 Vol.2, No.9, 1005-1014 (2010) Natural Science http://dx.doi.org/10.4236/ns.2010.29123 Copyright © 2010 SciRes. OPEN ACCESS Highly conserved domains in hemagglutinin of influenza viruses characterizing dual receptor binding Wei Hu Department of Computer Science, Houghton College, Houghton, USA; wei.hu@houghton.edu Received 4 June 2010; revised 15 July 2010; accepted 18 July 2010. ABSTRACT The hemagglutinin (HA) of influenza viruses in- itiates virus infection by binding receptors on host cells. Human influenza viruses preferenti- ally bind to receptors with α2,6 linkages to gala- ctose, avian viruses prefer receptors with α2,3 linkages to galactose, and swine viruses favor both types of receptors. The pandemic H1N1 2009 remains a global health concern in 2010. The novel 2009 H1N1 influenza virus has its ge- netic components from avian, human, and sw- ine viruses. Its pandemic nature is characterized clearly by its dual binding to the α2,3 as well as α2,6 receptors, because the seasonal human H1N1 virus only binds to the α2,6 receptor. In pr- evious studies, the informational spectrum me- thod (ISM), a bioinformatics method, was appli- ed to uncover highly conserved regions in the HA protein associated with the primary receptor binding preference in various subtypes. In the present study, we extended the previous work by discovering multiple domains in HA associa- ted with the secondary receptor binding prefer- ence in various subtypes, thus characterizing the distinct dual binding nature of these viruses. The domains discovered in the HA proteins were mapped to the 3D homology model of HA, which could be utilized as therapeutic and diag- nostic targets for the prevention and treatment of influenza infection. Keywords: Binding Specificity; Discrete Fourier Transform; Electron-Ion Interaction Potential; Hemagglutinin; Influenza; Informational Spectrum Method 1. INTRODUCTION Influenza A viruses have two surface proteins, hemagg- lutinin (HA) and neuraminidase (NA). HA is a hom- otrimer, in which each monomer comprises two subdo- mains, HA1 and HA2. HA1 initializes the contact with the cell membrane and HA2 mediates membrane fusion. In general, human influenza and avian viruses preferen- tially bind to the α2,3 sialylated and α2,6 sialylated gly- can receptors, respectively [1-3]. Pigs have receptors for both human and avian influenza viruses on their tracheal cells, thus they can serve as a mixing vessel to re-assort genes from different species to make new influenza vir- uses. The 2009 H1N1 pandemic was caused by a novel swine-origin influenza A virus. Using sequence analysis and homology modeling [4], the HA protein of 2009 H1N1 was found to have the sig- nature amino acid Asp190 and Asp225 known to confer binding affinity to α2,6 sialylated glycan receptors. The mutation Glu190Asp between avian and human H1 HA normally would lead to the loss of a critical contact with α2,3 glycans, which, however, was compensated by the presence of Lys145 in HA of 2009 H1N1. There were four loops in 2009 H1N1 HA, 130 loop, 140 loop, 150 loop, and 220 loop, each containing one Lys, to form a positively charged ‘lysine fence’ at the base of the bind- ing site to support optimal contacts with the α2,6 and α2,3 receptors. Based on this structural analysis, it was predicted that the HA protein of 2009 H1N1 virus can bind to the α2,6 as well as α2,3 sialylated glycan recap- tors, which was subsequently verified by the carbohyd- rate microarray analysis in [5]. There were several other efforts in expanding the kno- wledge on 2009 H1N1. One study [6] investigated the th- ree aspects of NA: the mutations and co-mutations, the stalk motifs, and the phylogenetic analysis. The potential mutations and strongly co-mutated positions of NA were found. A special NA stalk motif of high virulence, which was dominant in the past H5N1 strains, was