Paper Menu >>
Journal Menu >>
 A Journal of Software Engineering and Applications, 2012, 5, 84-87 doi:10.4236/jsea.2012.512b017 Published Online December 2012 (http://www.scirp.org/journal/jsea) Copyright © 2012 SciRes. JSEA Pharmacophore Model Generation of thrombin Inhibitors Xing Wang1, Yanling Zhang1, Yuhong Xiang2, Zhenzhen Ren1, Yanjiang Qiao1* 1School of Chinese Materia Medica, Beijing University of Chinese Medicine, Beijing 100102, China; 2Department of Chemistry, Capital Normal University, Beijing 100048, China. Email: yjqiao@263.net Received 2012 ABSTRACT Thrombin, an important factor of clotting system, take part in a variety of physiological actions, such as blood clotting, anticoagulation, thrombosis and fibrinolysis. Inhibiting thrombin is a pivotal and effective step for the prophylaxis of venous and arterial thrombosis, as well as prevent myocardial infarction for high-risk patients. In this study, a three di- mensional pharmacophore model was generated for the molecules which are responsible for vasodilation activities tar- geting thrombin. Ten compounds with known thrombin-inhibiting activity values were selected as training set to gener- ate the hypothesis using GALAHAD program in SYBYL 7.0 software. The best hypothesis comprises five pharma- cophore features: four hydrophobes groups and one positively charged group. It has been further validated towards a test set including known activity compounds obtained from Binding Database, the values of effectively active hit A% and comprehensive evaluation index CAI are respectively 63.33% and 2.34, the pharmacophore was proven to be suc- cessful in discriminating active and inactive inhibitors. Furthermore, the pharmacophore model was used as a 3D query to screen TCMD (Version 2009) database and 6 hit compounds of higher predicted activity were the reported cardiovascular activities, which may be useful for further study. Keywords: Cardiovascular Disease; Thrombin; Pharmacophore; Virtual Screening 1. Introduction Thrombin, a "trypsin-like" serine protease protein, is an important factor of clotting system. It take part in a vari- ety of physiological actions, such as blood clotting, anti- coagulation, thrombosis and fibrinolysis [1-3]. Moreover, previous studies have demonstrated that thrombin is one of the key factors for cancer developing, such as PAR1, an important type of thrombin, mediates the enhance- ment effects of thrombin in