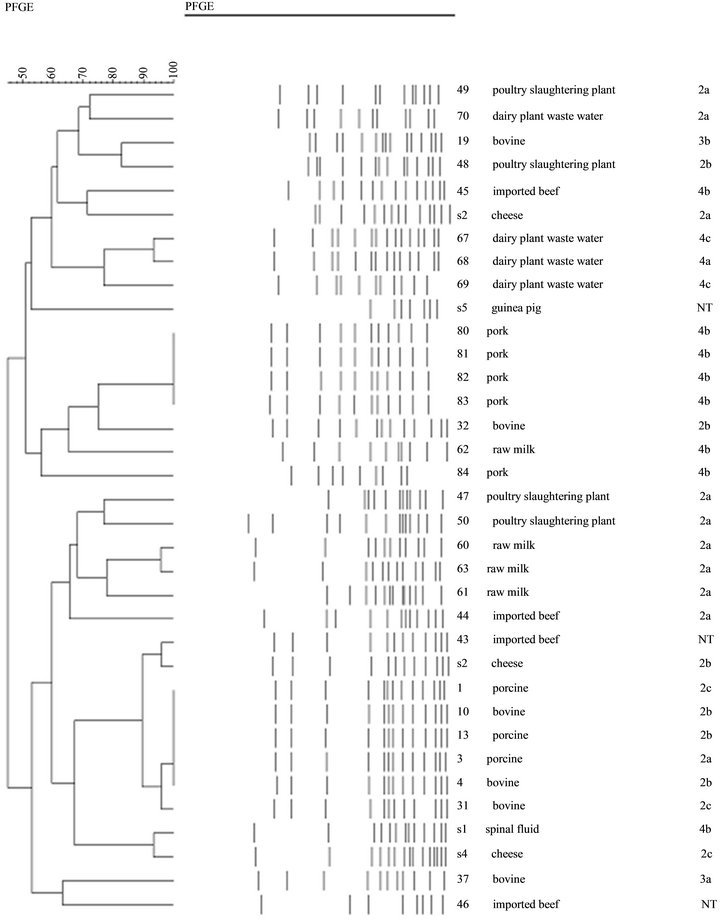
Figure 2. Dendrogram for 30 L. monocytogenes isolates and L. monocytogenes ATCC analyzed by PFGE (ApaI). Percentages of similarity are shown above the dendrogram. Perpendicular line was stand for 80% relative genetic similarity. The origins of strains are shown in Table 1.; 2a:1/2a, 2b:1/2b, 2c:1/2c.
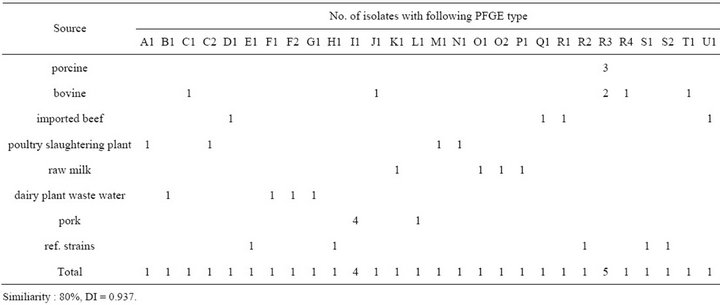
Table 5. Distribution of PFGE types determined with ApaI in L. monocytogenes isolates from different sources.
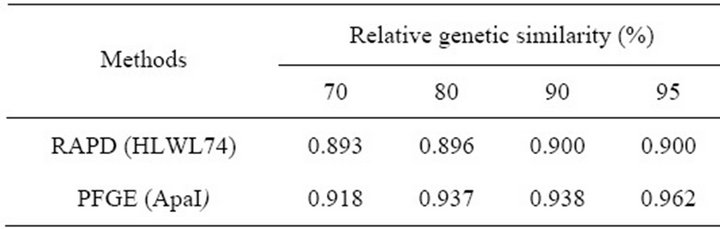
Table 6. Comparison of discriminative ability among RAPD (HLWL74) and PFGE (ApaI) methods used to subtyping of L. monocytogenes.
wild type [30]. Further studies are required to understand virulence among isolates compared to different molecular size of actA gene.
RAPD typing has been shown to be of value in resolving some specific epidemiological questions [31]. Some authors have been comparing different typing methods PFGE and RAPD showing the highest discriminatory efficiency on a set of L. monocytogenes [32,33].