discovered in H1N1 in 2009 for the first time. Another study [7] focused on HA and the interaction between HA and NA. The mutations of HA in 2009 H1N1 were found and ma- pped to the 3D homology model of H1, and the muta-  W. Hu / Natural Science 2 (2010) 1005-1014 Copyright © 2010 SciRes. OPEN ACCESS 1006 tions on the five epitope regions on H1 were identified. The distinct response patterns of HA to the changes of NA stalk motifs were discovered, illustrating the functio- nal dependence between HA and NA. With help from the results of the first study [6], two comutation netw- orks were uncovered, one in HA and one in NA, where each mutation in one network co-mutates with the mu- tations in the other network across the two proteins HA and NA. These two networks residing in HA and NA separately might provide a functional linkage between the mutations that could change the drug binding sites in NA and those that could affect the host immune respon- se or vaccine efficacy in HA. The genes of 2009 H1N1 were derived from avian, human, and swine viruses. Identification of host shift ma- rkers of this novel virus remains an urgent and important research topic. However, sequence survey of 2009 H1N1 suggested the absence of the well-known host switch ma- rkers. A new procedure was designed to locate a collec- tion of novel host markers in ten proteins/genes of 2009 H1N1 [8,9], which included, in addition to the SR polym- orphism found in [10], a set of markers in PB2 that mi- ght play compensatory roles in the efficient replication and transmission of this new virus. These novel markers of 2009 H1N1 offered new and ample opportunities for further investigation of their biological functions in host adaptation to humans experimentally. The informational spectrum method (ISM) [11] is a bioinformatics technique to study the biological functi- ons of proteins with their physicochemical properties, which first translates a protein sequence into a numerical sequence based on each amino acid’s electron-ion inter- action potential (EIIP) and then the discrete Fourier tra- nsformation (DFT) is applied to it to create an inform- ational spectrum. It is believed that the protein functions including the protein-protein interaction are encoded in the peaks of the informational spectrum. In [12,13] it was found that the CIS of HA1 of influe- nza strains have the primary characteristic dominant pea- ks at different IS frequencies as presented in Table 1. In this study, F(0.295) will be referred to as pandemic hu- man H1N1 receptor interaction frequency, F(0.055) as swine receptor interaction frequency, F(0.076) as avian receptor interaction frequency, and F(0.236) as seasonal human H1N1 receptor interaction frequency. Some infl- uenza strains display dual binding preference, one prim- ary and one secondary, as demonstrated in [12,13]. In [12,13] the ISM was applied to investigate the inte- raction between HA protein and its receptors, which sho- wed that HA1encodes highly conserved domains essenti- al for receptor binding affinity. Their analysis also found the following receptor recognition domains in HA prote- ins from H1N1, H3N2, H5N1, and H7N7 (Table 2). One region in HA1 of various strains was found to be connected with the primary receptor binding preference in [12] (Table 2). In [14], multiple such regions in HA1 responsible for the primary receptor binding preference in different subtypes were discovered (Table 3). In a stu- dy of receptor binding specificity of influenza viruses [14], the ISM was applied to find the mutations in HA1 and to elucidate the contribution to receptor binding switch from each mutation quantitatively. Two clusters of mutations in HA1 of 2009 H1N1 were uncovered, where the first was at residues 152-170 and the second at residues 257-278. The first cluster of mutations was contained in a