angiogenesis, invasion and metastasis, growth of cancer [4-6]. These facts have raised great interests to find an effective, safe, and orally available thrombin inhibitors, such as hirudin [7], dabi- gatran [8], rivaroxaban and apixaban [9], which could be useful anticoagulant drugs for the prophylaxis of venous and arterial thrombosis, as well as prevent myocardial infarction for high-risk patients [10]. In this paper, we have developed a pharmacophore model whose purpose is to identify the critical pharma- cophoric features necessary for potent thrombin inhibi- tors. Further, hypothesis was evaluated by a test set with known activity compounds, the effectively active hit A% and comprehensive evaluation index CAI were used to assess the model. Finally, the pharmacophore model was used as a 3D query to screen TCMD (Version 2009) da- tabase, 52 compounds were hit and the 6 hit compounds of higher predicted activity were the reported cardiovas- cular activities, which may be useful for further study. 2. Materials and Methods 2.1. Compounds and biological data Compounds 1~30, which can inhibit thrombin, were taken from the literatures [11-13] and served as the data- base in the pharmacophore modeling. The structures and inhibitory activities are listed in Figure 1. The chemical structures were drawn in ISIS-Draw software and saved in SYBYL mol2 format, then all the 2D structures were converted to 3D structures in SYBYL 7.0 software. N HN O H N SO O Compound 1 Compound 2 Compound 3 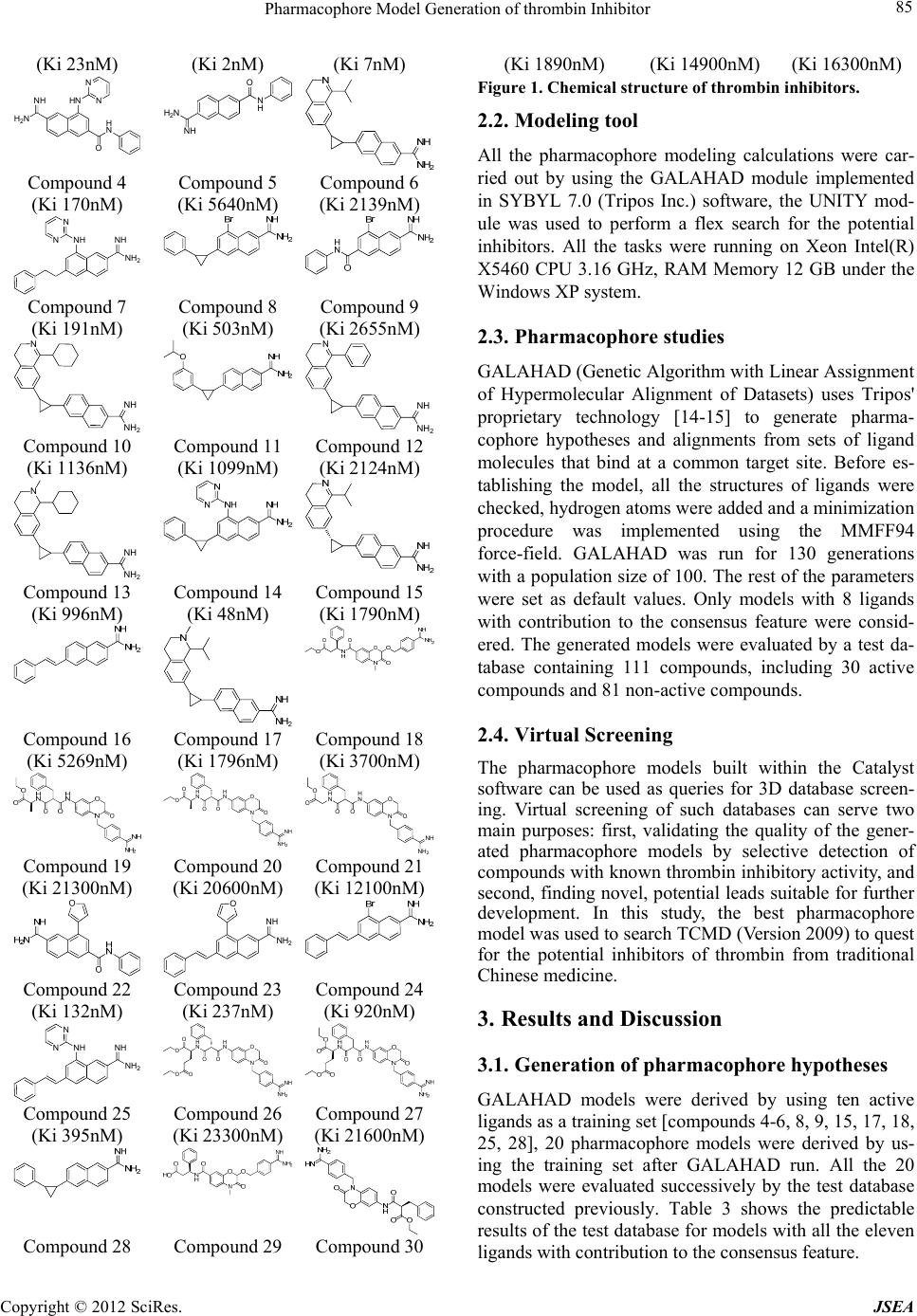 Pharmacophore Model Generation of thrombin Inhibitor Copyright © 2012 SciRes. JSEA 85 (Ki 23nM) (Ki 2nM) (Ki 7nM) HN H N N N NH O H2N NH H2N O N H Compound 4 (Ki 170nM) Compound 5 (Ki 5640nM) Compound 6 (Ki 2139nM) NH N NNH NH2 Compound 7 (Ki 191nM) Compound 8 (Ki 503nM) Compound 9 (Ki 2655nM) N NH NH2 N NH NH2 Compound 10 (Ki 1136nM) Compound 11 (Ki 1099nM) Compound 12 (Ki 2124nM) N NH NH2 Compound 13 (Ki 996nM) Compound 14 (Ki 48nM) Compound 15 (Ki 1790nM) Compound 16 (Ki 5269nM) Compound 17 (Ki 1796nM) Compound 18 (Ki 3700nM) N H NO H N NH OO O O NH2 O N O H N H N NH OO O O NH2 O Compound 19 (Ki 21300nM) Compound 20 (Ki 20600nM) Compound 21 (Ki 12100nM) NH O NH2 Compound 22 (Ki 132nM) Compound 23 (Ki 237nM) Compound 24 (Ki 920nM) NH N NNH NH2N H NO H N NH OO O O O NH2 O O Compound 25 (Ki 395nM) Compound 26 (Ki 23300nM) Compound 27 (Ki 21600nM) Compound 28 Compound 29 Compound 30 (Ki 1890nM) (Ki 14900nM) (Ki 16300nM) Figure 1. Chemical structure of thrombin inhibitors. 