Paper Menu >>
Journal Menu >>
 Journal of Software Engineering and Applications, 2012, 5, 172-174 doi:10.4236/jsea.2012.512b033 Published Online December 2012 (http://www.SciRP.org/journal/jsea) Copyright © 2012 SciRes. JSEA Biology Inspired Image Segmentation using Methods of Artificial Intelligence Radim Burget, Vaclav Uher, Jan Masek Departmentof Telecommunication, Brno University of Technology,Brno, Czech Republic. Email: burgetrm@feec.vutbr.cz Received 2012 ABSTRACT In recent years,many efforts have been devoted to image segmentation. Although for a man general image segmentation is considered an easy task, for computers it is still considered to be difficult, computationally intensive and still unre- solved task. This work presents an innovative algorithm combining theory of artificial intelligence and knowledge of human eye anatomy. The resulting algorithm has not ambitions to be universal like human brain but can be trained and perform on selected domain. The effectiveness of the algorithm is demonstrated on the selected examples. Keywords: Image Processing; Image Mining; Feature Extraction; Data Mining 1. Introduction In recent years, many efforts have been devoted to image segmentation [1-3]. Although for a human the segmenta- tion is relatively easy task, for computer it is still com- plex, difficult, and computationally intensive and it is still considered to be unresolved task. Recently, there have been several efforts, which were trying to create general segmentation algorithmthat is trying to perform like a human [4,5]. These approaches are computationally very intensive and also the accuracy is not very high for most of the cases. In many applications of the segmentation algorithms – e.g. in the production line cases - there is not necessary to perform well on general tasks. Specialized algorithms can offer better accuracy and often lower computation demands. The main contribution of this paper is a novel image segmentation method, which combines current state-of- he-art artificial intelligence algorithms with a finding in the field of biology of the human eye. The algorithm is demonstrated on a selected task and compared with thresholding and statistical region merging segmentation [6]. The resulting software[11-3] was published online and provided under open source license.1 The rest of the paper is structured as follows. Section 2 describes in more detail the problem being solved in this paper, section 3 describes proposed segmentation model, section 4 describes feature extraction from image data, section 6 demonstrates performance of the model on se- lected topics and section 7 concludes the paper. 2. Problem Definition As indicated earlier in the text, although the segmenta- tion is an easy task for a human, for computers it is quite difficult. In Figure 1 a correct segmentation by a human and incorrect segmentation performed by a statistical region merging [6] is depicted. When a man processes image, many aspects are taken into consideration includ- ing the known and expected shape of the object. In this paper a segmentation method based only on pixel information and some feature extraction methods will be used. 3. Segmentation Model The overall principle of the model is depicted in Figure 2. It can be divided into two parts: a training part and a performing part. An objective of the training part is to prepare a trained model, which can be then usedfor classification in the performing part in future. When the training part is com- pared to the performing part it is often relatively compu- tationally intensive.It can take from a few seconds to many hours of training. An input of the training part is some training data. These data are often called training data. On these data usually a domain expert (e.g. in case of medical or some specific domain data) has to classify the parts of training data which are positive and which are negative. The out- come