pandemic human H1N1 receptor recogni- tion domain (150:174) with the primary characteristic IS frequency at F(0.295) found in [15] (Table 3). Prompted by this finding, we searched for a similar domain in the vicinity of the second cluster, and found one such dom- ain (246:286) associated with swine binding preference at the secondary characteristic IS frequency F(0.055) [15] (Table 3). Our aim in this study was to extend these results by identifying multiple domains in HA1 of influ- enza viruses associated with the secondary receptor binding preference, thus providing the complete infor- mation about the dual binding of the HA proteins. These conserved domains in HA1 might be used to develop targets for new drugs and infection control. 2. MATERIALS AND METHODS 2.1. Sequence Data All HA sequences were retrieved from the Flu Resource Table 1. Primary characteristic IS frequencies of HA proteins in 2009 H1N1, swine H1N1/H1N2, avian H5N1, and seasonal human H1N1. Subtype 2009 H1N1 Swine H1N2/H1N1 Avian H5N1 Seasonal human H1N1 FrequencyF(0.295)F(0.055) F(0.076) F(0.236) Table 2. The receptor recognition domains of HA proteins in H1N1, H3N2, H5N1, and H7N7 influenza viruses. Strain Frequency Residues A/California/04/2009 (H1N1) F(0.295) 284-326 A/Hong Kong/213/03 (H5N1) F(0.076) 42-75 A/New Caledonia/20/99 (H1N1) F(0.236) 262-295 A/New York/383/2004 (H3N2) F(0.363) 57-90 A/equine/Prague/56 (H7N7) F(0.285) 28-61 A/Egypt/0636-NAMRU3/2007(H5N1) F(0.236) 99-132 A/South Carolina/1/18 (H1N1)) F(0.258) 87-120  W. Hu / Natural Science 2 (2010) 1005-1014 Copyright © 2010 SciRes. OPEN ACCESS 100 1007 Table 3. The receptor recognition domains of HA proteins in 2009 H1N1, swine H1N2, and avian H5N1 influenza viruses [14,15]. Subtype Frequency Residues Consensus HA1 Sequence 2009 H1N1 F(0.295) 106-130 SSVSSFERFEIFPKTSSWPNHDSNK 2009 H1N1 F(0.295) 150-174 WLVKKGNSYPKLSKSYINDKGKEVLV 2009 H1N1 F(0.295) 191-221 LYQNADAYVFVGSSRYSKKFKPEIAIRPKVR Swine H1N2 F(0.055) 1-29 DTLCIGYHANNSTDTVDTVLEKNVTVTHS Avian H5N1 F(0.076) 46-65 GVKPLILRDCSVAGWLLGNP Avian H5N1 F(0.076) 136-153 PYQGRSSFFRNVVWLIKK Avian H5N1 F(0.076) 269-286 LEYGNCNTKCQTPMGAIN (http://www.ncbi /nlm.nih.giv /geno mes/FLU/FLU.html) of the National Center for Biotechnology Information (NCBI) on Nov. 20, 2009. Only the full length and un- ique sequences were selected. There were 450 HA sequ- ences of human 2009 H1N1, 1228 HA sequences of av- ian H5N1 from 1959 to 2009, and 83 HA sequences of swine H1N2 from 1980 to 2009. All the sequences used in the study were aligned with MAFFT [16]. 2.2. Important Sites in HA Although there is a great variation due to high selection pressure in the HA1 sequences of various flu subtypes, the active site of HA1 is well conserved, which is loca- ted in a cleft composed of the residues 91, 150, 152, 180, 187, 191, and 192 [17]. The three amino acids at positi- ons 187, 191 and 192 are a part of the 190 helix. The ac- tive site cleft of HA is formed by its right edge (131_ GVTAA) and left edge (221_RGQAGR) (H1 number- ing), which are also commonly referred to as the 130 loop and 220 loop, respectively [18]. 2.3. Informational Spectrum Method The informational spectrum method is a bioinformatics technique that can be utilized to analyze protein sequen- ces. Prior to this analysis, the protein sequences have to be translated into numerical sequences. One such appro- ach is to assign each amino acid to its electron-ion inter- action potential (EIIP), which represents the average en- ergy of the valence electrons in the amino acid (Table 4). Table 4. The electron-ion interaction potential (EIIP) of amino acids used to encode amino acids. Amino acid EIIP Amino acid EIIP L 0.0000 Y 0.0516 I 0.0000 W 0.0548 N 0.0036 Q 0.0761 G 0.0050 M 0.0823 E 0.0057 S 0.0829 V 0.0058 C 0.0829 P 0.0198 T 0.0941 H 0.0242 F 0.0946 K 0.0371 R 0.0959 A 0.0373 D 0.1263 The