2.2. Modeling tool All the pharmacophore modeling calculations were car- ried out by using the GALAHAD module implemented in SYBYL 7.0 (Tripos Inc.) software, the UNITY mod- ule was used to perform a flex search for the potential inhibitors. All the tasks were running on Xeon Intel(R) X5460 CPU 3.16 GHz, RAM Memory 12 GB under the Windows XP system. 2.3. Pharmacophore studies GALAHAD (Genetic Algorithm with Linear Assignment of Hypermolecular Alignment of Datasets) uses Tripos' proprietary technology [14-15] to generate pharma- cophore hypotheses and alignments from sets of ligand molecules that bind at a common target site. Before es- tablishing the model, all the structures of ligands were checked, hydrogen atoms were added and a minimization procedure was implemented using the MMFF94 force-field. GALAHAD was run for 130 generations with a population size of 100. The rest of the parameters were set as default values. Only models with 8 ligands with contribution to the consensus feature were consid- ered. The generated models were evaluated by a test da- tabase containing 111 compounds, including 30 active compounds and 81 non-active compounds. 2.4. Virtual Screening The pharmacophore models built within the Catalyst software can be used as queries for 3D database screen- ing. Virtual screening of such databases can serve two main purposes: first, validating the quality of the gener- ated pharmacophore models by selective detection of compounds with known thrombin inhibitory activity, and second, finding novel, potential leads suitable for further development. In this study, the best pharmacophore model was used to search TCMD (Version 2009) to quest for the potential inhibitors of thrombin from traditional Chinese medicine. 3. Results and Discussion 3.1. Generation of pharmacophore hypotheses GALAHAD models were derived by using ten active ligands as a training set [compounds 4-6, 8, 9, 15, 17, 18, 25, 28], 20 pharmacophore models were derived by us- ing the training set after GALAHAD run. All the 20 models were evaluated successively by the test database constructed previously. Table 3 shows the predictable results of the test database for models with all the eleven ligands with contribution to the consensus feature. 