Paper Menu >>
Journal Menu >>
 Vol.2, No.9, 1065-1071 (2010) Health doi:10.4236/health.2010.29156 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ P53 pseudogene: potential role in heat shock induced apoptosis in a rat histiocytoma Amere Subbarao Sreedhar Centre for Cellular and Molecular Biology, Hyderabad, India; assr@ccmb.res.in Received 1 June 2010; revised 2 July 2010; accepted 5 July 2010. ABSTRACT The p53 tumor suppressor gene is either non- functional or highly and frequently mutated in majority of cancers. In our study towards un- derstanding cellular adaptations to stress using a rat histiocytic tumor model, we have identified mis-sense mutation in p53 that led to premature termination of translation at the carboxyl-termi- nus. Further, the cDNA isolated from heat stre- ssed cells producing two amplicons with cDNA specific primers (N-terminus) suggested oc- currence of possible pseudogene(s). A com- parative analysis between different tumor cell lines of rat origin and rat genomic DNA using p53 gene specific primers resulted in the ampli- fication of a processed pseudogene and its positive interaction with wild type p53 probe on Southern blot analysis. The genomic DNA se- quence analysis, and sequence comparison with cDNA discovered that the processed pseu- dogene lacks DNA binding domain and nuclear localization signal, however, contains the ribo- somal entry and stop signals. Rat genome BLAST analysis of the pesudogene suggested chromosome-18 localization which was in addi- tion to 14, 13, 10, 9 localization of the cDNA. In the interest of unraveling hidden dimensions of p53 tumor suppressor gene, our study explores the probability of p53 functional pseudogenes in rat histiocytoma. Keywords: Pseudogene; P53; Tumor; Rat Histiocytoma 1. INTRODUCTION The tumor suppressor p53 is a multifunctional protein that is involved in a variety of biological processes such as growth arrest, apoptosis, differentiation and senes- cence [1,2]. Aberrant expression of this gene results in either a gain of transforming potential or a loss in tumor suppressor activity [3,4]. The p53 gene mutation, dele- tion, insertion or protein sequestration etc are often found in many cancers [5,6] and these mutations affect the p53 binding to DNA [7]. Analysis of the degeneracy of p53 DNA-binding site suggests that there may be as many as 200-400 p53 target sequences or perhaps more [8]. Despite the high frequency with which p53 is mutated during tumor development, a substantial proportion of tumors still express the wild type p53 [9]. This could be the reason in spite of exhaustive information on p53 modi- fications the corresponding role of p53 modification in experimental animal tumor models is poorly understood. We are investigating the role of p53 in heat stress-in- duced rat histiocytic tumors models. In the process of elucidating heat stress induced cell death pathways and evaluating the functional significance of p53 in heat shock induced cell death in tumor cells, we have identi- fied mutated form of p53 with two functional alleles by reverse transcriptase polymerase chain reaction, and the deletion and addition of nucleotides had resulted in C-terminal deletion of 50 amino acids. We demonstrated that Fas/CD95 induced apoptosis requires p53, and hy- pothesized that C-terminal deletion and loss of oligo- merization domain and nuclear localization signal proba- bly are responsible for p53-transcritpion independent apoptosis as suggested [10,11]. In the present study we show that there are two processed pseudogenes for p53 in this tumor model and one of them also has ribosomal entry site. A comparative genome analysis further re- vealed that the processed pseudogene is predominantly present in all the rat and mouse species but absent in humans. 