application of EIIP to protein function analysis assu- mes that the strength of the electromagnetic field surrou- nding the protein is indicative of its biological function. This method was successful in revealing various protein properties [11]. The numerical sequence x(m) of a protein sequence is transformed into the frequency domain using DFT. The DFT coefficients X(n) are defined as 2 ()( )1,2,2 jnm N nxme n N where N is the length of sequence x(m). The energy density spectrum is defined as 2,1,2,,2SnXnX nXnnN The informational spectrum (IS) of a sequence x(m) comprises the frequencies and the amplitudes of its DFT. Peak frequencies of IS of a protein sequence reflect its biological or biochemical functions. To determine the sa- me biological or biochemical functions of a group of pro- tein sequences, a consensus informational spectrum (CIS) can be used, which is defined as the product of energy de- nsity spectrum S(n) of each sequence in the group. A me- asure of similarity for each peak is a signal-to-noise ratio (S/N), which is defined as a ratio of signal density to the mean value of the whole spectrum [11]. 2.4. Consensus HA1 Sequences We employed MAFFT to align the three consensus HA1 sequences of 2009 H1N1, avian H5N1, and swine H1N2 (Figure 1). Each consensus sequence was then used in the ISM analysis to find the highly conserved domains in HA1 of different influenza subtypes. 3. RESULTS As demonstrated in [12-15], the HA1 sequences in vari- ous influenza subtypes had a distinct inclination to inte- ract with a specific primary receptor as well as a specific secondary receptor, and there were regions in HA1 enco- ding highly conserved information associated with the  W. Hu / Natural Science 2 (2010) 1005-1014 Copyright © 2010 SciRes. OPEN ACCESS 1008 Figure 1. Multiple sequence alignment of three consensus HA1 sequences of 2009 H1N1, swine H1N2, and avian H5N1 (pre- sented in this order as well). The binding sites in HA are col- ored in red, the left and right edges of the binding cleft in blue. primary binding preference. The main task of the present study is to explore the other parts of the HA1 sequences to find domains associated with the secondary binding preference. The entropy of HA1 of 2009 H1N1, swine H1N2, and avian H5N1 was calculated in [14], illustrat- ing the most conserved positions in the HA1 sequences of each subtype. The ISM bioinformatics technique was applied to the three consensus HA1 sequences as prese- nted in Figure 1 to uncover the conserved domains in HA1, which might affect the secondary receptor speci- ficity in each subtype. In contrast, the ISM analysis in [12,13] was applied to a particular selected strain in a subtype such as A/California/04/2009 in 2009 H1N1 to find a conserved region. Overall, the conserved domains discovered by our approach using a consensus sequence had more coverage to different strains in a subtype than the single strain approach used in [12,13]. 3.1. Conserved Domains in HA1 of 2009 H1N1 Using the ISM, we discovered seven conserved domains in HA1 of 2009 H1N1 responsible for swine binding pr- eference, which were located at residues 5-23, 63-81, 142-158, 178-195, 201-216, 246-269, and 277-287, re- spectively (Figure 2). The ISs of these domains and the consensus HA1 sequence in 2009 H1N1 were plotted in Figure 3. The IS of the consensus HA1 sequence in 2009 H1N1displayed a primary dominant peak of frequency F(0.295) and a secondary prominent peak of frequency F(0.055), demonstrating its dual binding preference. The domain 142:158 contained two binding sites 150 and 152 Figure 2. The two plots show in 3D structure the seven highly conserved domains related to swine binding characteristic fou- nd in HA of 2009 H1N1. Domain 5:23 is colored in red, dom- ain 63:81 in blue, domain 142:158 in green, domain 178:195 in orange, domain 201:216 in