In this study, we have identified 19 different banding types at a relative genetic similarity of 80%. 19 patterns were found after RAPD typing L. monocytogenes using primer HLWL74, of which Q type accounted for 75.0% (3/4) of the isolates from dairy plant waste water. Of the animal isolates, 7 (77.8%) belonged to RAPD type S. Pork samples were found to type O (2/5, 40%) and P type (3/5, 60%). Recently, Luca C. et al. published a study on the characterization of L. monocytogenes isolated from various sources (food, environment, and animals), and investigate their capability to define the strain origin [11]. Mazurier SI et al. found that identical patterns were found with the primer HLWL74 of L. monocytogenes and suggested that RAPD offers an attractive alternative to phage typing [34]. Niederhauser C et al. used a 19-mer primer to subtype 57 L. monocytogenes isolates and reported that the method allowed the tracing of L. monocytogenes contamination in several food outlets to be traced back to a food processing plant. They found RAPD to be highly discriminating for subtyping [35].
PFGE has been considered as a standard subtyping method for L. monocytogenes [36]. And PFGE is a method with high discriminatory power and it has shown to be very accurate and reproducible for fine structure comparison and molecular typing of L. monocytogenes [2, 37]. Types of PFGE did not show any significant match to bacterial serotypes. It is generally considered laborious and time-consuming and thus also too expensive for preliminary screening of hundreds of isolates as required in extensive hygiene surveys for tracing contamination sources [38]. Our data indicate that the PFGE molecular subtypes of the 35 L. monocytogenes strains including ATCC reference strains are presented in a dendrogram that utilized the PFGE data following ApaI digestion. Results indicate that PFGE (ApaI) digestions provided high levels of discrimination between isolates from animal, meat, milk, and environment samples with discriminatory indexes 0.937 at 80% relative genetic similarity. The two enzymes have also been recommended and used in subtype differentiating and epidemiologic studies for L. monocytogenes [39]. Our result showed that the 35 L. monocytogenes isolates from various different PFGE profiles (21 pulsotypes) with discriminatory indexes 0.937 at 80% relative genetic similarity. We have identical pattern from same source (F type: dairy plant waste water, I type: pork, O type: raw milk, R type: animal isolates). We noted that animal isolates (R type) unrelated imported beef (No. 43) shared identical patterns, suggesting that they are related. This result was consistent with RAPD. In many cases, the genetic patterns of strains showed no association with any of the properties that correlated with their origin [40]. It was also demonstrated that similar subtypes of strains can be found in different product types and in different processing environments and that, therefore, the recovery of identical patterns form various food and patient strains does not prove that a particular food is the vehicle of infection [41]. It was reported a similar finding with an identical PFGE pattern that was shared by different serotypes, even from different flagella antigen groups [42]. It was indicated that the poor correlation between serotyping and molecular subtyping may be due to horizontal gene transfers or point mutations in genomic DNA resulting in phenotypic shifts that affect serotyping [43]. In the results of the present study, specific PFGE types could not be connected with serotype, especially most animal isolates could differentiated with Q type showing serotypes 1/2a, 1/2b, and 1/2c which is in agreement with the results of other studies having examined the relation of L. monocytogenes between subtypes and genotypes. For example, a study was carried in pork slaughtering and cutting plants inspected 287 isolates that produced 17 ApaI PFGE types and four serotypes (serotypes 1/2a, 3a, 1/2c, and 3c) [44]. This study showed that there was overrepresentation of one L. monocytogenes PFGE type that accounted for 90% of the isolates (1/2a). A report that investigated the prevalence of L. monocytogenes in frankfurter packages from seven plants showed that 90% had the same ribotype profile and serotype [45]. Other investigators have also shown that PFGE and serotyping showed that L. monocytogenes is heterogeneous serologically and genetically as observed herein [13]. Correlations between molecular subtyping and serotyping of L. monocytogenes have been reported previously [43]. There is currently no thorough knowledge of the molecular basis for the relationship between serotypes and molecular subtyping of L. monocytogenes [13]. But, genomic DNA isolates of serotype 4b digested by ApaI enzyme in our study showed relatively distinguishable patterns not including No.45 sample and reference strain s1. It was indicated that identical PFGE patterns belonged to the same serotype [20,41]. The use of both RAPD and PFGE for typing L. monocytogenes has previously been reported to identify similar group and isolates from different sources [11,44]. However, Destro MT et al. insisted that using more than one method may increase the discriminatory ability [46].