of this process is so called labelled data. In order 1http://splab.cz/ts 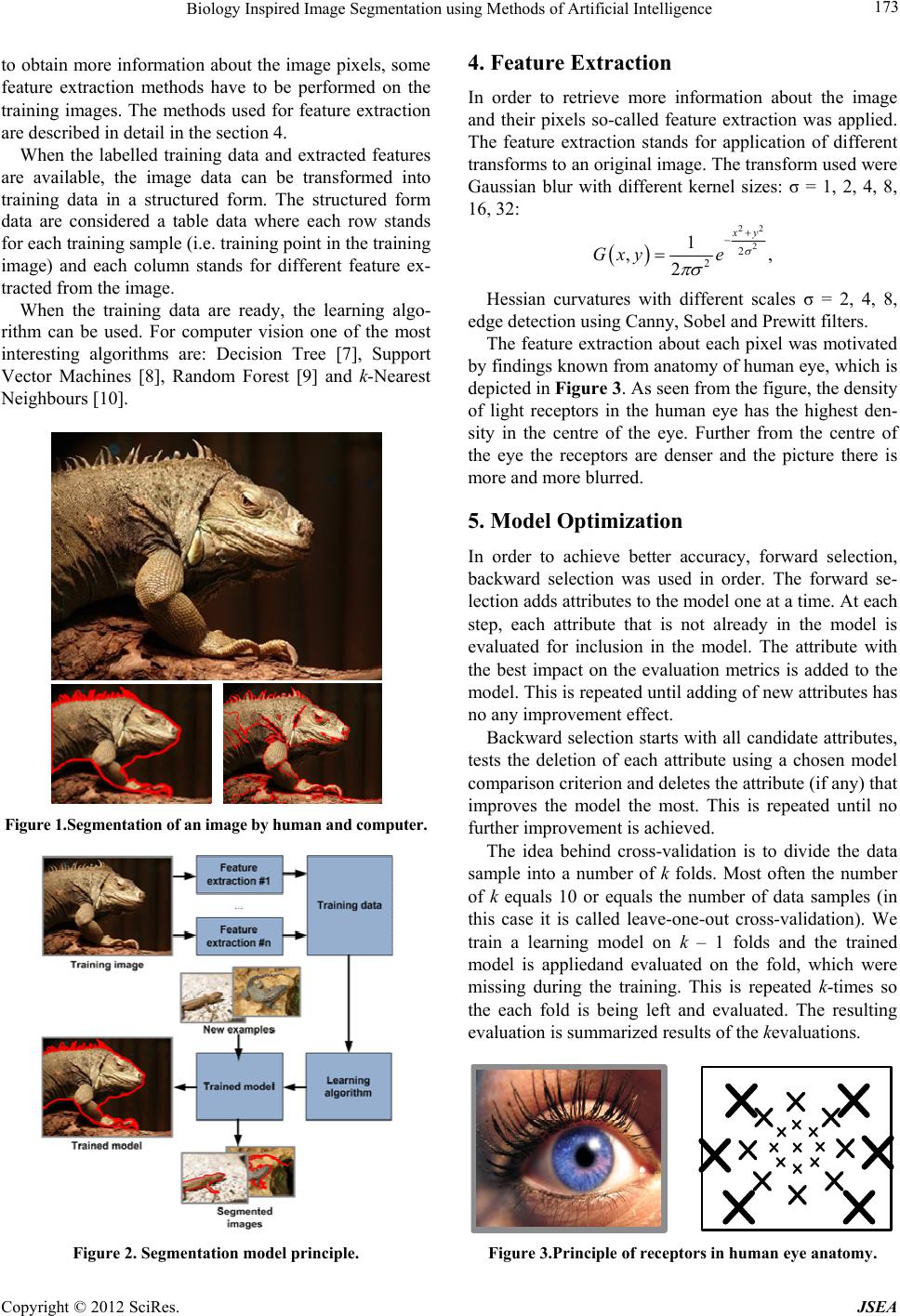 Biology Inspired Image Segmentation using Methods of Artificial Intelligence Copyright © 2012 SciRes. JSEA 173 to obtain more information about the image pixels, some feature extraction methods have to be performed on the training images. The methods used for feature extraction are described in detail in the section 4. When the labelled training data and extracted features are available, the image data can be transformed into training data in a structured form. The structured form data are considered a table data where each row stands for each training sample (i.e. training point in the training image) and each column stands for different feature ex- tracted from the image. When the training data are ready, the learning algo- rithm can be used. For computer vision one of the most interesting algorithms are: Decision Tree [7], Support Vector Machines [8], Random Forest [9] and k-Nearest Neighbours [10]. Figure 1.Segmentation of an image by human and computer. Figure 2. Segmentation model principle. 4. Feature Extraction In order to retrieve more information about the image and their pixels so-called feature extraction was applied. The feature extraction stands for application of different transforms to an original image. The transform used were Gaussian blur with different kernel sizes: σ = 1, 2, 4, 8, 16, 32: 22 2 2 2 1 ,, 2 xy Gxy e Hessian curvatures with different scales σ = 2, 4, 8, edge detection using Canny, Sobel and Prewitt filters. The feature extraction about each pixel was motivated by findings known from anatomy of human eye, which is depicted in Figure 3. As seen from the figure, the density of light receptors in the human eye has the highest den- sity in the centre of the eye. Further from the centre of the eye the receptors are denser and the picture there is more and more blurred. 5. Model Optimization In order to achieve better accuracy, forward selection, backward selection was used in order. The forward se- lection adds attributes to the model one at a time. At each step, each attribute that is not already in the model is evaluated for inclusion in the model. The attribute with the best impact on the evaluation metrics is added to the model. This is repeated until adding of new attributes has no any improvement effect. Backward selection starts with all candidate attributes, tests the deletion of each attribute using a chosen model comparison criterion and deletes the attribute (if any) that improves the model the most. This is repeated until no further improvement is achieved. The idea behind cross-validation is to divide the data sample into a number of k folds. Most often the number of k equals 10 or equals the number of data samples (in this case it is called leave-one-out cross-validation). We train a learning model on k – 1 folds and the trained model is appliedand evaluated on the fold, which were missing during the training. This is repeated k-times so the each fold is being left and evaluated. The resulting evaluation is summarized results of the kevaluations. Figure 3.Principle of receptors in human eye anatomy.  Biology Inspired Image Segmentation using Methods of Artificial Intelligence Copyright © 2012 SciRes. JSEA 174 The evaluation metrics can be arbitrary. In case of image segmentation most often metrics are accuracy: TP TN TP TN FP FN , nn Annnn precision: TP TP TN FP FN , n Pnnnn or root mean square error 02 TP TN FP FN () . in i i yy Ennnn TP n stands for number of true positively classified pixels, TN n stands for number of true negatively classi- fied pixels, FP n stands for number of false positively- classified pixels, FN n stands for number of false posi- tively classified pixels, n stands for number of all classi- fied pixels, ˆi y stands for true value (-1 or +1) and stands for classification and the confidence about the classification (real number in interval -1 to +1). 