yellow, domain 246:269 in pink, and domain 277:287 in gray (PDB code: 1RU7). and was near the right edge of the active site. The dom- ain 178:195 included four binding sites 180, 187, 191, and 192. The domains 201:216 and 246:269 were close to the left edge of the active site, illustrating their important roles in receptor binding preference. As seen from the entropy distribution of HA1 of 2009 H1N1 in [14], these seven domains were highly conserved. In [12], a similar domain with human receptor binding characteristic was found in the C-terminus of the HA protein consisting of residues 284-326. Multiple such domains were identified in [14], and one domain with swine receptor binding characteristic was found in [15] (Table 1). 3.2. Conserved Domains in HA1 of Swine H1N2 In [12], the sequences of swine H1N1 and H1N2 were combined into a single dataset for analysis. Here the swine H1N2 was treated as a single dataset. The con- served domains found in this subtype were separated in two categories, i.e., one had swine binding preference (primary) and one had human binging preference (sec- ondary). The four domains with swine binding were at residues 63-81, 178-189, 241-271, and 277-287, respec- tively (Figure 4), and the four domains with human binding were at residues 104-129, 189-213, 240-265, and 271-301, respectively (Figure 5). The ISs of these sequence in swine H1N2 revealed two dominant peaks at 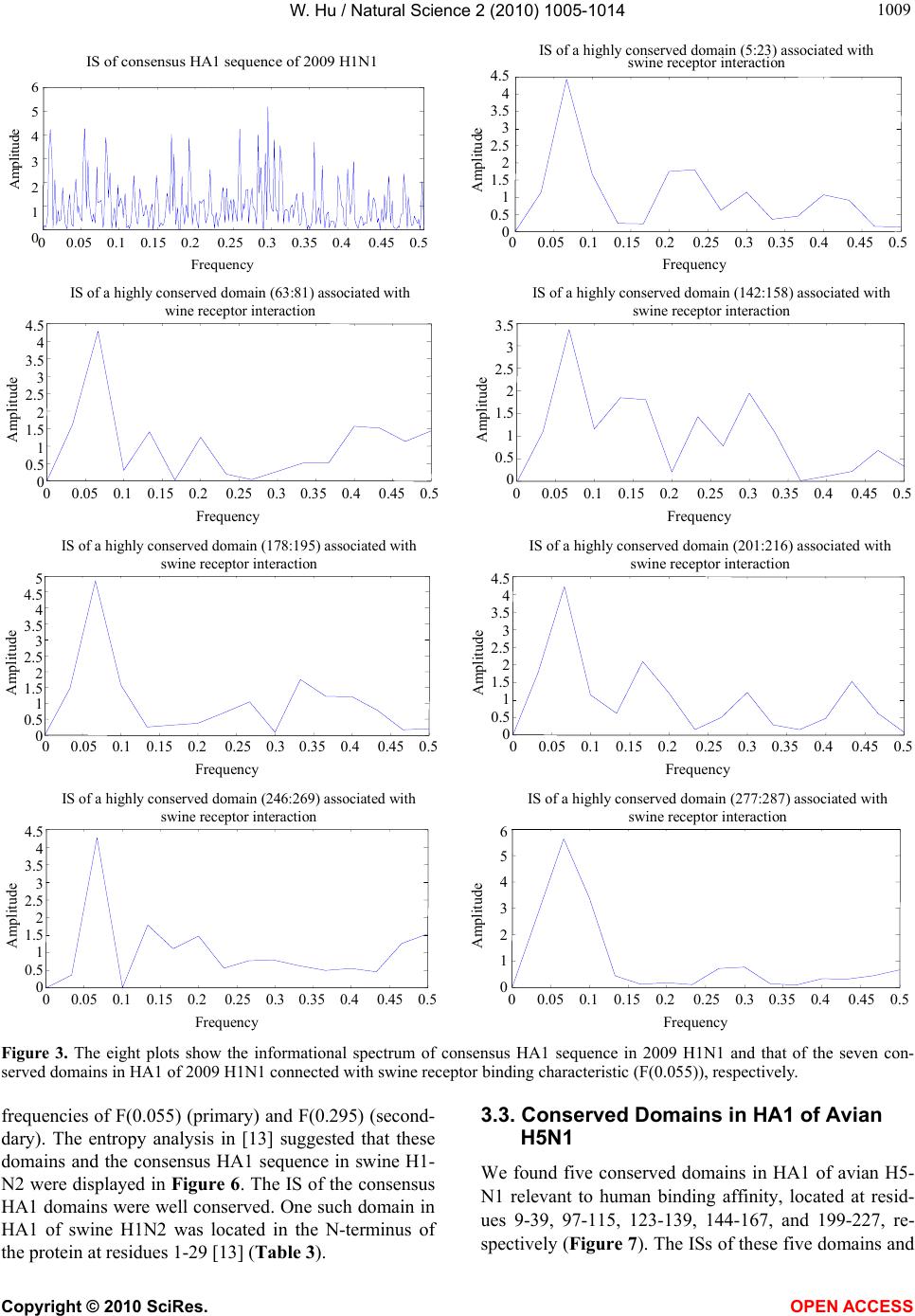 W. Hu / Natural Science 2 (2010) 1005-1014 Copyright © 2010 SciRes. OPEN ACCESS 100 1009 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 6 5 4 3 2 1 0 Fre uenc Amplitude IS of consensus HA1 sequence of 2009 H1N1 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequenc Am litude 4.5 4 3.5 3 2.5 2 1.5 1 0.5 0 IS of a highly conserved domain (5:23) associated with swine receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequency Am litude 4.5 4 3.5 3 2.5 2 1.5 1 0.5 0 IS of a highly conserved domain (63:81) associated with wine receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequency Am litude 3.5 3 2.5 2 1.5 1 0.5 0 IS of a highly conserved domain (142:158) associated with swine receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequency Am litude 5 4.5 4 3.5 3 2.5 2 1.5 1 0.5 0 IS of a highly conserved domain (178:195) associated with swine receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequency Am litude 4.5 4 3.5 3 2.5 2 1.5 1 0.5 0 IS of a highly conserved domain (201:216) associated with swine receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequency Am litude 4.5 4 3.5 3 2.5 2 1.5 1 0.5 0 IS of a highly conserved domain (246:269) associated with swine receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequency Am litude 6 5 4 3 2 1 0 IS of a highly conserved domain (277:287) associated with swine receptor interaction Figure 3. The eight plots show the informational spectrum of consensus HA1 sequence in 2009 H1N1 and that of the seven con- served domains in HA1 of 2009 H1N1 connected with swine receptor binding characteristic (F(0.055)), respectively. frequencies of F(0.055) (primary) and F(0.295) (second- dary). The entropy analysis in [13] suggested that these domains and the consensus HA1 sequence in swine H1- N2 were displayed in Figure 6. The IS of the consensus HA1 domains were well conserved. One such domain in HA1 of swine H1N2 was located in the N-terminus of the protein at residues 1-29 [13] (Table 3). 