Our results showed that DI values varied at different relative genetic similarity (70% - 95%). The most discriminate DI value was 0.900 at 95% relative genetic similarity. It was proposed that the higher the DI, the more discriminatory is the procedure [47]. But, it was insisted that DI values must be regarded with caution for small samples and typing schemes should not be validated with limited sample sizes [16]. So, we choose 80% relative genetic similarity for differentiating L. monocytogenes isolated various sources. Most of the serotype showed not consistent with RAPD type but 4b serotype had same RAPD type (except s1 and No.45). And these results were same as PFGE.
REFERENCES
- J. Bille and J. Rocourt, “WHO International Multicenter Listeria monocytogenes Subtyping Study-Rationale and Set-Up of the Study,” International Journal of Food Microbiology, Vol. 32, No. 3, 1996, pp. 251-262. doi:10.1016/S0168-1605(96)01140-3
- A. Vines and B. Swaminathan, “ Identification and Characterization of Nucleotide Sequence Differences in Three Virulence-Associated Genes of Listeria monocytogenes Strains Representing Clinically Important Serotypes,” Current Microbiology, Vol. 36, No. 5, 1998, pp. 309-318. doi:10.1007/s002849900315
- J. M. Farber and P. I. Peterkin, “Listeria monocytogenes, a Food-Borne Pathogen,” Microbiological Reviews, Vol. 55, No. 3, 1991, pp. 467-511.
- M. K. Miettinen, K. J. Bjorkroth and H. J. Korkeala, “Characterization of Listeria monocytogenes from an Ice Cream Plant by Serotyping and Pulsed-Field Gel Electrophoresis,” International Journal of Food Microbiology, Vol. 46, No. 3, 1999, pp. 187-192. doi:10.1016/S0168-1605(98)00185-8
- H. Even, A. L. Bjorn, J. R. Ole, V. Traute, K. Georg and N. Truls, “Molecular Epidemiology and Disinfectant Susceptibility of Listeria monocytogenes from Meat Processing Plants and Human Infections,” International Journal of Food Microbiology, Vol. 96, No. 1, 2004, pp. 85- 96. doi:10.1016/j.ijfoodmicro.2004.03.014
- F. X. Riedo, R. E. Weaver, B. D. Plikaytis and C. V. Broome, “A Point-Source Foodborne Listeriosis Outbreak: Documented Incubation Period and Possible Mild Illness,” The Journal of Infectious Diseases, Vol. 170, No. 3, 1994, pp. 693-696. doi:10.1093/infdis/170.3.693
- S. J. Yong, F. Joseph, R. Frank, E. Brackett and C. Jinru, “Polymerase Chain Reaction Dection of Listeria monocytogenes on Frankfurters Using Oligonucleotide Primers Targeting the Genes Encoding Internalin AB,” Journal of Food Protection, Vol. 66, No. 2, 2003, pp. 237-241.
- M. Gianfranceschi, G. Fransiosa, A. Gattuso and P. Aureli, “Detection of Two Phospholipases C by Means of Plate Tests for the Rapid Identification of Pathogenic Listeria monocytogenes,” Archiv fur Lebensmittelhygiene, Vol. 49, No. 3, 1998, pp. 54-57.
- D. K. Winters, T. P. Maloney and M. G. Johnson, “Rapid Detection of Listeria monocytogenes by a PCR Assay Specific for an Aminopeptidase,” Molecular and Cellular Probes, Vol. 13, No. 2, 1999, pp. 127-131. doi:10.1006/mcpr.1999.0224
- B. Jersek, E. Tcherneva, N. Rijpens and L. Herman, “Repetitive Element Sequence-Based PCR for Species and Strain Discrimination in the Genus Listeria,” Letters in Applied Microbiology, Vol. 23, No. 1, 1996, pp. 55-60.