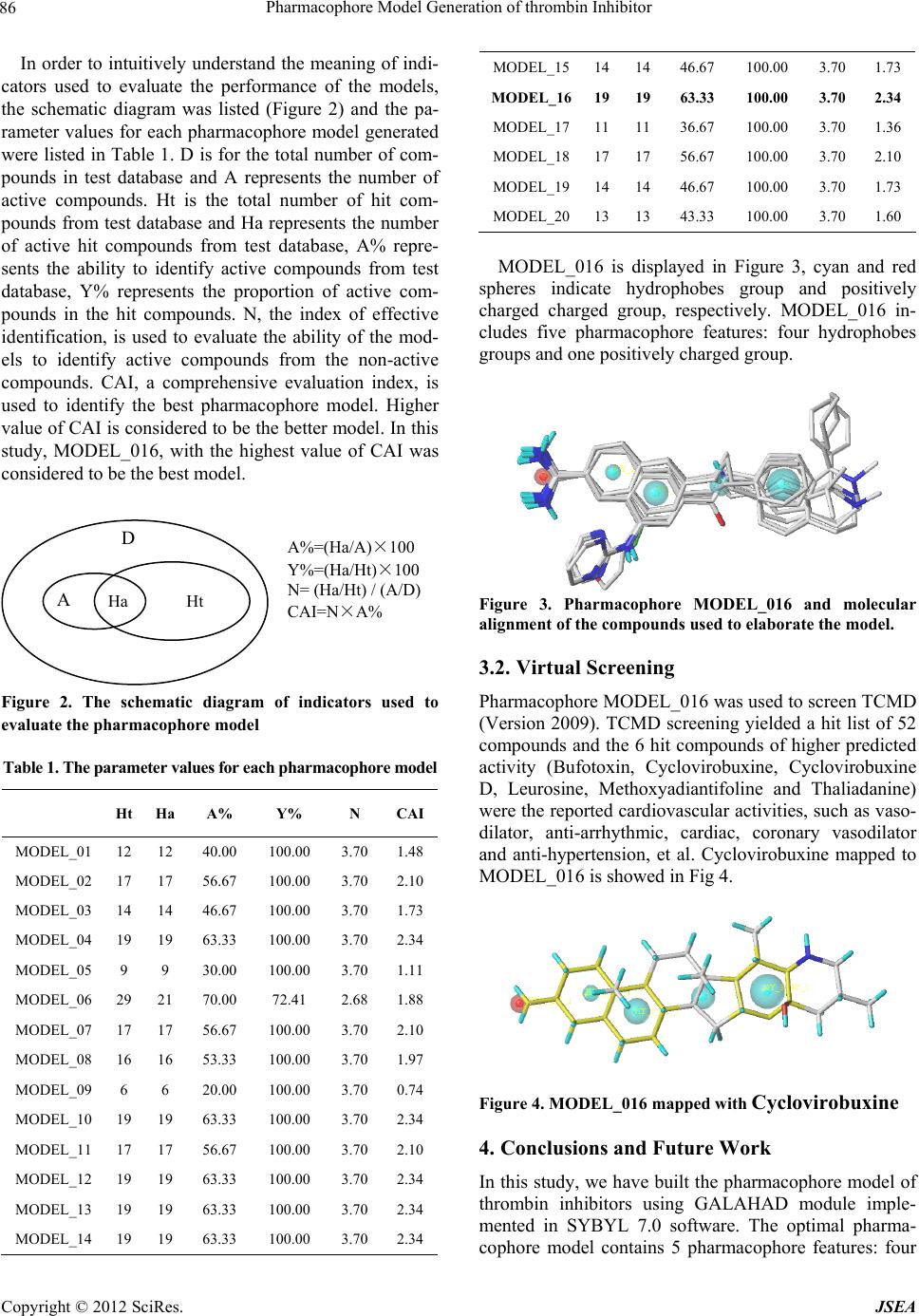 Pharmacophore Model Generation of thrombin Inhibitor Copyright © 2012 SciRes. JSEA 86 In order to intuitively understand the meaning of indi- cators used to evaluate the performance of the models, the schematic diagram was listed (Figure 2) and the pa- rameter values for each pharmacophore model generated were listed in Table 1. D is for the total number of com- pounds in test database and A represents the number of active compounds. Ht is the total number of hit com- pounds from test database and Ha represents the number of active hit compounds from test database, A% repre- sents the ability to identify active compounds from test database, Y% represents the proportion of active com- pounds in the hit compounds. N, the index of effective identification, is used to evaluate the ability of the mod- els to identify active compounds from the non-active compounds. CAI, a comprehensive evaluation index, is used to identify the best pharmacophore model. Higher value of CAI is considered to be the better model. In this study, MODEL_016, with the highest value of CAI was considered to be the best model. Figure 2. The schematic diagram of indicators used to evaluate the pharmacophore mode l Table 1. The parameter values for each pharmacophore model Ht Ha A% Y% N CAI MODEL_01 12 12 40.00 100.00 3.70 1.48 MODEL_02 17 17 56.67 100.00 3.70 2.10 MODEL_03 14 14 46.67 100.00 3.70 1.73 MODEL_04 19 19 63.33 100.00 3.70 2.34 MODEL_05 9 9 30.00 100.00 3.70 1.11 MODEL_06 29 21 70.00 72.41 2.68 1.88 MODEL_07 17 17 56.67 100.00 3.70 2.10 MODEL_08 16 16 53.33 100.00 3.70 1.97 MODEL_09 6 6 20.00 100.00 3.70 0.74 MODEL_10 19 19 63.33 100.00 3.70 2.34 MODEL_11 17 17 56.67 100.00 3.70 2.10 MODEL_12 19 19 63.33 100.00 3.70 2.34 MODEL_13 19 19 63.33 100.00 3.70 2.34 MODEL_14 19 19 63.33 100.00 3.70 2.34 MODEL_15141446.67 100.00 3.70 1.73 MODEL_16191963.33 100.00 3.70 2.34 MODEL_17111136.67 100.00 3.70 1.36 MODEL_18171756.67 100.00 3.70 2.10 MODEL_19141446.67 100.00 3.70 1.73 MODEL_20131343.33 100.00 3.70 1.60 MODEL_016 is displayed in Figure 3, cyan and red spheres indicate hydrophobes group and positively charged charged group, respectively. MODEL_016 in- cludes five pharmacophore features: four hydrophobes groups and one positively charged group. Figure 3. Pharmacophore MODEL_016 and molecular alignment of the compounds used to elaborate the model. 