3.3. Conserved Domains in HA1 of Avian H5N1 We found five conserved domains in HA1 of avian H5- N1 relevant to human binding affinity, located at resid- ues 9-39, 97-115, 123-139, 144-167, and 199-227, re- spectively (Figure 7). The ISs of these five domains and  W. Hu / Natural Science 2 (2010) 1005-1014 Copyright © 2010 SciRes. OPEN ACCESS 1010 Figure 4. The two plots show in 3D structure the four highly conserved domains of swine binding character- istic found in HA of swine H1N2. Domain 63:81 is co- lored in red, domain 178:189 in blue, domain 241:271 in green, and domain 277:287 in orange. Figure 5. The two plots show in 3D structure the four highly conserved domains of human binding characte- ristic found in HA of swine H1N2. Domain 104:129 is co- lored in red, domain 189:213 in blue, domain 240:265 in orange, and domain 271:301 in yellow. 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 7 6 5 4 3 2 1 0 Fre uenc Amplitude IS of consensus HA1 sequence of swine H1N2 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequency Amplitude 4.5 4 3.5 3 2.5 2 1.5 1 0.5 0 IS of a highly conserved domain (63:81) associated with swine receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequenc Am litude 5 4.5 4 3.5 3 2.5 2 1.5 1 0.5 0 IS of a highly conserved domain (178:189) associated with swine receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequency Am litude 4 3.5 3 2.5 2 1.5 1 0.5 0 IS of a highly conserved domain (241:271) associated with swine receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequ e nc Am litude 6 5 4 3 2 1 0 IS of a highly conserved domain (277:287) associated with swine receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequenc Am litude 4 3.5 3 2.5 2 1.5 1 0.5 0 IS of a highly conserved domain (104:129) associated with pandemic human H1N1 receptor interaction  W. Hu / Natural Science 2 (2010) 1005-1014 Copyright © 2010 SciRes. OPEN ACCESS 101 1011 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequency Am litude 9 8 7 6 5 4 3 2 1 0 IS of a highly conserved domain (189:213) associated with pandemic human H1N1 receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequency Am litude 7 6 5 4 3 2 1 0 IS of a highly conserved domain (271:301) associated with pandemic human H1N1 receptor interaction IS of a highly conserved domain (240:265) associated with pandemic human H1N1 receptor interaction Freq u enc y Am litude 5 4.5 4 3.5 3 2.5 2 1.5 1 0.5 0 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Figure 6. The nine plots show the informational spectrum of consensus HA1 sequence of swine H1N2, and that of seven conserved domains in swine H1N2 associated with human and swine binding (F(0.295) and F(0.055)), respectively. Figure 7. The two plots show in 3D structure the five highly conserved domains found in HA of avian H5N1 related to hu- man binding. Domain 9:39 is colored in red, domain 97:115 in blue, domain 123:139 in green, domain 144:167 in orange, and domain 199:227 in yellow (PDB code: 2IBX). the consensus HA1 sequence in avian H5N1 were illus- trated in Figure 8. The IS of the consensus HA1 seque- nce in avian H5N1 demonstrated two dominant peaks of frequencies F(0.076) (primary) and F(0.236) (secondary). The entropy analysis in [14] implied that the five doma- ins were well conserved, and the HA1 sequences of av- ian H5N1 were quite stable, in contrast to those of 2009 H1N1 and swine H1N2. In [13], a similar domain with avian binding was found in the N-terminus of the HA pr- otein comprising residues 42-75. In [14], multiple such domains with avian binding were uncovered (Table 3). 4. CONCLUSIONS Identifying the conserved characteristics of influenza vi- ruses relevant to receptor binding preference is of great importance in flu research. The informational and struct- ural properties of HA1 of influenza viruses associated with the primary receptor affinity were investigated in [12-15]. To extend the previous results, we sought to un- cover multiple domains in HA1 associated with the seco- ndary binding preference, thus to expand our repertoire of these key regions in HA1 of influenza viruses. The main findings of this study were presented in Ta- ble 5, which showed the locations, the characteristic IS frequencies, and the consensus sequences of the seven