6. Experiments and Evaluation The evaluation examples of the proposed algorithm are depicted in the figure 4. In the first example there is de- picted segmentation of data taken from Google Earth application in order to automate a measurement of build- up area. The measurement takes about 30 seconds for an image of dimension 1128x661 pixels. The second example is biomedical data taken from high resolution microscope depicting brain structure of Drosophila[13]. The task here is to segment membrane from the rest of the images in order to be able to analyse micro- and macro- electrical circuits in the brain. Since a single cubic millimetre contains for about 50 thousands connection, for a human it is very time consuming task. There is also another successful work that uses the presented algorithms such as [14,15]. 7. Conclusion In this paper an innovative work dealing with trainable image segmentation was presented. The work uses dif- ferent feature extraction, machine learning and optimiza- tion methods. The work [11-13|] is able produce high accuracy segmentation models for variety of specific tasks and was released as an open-source model2. 8. Acknowledgements This work was supported by projects FR-TI4/151 and VUT BD18116001, Czech Republic. REFERENCES [1] J. Tang, A color image segmentation algorithm based on region growing, 2nd International Conference on Com- puter Engineering and Technology (ICCET), 2010 [2] ChuanLong Li; Ying Li; XueRui Wu, Novel Fuzzy C-Means Segmentation Algorithm for Image with the Spatial Neighborhoods, 2nd International Conference on Remote Sensing, Environment and Transportation Engi- neering (RSETE), 2012 [3] Hasanzadeh, M.; Kasaei, S.; Mohseni, H., A New Fuzzy Connectedness Relation for Image Segmentation, 3rd In- ternational Conference on Information and Communica- tion Technologies: From Theory to Applications, 2008. ICTTA 2008. [4] M Rousson, C Xu, A general framework for image seg- mentation using ordered spatial dependency, Medical Image Computing and Computer-Assisted Intervention – MICCAI 2006, Lecture Notes in Computer Science, 2006, Volume 4191/2006 [5] Yong Xia, A General Image Segmentation Model and its Application, Conference on Image and Graphics, 2009. ICIG '09. Fifth International [6] R. Nock, F. Nielsen (2004), "Statistical Region Merging", IEEE Trans. Pattern Anal. Mach. Intell. 26 (11): 1452-1458 [7] J. R. Quinlan, Induction of Decision Trees, Journal Ma- chine Learning, 1986 [8] Cortes, Corinna; and Vapnik, Vladimir N.; "Sup- port-Vector Networks", Machine Learning, 20, 1995. [9] L. Breiman: Random Forests. Machine Learning 45(1): 5-32 (2001) [10] Cover TM, Hart PE, "Nearest neighbor pattern classifica- tion". IEEE Transactions on Information Theory 13 (1): 21–27. [11] RadimBurget, PetrCika, Martin Zukal, Jan Masek: Auto- mated localization of Temporomandibular Joint Disc in MRI images. Telecommunications and Signal Processing (TSP), 2011 34th International Conference on: p.: 413-41 [12] Burget, R., Uher, V., Masek, J., Trainable Segmentation Based on Local-level and Segment-level Feature Extrac- tion, 2012 IEEE International Symposium on Biomedi- cal Imaging: From nano to Macro, 2012 [13] Uher, V., Burget, R. , Automatic 3D Segmentation of Human Brain Images Using Data-mining Techniques, 35th International Conference on Telecommunications and Signal Processing (TSP 2012), 2012 [14] Burget, R., Karasek, J., Smekal, Z., et al., RapidMiner Image Processing Extension: A Platform for Collabora- tive Research, 33rd International Conference on Tele- communication and Signal Processing Location: Vienna, AUSTRIA Date: AUG 17-20, 2010 [15] Burget, R.; Cika, P.; Zukal, M.,Automated Localization of Temporomandibular Joint Disc in MRI Images 34th International Conference on Telecommunications and Signal Processing (TSP), Budapest, HUNGARY, AUG 18-20, 2011 2http://splab.cz/ts |