3.2. Virtual Screening Pharmacophore MODEL_016 was used to screen TCMD (Version 2009). TCMD screening yielded a hit list of 52 compounds and the 6 hit compounds of higher predicted activity (Bufotoxin, Cyclovirobuxine, Cyclovirobuxine D, Leurosine, Methoxyadiantifoline and Thaliadanine) were the reported cardiovascular activities, such as vaso- dilator, anti-arrhythmic, cardiac, coronary vasodilator and anti-hypertension, et al. Cyclovirobuxine mapped to MODEL_016 is showed in Fig 4. Figure 4. MODEL_016 mapped with Cyclovirobuxine 4. Conclusions and Future Work In this study, we have built the pharmacophore model of thrombin inhibitors using GALAHAD module imple- mented in SYBYL 7.0 software. The optimal pharma- cophore model contains 5 pharmacophore features: four D A Ha Ht A%=(Ha/A)×100 Y%=(Ha/Ht)×100 N= (Ha/Ht) / (A/D) CAI=N×A%  Pharmacophore Model Generation of thrombin Inhibitor Copyright © 2012 SciRes. JSEA 87 hydrophobes group and one positively charged charged group. The external validation shows that the values of effectively active hit A% and comprehensive evaluation index CAI are respectively 63.33% and 2.34, which can prove that the pharmacophore model is reliable and available. The virtual screen results of TCMD (Version 2009) database shows that the pharmacophore model has the ability of determining vasoactive compound related to thrombin, and the validation experiments needs to be further studied. 5. Acknowledgements This work is financially supported by the Foundation of National Natural Science Foundation of China (No. 81173522) and National Key Technology R&D Program (No. 2008BAI51B01) in Beijing University of Chinese Medicine. REFERENCES [1] Sambrano GR, Weiss EJ, Zheng YW, et al. “Role of thrombin signalling in platelets in haemostasis and thrombosis”. Nature, Vol. 413, No. 6, 2011, pp. 74-78. doi:10.1038/35092573 [2] H LUO, Y LIANG. “Thrombin-activatable fibrinolysis inhibitor: a new modulator linking coagulation and fibri- nolysis”. Basic Medical Sciences and Clinics, Vol. 23, No. 5, 2003, pp. 484-489. doi:6325(2003)05-0484-06 [3] Weitz JI. “Factor Xa and thrombin as targets for new oral anticoagulants”. Thrombosis Research, Vol. 127, No. 2, 2011, pp. S5-S12. doi:10.1016/S0049-3848(10)70147-X [4] Crawley JT, Zanardelli S, Chion CK, et al. “The central role of thrombin in hemostasis”. Journal of Thrombosis and Haemostasis,Vol. 5, Sup. s1, 2007,pp. 95-101. doi:10.1111/j.1538-7836.2007.02500.x [5] Nierodzik ML,Karpatkin S. “Thrombin induces tumor growth, metastasis, and angiogenesis: Evidence for a thrombin-regulated