highly conserved domains in HA1 discovered in differ- ent subtypes. Combined with the results in Table 3 and those in [12,13], these findings provided the complete  W. Hu / Natural Science 2 (2010) 1005-1014 Copyright © 2010 SciRes. OPEN ACCESS 1012 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 6 5 4 3 2 1 0 Fre uenc Amplitude IS of consensus HA1 sequence of avian H5N1 Frequenc Am litude 4 3.5 3 2.5 2 1.5 1 0.5 0 IS of a highly conserved domain (9:39) associated with seasonal human H1N1 receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequency Am litude 6 5 4 3 2 1 0 IS of a highly conserved domain (97:115) associated with seasonal human H1N1 receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequency Am litude 5 4.5 4 3.5 3 2.5 2 1.5 1 0.5 0 IS of a highly conserved domain (123:139) associated with seasonal human H1N1 receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequency Am litude 6 5 4 3 2 1 0 IS of a highly conserved domain (97:115) associated with seasonal human H1N1 receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Frequency Am litude 4.5 4 3.5 3 2.5 2 1.5 1 0.5 0 IS of a highly conserved domain (199:227) associated with seasonal human H1N1 receptor interaction 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 IS of a highly conserved domain (240:265) associated with pandemic human H1N1 receptor interaction Frequenc Am litude 5 4.5 4 3.5 3 2.5 2 1.5 1 0.5 0 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Figure 8. The seven plots show the informational spectrum of consensus HA1 sequence in avian H5N1 and the informational spec- trum of the six conserved domains in avian H5N1 associated with human binding (F(0.236)), respectively. information about the conserved domains in HA1 that might modulate the dual receptor binding preference. The domains in Table 5 are shorter than those discov- ered in [12,13], implying that they were more easily co- nserved by the HA sequences than their longer counterp- arts. Furthermore, the identified multiple domains in HA1 related to dual receptor binding preference could provide ample options to design new therapeutic targets for drug development. Finally, because a consensus sequence of each subtype was employed to find these multiple dom- ains in HA1, they are more representative of the strains in a given subtype than those obtained in [12,13].  W. Hu / Natural Science 2 (2010) 1005-1014 Copyright © 2010 SciRes. OPEN ACCESS 101 1013 Table 5. The receptor recognition domains in HA protein in 2009 H1N1, avian H5N1, and swine H1N2 influenza viruses. Subtype Frequency Residues Consensus HA1 Sequence 2009 H1N1 F(0.055) 5-23 IGYHANNSTDTVDTVLEKN 2009 H1N1 F(0.055) 63-81 GNPECESLSTASSWSYIVE 2009 H1N1 F(0.055) 142-158 KSFYKNLIWLVKKGNSY 2009 H1N1 F(0.055) 178-195 GIHHPSTSADQQSLYQNA 2009 H1N1 F(0.055) 201-216 VGSSRYSKKFKPEIAI 2009 H1N1 F(0.055) 246-269 GNLVVPRYAFAMERNAGSGIIISD 2009 H1N1 F(0.055) 277-287 TTCQTPKGAIN Swine H1N2 F(0.295) 104-129 QLSSVSSFERFEIFPKESSWPNHDTN Swine H1N2 F(0.295) 189-213 QSLYQNADAYVFVGSSHYSKKFTPE Swine H1N2 F(0.295) 240-265 ITFEATGNLVVPRYAFALKRGSGSGI Swine H1N2 F(0.295) 271-301 SVHDCNTTCQTPKGAINTSLPFQNIHPVTIG Swine H1N2 F(0.055) 63-81 GNPECESLFTASSWSYIVE Swine H1N2 F(0.055) 178-189 GIHHPSTSADQQ Swine H1N2 F(0.055) 241-271 TFEATGNLVVPRYAFALKRGSGSGIIISDTS Swine H1N2 F(0.055) 277-287 TTCQTPKGAIN Avian H5N1 F(0.236) 9-39 ANNSTEQVDTIMEKNVTVTHAQDILEKTHNG Avian H5N1 F(0.236) 97-115 DYEELKHLLSRINHFEKIQ Avian H5N1 F(0.236) 123-139 SDHEASSGVSSACPYQG Avian H5N1 F(0.236) 144-167 FRNVVWLIKKNNAYPTIKRSYNNT Avian H5N1 F(0.236) 199-227 SVGTSTLNQRLVPKIATRSKVNGQSGRME It was suggested in [12,13] that the 2009 H1N1 strains will continue to mutate in their HA gene, which could further favor the human interaction pattern by increasing the amplitude at frequency F(0.295) and at the same time decreasing that at frequency