- C. Luca, S. Simone, N. Raffaella, B. Elena, C. Carlo and C. Giuseppe, “Analysis of PCR-Based Methods for Characterization of Listeria monocytogenes Strains Isolated from Different Sources,” International Journal of Food Microbiology, Vol. 103, No. 2, 2005, pp. 167-178. doi:10.1111/j.1472-765X.1996.tb00028.x
- Z. Xiaohui, J. Xinan and M. Wiedmann, “Listeria monocytogenes in the Chinese Food System: Strain Characterization through Partial actA Sequencing and Tissue-Culture Pathogenicity Assays,” Journal of Medical Virology, Vol. 54, No. 3, 2005, pp. 217-224. doi:10.1016/j.ijfoodmicro.2004.12.027
- O. Okwumabua, M. O’Connor, E. Shull, K. Strelow, M. Hamacher, T. Kurzynski and D. Warshauer, “Characterization of Listeria monocytogenes Isolates from Food Animal Clinical Cases: PFGE Pattern Similarity to Strains from Human Listeriosis Cases,” FEMS Microbiology Letters, Vol. 249, No. 2, 2005, pp. 275-281. doi:10.1016/j.femsle.2005.06.018
- K. Wernars, P. Boerlin, A. Audurier, E. G. Russell, G. D. Curtis, L. Herman and N. van der Mee-Marquet, “The WHO Multicenter Study on Listeria monocytogenes Subtyping: Random Amplification of Polymorphic DNA (RAPD),” International Journal of Food Microbiology, Vol. 32, No. 3, 1996, pp. 325-341. doi:10.1016/S0168-1605(96)01146-4
- L. M. Graves and B. Swaminathan, “PulseNet Standardized Protocol for Subtyping Listeria monocytogenes by Macrorestriction and Pulsed-Field Gel Electrophoresis,” International Journal of Food Microbiology, Vol. 65, No. 1-2, 2001, pp. 55-62. doi:10.1016/S0168-1605(00)00501-8
- P. R. Hunter and M. A. Gaston, “Numerical Index of the Discriminatiory Ability of Typing Systems and Application of Simpson’s Index of Diversity,” Journal of Clinical Virology, Vol. 26, No. 11, 1988, pp. 2456-2466.
- P. D. MacDonald, R. E. Whitwam, J. D. Boggs, J. N. MacCormack, K. L. Anderson, J. W. Reardon, J. R. Saah, L. M. Graves, S. B. Hunter and J. Sobel, “Outbreak of Listeriosis among Mexican Immigrants as a Result of Consumption of Illicitly Produced Mexican-Style Cheese,” Clinical Infectious Diseases, Vol. 40, No. 5, 2005, pp. 677-682. doi:10.1086/427803
- K. Sophia, “Listeria monocytogenes Virulence and Pathogenicity, a Food Safety Perspective,” Journal of Food Protection, Vol. 65, No. 11, 2002, pp. 1811-1829.
- S. S. Jang, C. Euiyoung, H. Kiseon, M. Takahisa, H. Sunggi and R. Sangryeol, “Antibiotic Resistance and Genetic Diversity of Listeria monocytogenes Isolated from Chicken Carcasses in Korea,” Journal of Microbiology and Biotechnology, Vol. 16, No. 8, 2006, pp. 1276-1284.
- H. C. Chung and C. Wang, “Genetic Relatedness between Listeria monocytogenes Isolates from Seafood and Humans Using PFGE and REP-PCR,” International Journal of Food Microbiology, Vol. 110, No. 2, 2006, pp. 135- 148. doi:10.1016/j.ijfoodmicro.2006.02.003
- J. Nicolas and N. Vidaud, “Problems of Listeriosis,” Proceedings of the 9th International Symposium, Nantes, University of Nantes, 1986, pp. 330-338.
- B. Swaminathan and P. Gerner-Smidt, “The Epidemiology of Human Listeriosis,” Microbes and Infection, Vol. 9, No. 10, 2007, pp. 1236-1243. doi:10.1016/j.micinf.2007.05.011
- Z. W. Jaradat, G. E. Schutze and A. K. Bhunia, “Genetic Homogeneity among Listeria monocytogenes Strains from Infected Patients and Meat Products from Two Geographic Locations Determined by Phenotyping, Ribotyping and PCR Analysis of Virulence Genes,” International Journal of Food Microbiology, Vol. 76, No. 1-2, 2002, pp. 1-10. doi:10.1016/S0168-1605(02)00050-8
- M. Wiedmann, J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough and C. A. Batt, “Ribotypes and Virulence Gene Polymorphisms Suggest Three Distinct Listeria monocytogenes Lineages with Differences in Pathogenic Potential,” Infection and Immunity, Vol. 65, No. 7, 1997, pp. 2707-2716.