dormant tumor phenotype”. Cancer Cell, vol.10,No.5,2006, pp.355-362. doi.org/10.1016/j.ccr.2006.10.002 [6] Bajzar L, Chan AK, Massicotte MP, et al. “Thrombosis in children with malignancy”. Current Opinion in Pe- diatrics,vol.18,No.1, 2006 ,pp.1-9. doi:10.1097/01.mop.0000193270.09001.ea [7] Eriksson BI, Quinlan DJ, Eikelboom JW. “Novel Oral Factor Xa and Thrombin Inhibitors in the Management of Thromboembolism”. Annual Review of Medicine, vol.62,2011,pp. 41-57. doi:10.1146/annurev-med-062209-095159 [8] Bendel SD, Bona R, Baker WL. “Dabigatran: an oral direct thrombin inhibitor for use in atrial fibrillation”. Advances in Therapy,vol.28,No.6,2011,pp.460-472. doi:10.1007/s12325-011-0025-1 [9] A. Kobsar , J. Koessler , L. Kehrer,etal. “The thrombin inhibitors hirudin and Refludan® activate the soluble guanylyl cyclase and the cGMP pathway in washed hu- man platelets”. ThrombosisandHaemosta- sis.Vol.107.No.3,2012,pp.521-529. doi:10.1160/TH11-07-0461 [10] Steinmetzer T, Sturzebecher J. “Progress in the Devel- opment of Synthetic Thrombin Inhibitors as New Orally Active Anticoagulants”.Current Medicinal Chemis- try,Vol.11,No.17, 2004,pp.2297-2321. doi:10.2174/0929867043364540 [11] Santosh P, Gregory A. L., Julie E. P. “Conformation Mining: An Algorithm for Finding Biologically Rele- vant Conformations”. J. Med. Chem., Vol. 48, No.9, 2005, pp. 3313-3318. doi:10.1021/jm049066l. [12] Milan Bruncko, William J. McClellan, Michael D. Wendt, et al. “Naphthamidine urokinase plasminogen activator inhibitors with improved pharmacokinetic properties”. Bioorganic & Medicinal Chemistry Letters, Vol.15,No.1, 2005, pp. 93-98. doi:10.1016/j.bmcl.2004.10.026 [13] Petra Štefanic Anderluh, Marko Anderluh, Janez Ilaš. “Toward a Novel Class of Antithrombotic Compounds with Dual Function. Discovery of 1,4-Benzoxazin-3(4H)-one Derivatives Possessing Thrombin Inhibitory and Fibrinogen Receptor Antagonis- tic Activities”, J. Med. Chem., 2005, 48 (9), pp 3110–3113. doi:10.1021/jm048984g [14] Richmond, N.J., Abram, C.A., Wolohan, P.R., et al. “GALAHAD: 1 Pharmacophore Identification by Hy- permolecular Alignment of Ligands in 3D”. J COMPUT AID MOL DES, Vol. 20, No. 9, 2006, pp. 567-587. doi:10.1007/s10822-006-9082-y. [15] N.J. Richmond, P.Willett, R.D. Clark. “Alignment of three-dimensionalmolecules using an image recognition algorithm”. J.Mol.Graph.Model. Vol. 23, No. 2, 2004, pp. 199-209. doi:10.1016/j.bbr.2011.03.031. |