F(0.055). Identification of multiple domains in HA of influenza viruses related to dual receptor binding preference provided relevant info- rmation for the development of potential targets for new drugs and treatment of influenza infection 5. ACKNOWLEDGEMENTS We thank Houghton College for its financial support. REFERENCES [1] Gambaryan, A., Tuzikov, A., Pazynina, G. et al. (2006) Evolution of the receptor binding phenotype of influenza A (H5) viruses. Virology, 344(2), 432-438. [2] Iwata, T., Fukuzawa, K., Nakajima, K., et al. (2008) Theoretical analysis of binding specificity of influenza viral hemagglutinin to avian and human receptors based on the fragment molecular orbital method. Computa- tional Biology and Chemistry, 32(3), 198-211. [3] Yamada, S., Suzuki, Y., Suzuki, T., et al. (2006) Hae- magglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Na- ture, 444(7117), 378-382. [4] Soundararajan, V., Tharakaraman, K., et al. (2009) Ex- trapolating from sequence—the 2009 H1N1 ‘swine’ in- fluenza virus. Nature Biotechnology, 27(6), 510-513. [5] Childs, R.A., Palma, A.S., Wharton, S., et al. (2009) Re- ceptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microar- ray. Nature Biotechnology, 27(9), 797-799. [6] Hu, W. (2009) Analysis of Correlated Mutations, Stalk Motifs, and Phylogenetic Relationship of the 2009 Influ- enza A Virus Neuraminidase Sequences. Journal of Bio- medical Science and Engineering, 2(7), 550-558. [7] Hu, W. (2010) The Interaction between the 2009 H1N1 Influenza A Hemagglutinin and Neuraminidase: Mutati- ons, Co-mutations, and the NA Stalk Motifs. Journal of Biomedical Science and Engineering, 3(1), 1-12. [8] Hu, W. (2010) Novel host markers in the 2009 pandemic H1N1 influenza A virus. Journal of Biomedical Science and Engineering, 3(6), 584-601. [9] Hu, W. (2010) Nucleotide host markers in the influenza a viruses. Journal of Biomedical Science and Engineering, 3(7), 684-699. [10] Mehle, A. and Doudna, J.A. (2009) Adaptive strategies of the influenza virus polymerase for replication in humans, Proceedings of the National Academy of Sciences of the United States of America, 106(50), 21312-21316. [11] Cosic, I. (1997) The resonant recognition model of macro- molecular bioreactivity, theory and application. Birk- hauser Verlag, Berlin. [12] Veljkovic, V., Niman, H.L., Glisic, S., et al. (2009) Identi- fication of hemagglutinin structural domain and poly- morphisms which may modulate swine H1N1 interac- tions with human receptor. BMC Structural Biology, 9,  W. Hu / Natural Science 2 (2010) 1005-1014 Copyright © 2010 SciRes. OPEN ACCESS 1014 62. [13] Veljkovic, V., Veljkovic, N., Muller, C.P., Müller, S., Glisic, S., Perovic, V. and Köhler, H. (2009) Characteri- zation of conserved properties of hemagglutinin of H5N1 and human influenza viruses: possible consequences for therapy and infection control. BMC Structural Biology, 7, 9-21. [14] Hu, W. (2010) Identification of highly conserved dom- ains in hemagglutinin associated with the receptor bind- ing specificity of influenza viruses: 2009 H1N1, avian H5N1, and swine H1N2. Journal of Biomedical Science and Engineering, 3(2), 114-123. [15] Hu, W. (2010) Quantifying the effects of mutations on re- ceptor binding specificity of influenza viruses. Journal of Biomedical Science and Engineering, 3(3), 227-240. [16] Katoh, K., Kuma, K., Toh, H. and Miyata, T. (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Research, 33(2), 511-518. [17] KováccaronOVá, A., Ruttkay-Nedecký, G., HaverlíK, I.K., and Janecccaronek, S. (2002) Sequence Similarities and Evolutionary Relationships of Influenza Virus A Hemag- glutinins. Virus Genes, 24(1), 57- 63. [18] Gamblin, S.J., Haire, L.F., Russell, R.J., Stevens, D.J., Xiao, B., Ha, Y. et al. (2004) The Structure and Receptor Binding Properties of the 1918 Influenza Hemagglutinin. Science, 303(5665), 1838-1842.
|