- O. F. Rasmussen, T. Beck, J. E. Olsen, L. Dons and L. Rossen, “Listeria monocytogenes Isolates Can Be Classified into Two Major Types According to the Sequence of the Listeriolysin Gene,” Infection and Immunity, Vol. 59, 1991, pp. 3945-3951.
- A. Bubert, I. Hein, M. Rauch, A. Lehner, B. Yoon, W. Goebel and M. Wagner, “Detection and Differentiation of Listeria spp. by a Single Reaction Based on Multiplex PCR,” Applied and Environmental Microbiology, Vol. 65, No. 10, 1999, pp. 4688-4692.
- H. Ericsson, P. Stalhandske, M. L. Danielsson-Tham, E. Bannerman, J. Bille, C. Jacquet, J. Rocourt and W. Tham, “Division of Listeria monocytogenes Serovar 4b Strains into Two Groups by PCR and Restriction Enzyme Analysis,” Applied and Environmental Microbiology, Vol. 61, No. 11, 1995, pp. 3872-3874.
- C. Jacquet, E. Gouin, D. Jeannel, P. Cossart and J. Rocourt, “Expression of ActA, Ami, InlB, and Listeriolysin O in Listeria monocytogenes of Human and Food Origin,” Applied and Environmental Microbiology, Vol. 68, No. 2, 2002, pp. 616-622. doi:10.1128/AEM.68.2.616-622.2002
- I. Lasa, V. David, E. Gouin, J. B. Marchand and P. Cossart, “The Amino-Terminal Part of ActA Is Critical for the Actin-Based Motility of Listeria monocytogenes; The Central Proline-Rich Region Acts as a Stimulator,” Molecular Microbiology, Vol. 18, No. 3, 1995, pp. 425-436. doi:10.1111/j.1365-2958.1995.mmi_18030425.x
- G. A. Smith, J. A. Theriot and D. A. Portnoy, “The Tandem Repeat Domain in the Listeria monocytogenes Act a Protein Controls the Rate of Actin-Based Motility, the Percentage of Moving Bacteria, and the Localization of Vasodilator-Stimulated Phosphoprotein and Profiling,” The Journal of Cell Biology, Vol. 135, No. 3, 1996, pp. 647-660. doi:10.1083/jcb.135.3.647
- J. G. Williams, A. R. Kubelik, K. J. Livak, J. A. Rafalski and S. V. Tingey, “DNA Polymorphisms Amplified by Arbitrary Primers Are Useful as Genetic Markers,” Nucleic Acids Research, Vol. 25, No. 22, 1990, pp. 6531- 6535. doi:10.1093/nar/18.22.6531
- A. Kerouanton, A. Brisabois, E. Denoyer, F. Dilasser, J. Grout, G. Salvat and B. Picard, “Comparison of Five Typing Methods for the Epidemiological Study of Listeria monocytogenes,” International Journal of Food Microbiology, Vol. 43, No. 1-2, 1998, pp. 61-71. doi:10.1016/S0168-1605(98)00098-1
- M. Louie, P. Jayaratne, I. Luchsinger, J. Devenish, J. Yao, W. Schlech and A. Simor, “Comparison of Ribotyping, Arbitrarily Primed PCR, and Pulsed-Field Gel Electrophoresis for Molecular Typing of Listeria monocytogenes,” Journal of Clinical Virology, Vol. 34, No. 1, 1996, pp. 15-19.
- S. I. Mazurier and K. Wernars, “Typing of Listeria Strains by Random Amplification of Polymorphic DNA,” Research in Microbiology, Vol. 143, No. 5, 1992, pp. 499- 505. doi:10.1016/0923-2508(92)90096-7
- C. Niederhauser, C. Höfelein, M. Allmann, P. Burkhalter, J. Lüthy and U. Candrian, “Random Amplification of Polymorphic Bacterial DNA: Evaluation of 11 Oligonucleotides and Application to Food Contaminated with Listeria monocytogenes,” Journal of Applied Bacteriology, Vol. 77, No. 5, 1994, pp. 574-582. doi:10.1111/j.1365-2672.1994.tb04404.x
- L. M. Graves, S. B. Hunter, A. R. Ong, D. Schoonmaker-Bopp, K. Hise, L. Kornstein, W. E. DeWitt, P. S. Hayes, E. Dunne, P. Mead and B. Swaminathan, “Microbiological Aspects of the Investigation That Traced the 1998 Outbreak of Listeriosis in the United States to Contaminated Hot Dogs and Establishment of Molecular Subtyping-Based Surveillance for Listeria monocytogenes in the PulseNet Network,” Journal of Clinical Virology, Vol. 43, No. 5. 2005, pp. 2350-2355.
- R. Brosch, M. Brett, B. Catimel, J. B. Luchansky, B. Ojeniyi and J. Rocourt, “Genomic Fingerprinting of 80 Strains from the WHO Multicenter International Typing Study of Listeria monocytogenes via Pulsed-Field Gel Electrophoresis (PFGE),” International Journal of Food Microbiology, Vol. 32, No. 3, 1996, pp. 343-355. doi:10.1016/S0168-1605(96)01147-6
- M. L. Suihko, S. Salo, O. Niclasen, B. Gudbjörnsdóttir, G. Torkelsson, S. Bredholt, A. M. Sjöberg and P. Gustavsson, “Characterization of Listeria monocytogenes Isolates from the Meat, Poultry and Seafood Industries by Automated Ribotyping,” International Journal of Food Microbiology, Vol. 72, No. 1-2, 2002, pp. 137-146. doi:10.1016/S0168-1605(01)00631-6
- E. Heir, B. A. Lindstedt, O. J. Røtterud, T. Vardund, G. Kapperud and T. Nesbakken, “Molecular Epidemiology and Disinfectant Susceptibility of Listeria monocytogenes from Meat Processing Plants and Human Infections,” International Journal of Food Microbiology, Vol. 96, No. 1, 2004, pp. 85-96.
- I. Martinez, L. M. Rorvik, V. Brox, J. Lassen, M. Seppola, L. Gram and V. B. Fonnesbech, “Genetic Variability among Isolates of Listeria monocytogenes from Food Products, Clinical Samples and Processing Environments, Estimated by RAPD Typing,” International Journal of Food Microbiology, Vol. 84, No. 3, 2003, pp. 285-297. doi:10.1016/S0168-1605(02)00423-3
- T. Autio, J. Lundon, A. M. Fredriksson, J. Bjorkroth, A. M. Sjoberg and H. Korkeala, “Similar Listeria monocytogenes Pulsotypes Detected in Several Foods Originating from Different Sources,” International Journal of Food Microbiology, Vol. 77, No. 1-2, 2002, pp. 83-90. doi:10.1016/S0168-1605(02)00055-7
- A. Margolles, B. Mayo and C. G. de los Reyes-Gavilan, “Polymorphism of Listeria monocytogenes and Listeria Innocua Strains Isolated from Short-Ripened Cheeses,” Journal of Applied Microbiology, Vol. 84, No. 2, 1998, pp. 255-262. doi:10.1046/j.1365-2672.1998.00339.x
- C. A. Nadon, D. L. Woodward, C. Young, F. G. Rodgers and M. Wiedmann, “Correlations between Molecular Subtyping and Serotyping of Listeria monocytogenes,” Journal of Clinical Virology, Vol. 39, No. 7, 2001, pp. 2704- 2707.
- I. Giovannacci, C. Ragimbeau, S. Queguiner, G. Salvat, J. L. Vendeuvre, V. Carlier and G. Ermel, “Listeria monocytogenes in Pork Slaughtering and Cutting Plants. Use of RAPD, PFGE and PCR-REA for Tracing and Molecular Epidemiology,” International Journal of Food Microbiology, Vol. 53, No. 2-3, 1999, pp. 127-140. doi:10.1016/S0168-1605(99)00141-5
- F. M. Wallace, J. E. Call, A. C. Porto, G. J. Cocoma and J. B. Luchansky, “Recovery Rate of Listeria monocytogenes from Commercially Prepared Frankfurters during Extended Refrigerated Storage,” Journal of Food Protection, Vol. 66, No. 4, 2003, pp. 584-591.
- M. T. Destro, M. Leitao and J. M. Farber, “Use of Molecular Typing Methods to Trace the Dissemination of Listeria monocytogenes in a Shrimp Processing Plant,” Applied and Environmental Microbiology, Vol. 62, No. 5, 1996, pp. 1852-1853.
- P. Boerlin, E. Bannerman, F. Ischer, J. Rocourt and J. Bille, “Typing Listeria monocytogenes: A Comparison of Random Amplification of Polymorphic DNA with 5 Other Methods,” Research in Microbiology, Vol. 146, No. 1, 1995, pp. 35-49. doi:10.1016/0923-2508(96)80269-5
NOTES
*Corresponding author.