2. MATERIALS AND METHODS 2.1. Animal Handling All animal maintenance and handling was accomplished as per the institutional ethical committee approval at Centre for Cellular and Molecular Biology, Hyderabad, India. 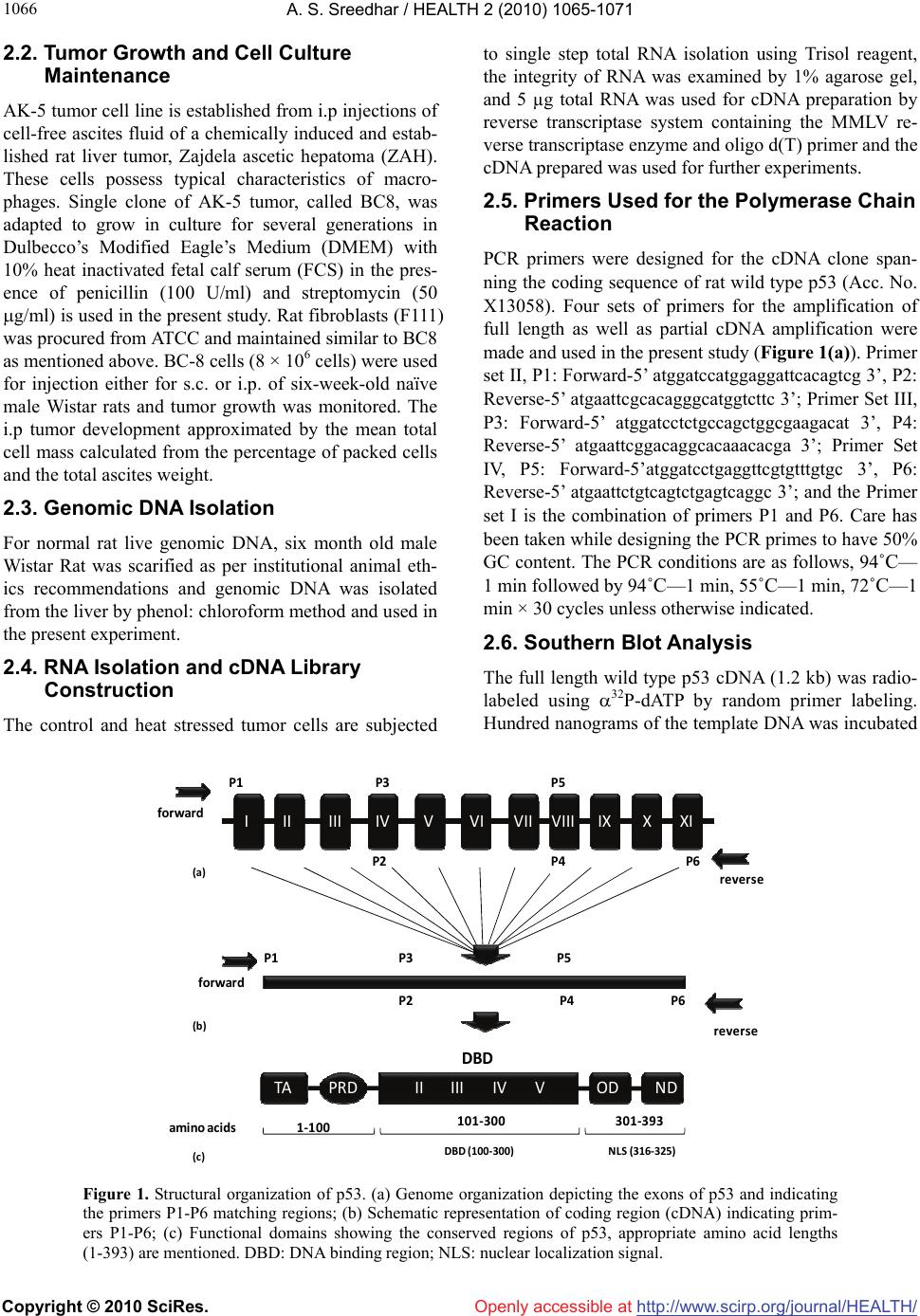 A. S. Sreedhar / HEALTH 2 (2010) 1065-1071 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 1066 2.2. Tumor Growth and Cell Culture Maintenance AK-5 tumor cell line is established from i.p injections of cell-free ascites fluid of a chemically induced and estab- lished rat liver tumor, Zajdela ascetic hepatoma (ZAH). These cells possess typical characteristics of macro- phages. Single clone of AK-5 tumor, called BC8, was adapted to grow in culture for several generations in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% heat inactivated fetal calf serum (FCS) in the pres- ence of penicillin (100 U/ml) and streptomycin (50 g/ml) is used in the present study. Rat fibroblasts (F111) was procured from ATCC and maintained similar to BC8 as mentioned above. BC-8 cells (8 × 106 cells) were used for injection either for s.c. or i.p. of six-week-old naïve male Wistar rats and tumor growth was monitored. The i.p tumor development approximated by the mean total cell mass calculated from the percentage of packed cells and the total ascites weight. 2.3. Genomic DNA Isolation For normal rat live genomic DNA, six month old male Wistar Rat was scarified as per institutional animal eth- ics recommendations and genomic DNA was isolated from the liver by phenol: chloroform method and used in the present experiment. 2.4. RNA Isolation and cDNA Library Construction The control and heat stressed tumor cells are subjected to single step total RNA isolation using Trisol reagent, the integrity of RNA was examined by 1% agarose gel, and 5 µg total RNA was used for cDNA preparation by reverse transcriptase system containing the MMLV re- verse transcriptase enzyme and oligo d(T) primer and the cDNA prepared was used for further experiments. 2.5. Primers Used for the Polymerase Chain Reaction PCR primers were designed for the cDNA clone span- ning the coding sequence of rat wild type p53 (Acc. No. X13058). Four sets of primers for the amplification of full length as well as partial cDNA amplification were made and used in the present study (Figure 1(a)). Primer set II, P1: Forward-5’ atggatccatggaggattcacagtcg 3’, P2: Reverse-5’ atgaattcgcacagggcatggtcttc 3’; Primer Set III, P3: Forward-5’ atggatcctctgccagctggcgaagacat 3’, P4: Reverse-5’ atgaattcggacaggcacaaacacga 3’; Primer Set IV, P5: Forward-5’atggatcctgaggttcgtgtttgtgc 3’, P6: Reverse-5’ atgaattctgtcagtctgagtcaggc 3’; and the Primer set I is the combination of primers P1 and P6. Care has been taken while designing the PCR primes to have 50% GC content. The PCR conditions are as follows, 94˚C— 1 min followed by 94˚C—1 min, 55˚C—1 min, 72˚C—1 min × 30 cycles unless otherwise indicated. 2.6. Southern Blot Analysis The full length wild type p53 cDNA (1.2 kb) was radio- labeled using 32P-dATP by random primer labeling. Hundred nanograms of the template DNA was incubated IIIIIIIVVVIVII VIIIIXXXI II IIIIVVTA PRD DBD OD ND 1‐100 101‐300 301‐393 P1 P3P5 P2P4 P6 forward reverse P1 P3P5 P2P4 P6 forward reverse ami noacids NLS(316‐325)DBD(100‐300) (a) (b) (c) Figure 1. Structural organization of p53. (a) Genome organization depicting the exons of p53 and indicating the primers P1-P6 matching regions; (b) Schematic representation of coding region (cDNA) indicating prim- ers P1-P6; (c) Functional domains showing the conserved regions of p53, appropriate amino acid lengths (1-393) are mentioned. DBD: DNA binding region; NLS: nuclear localization signal. 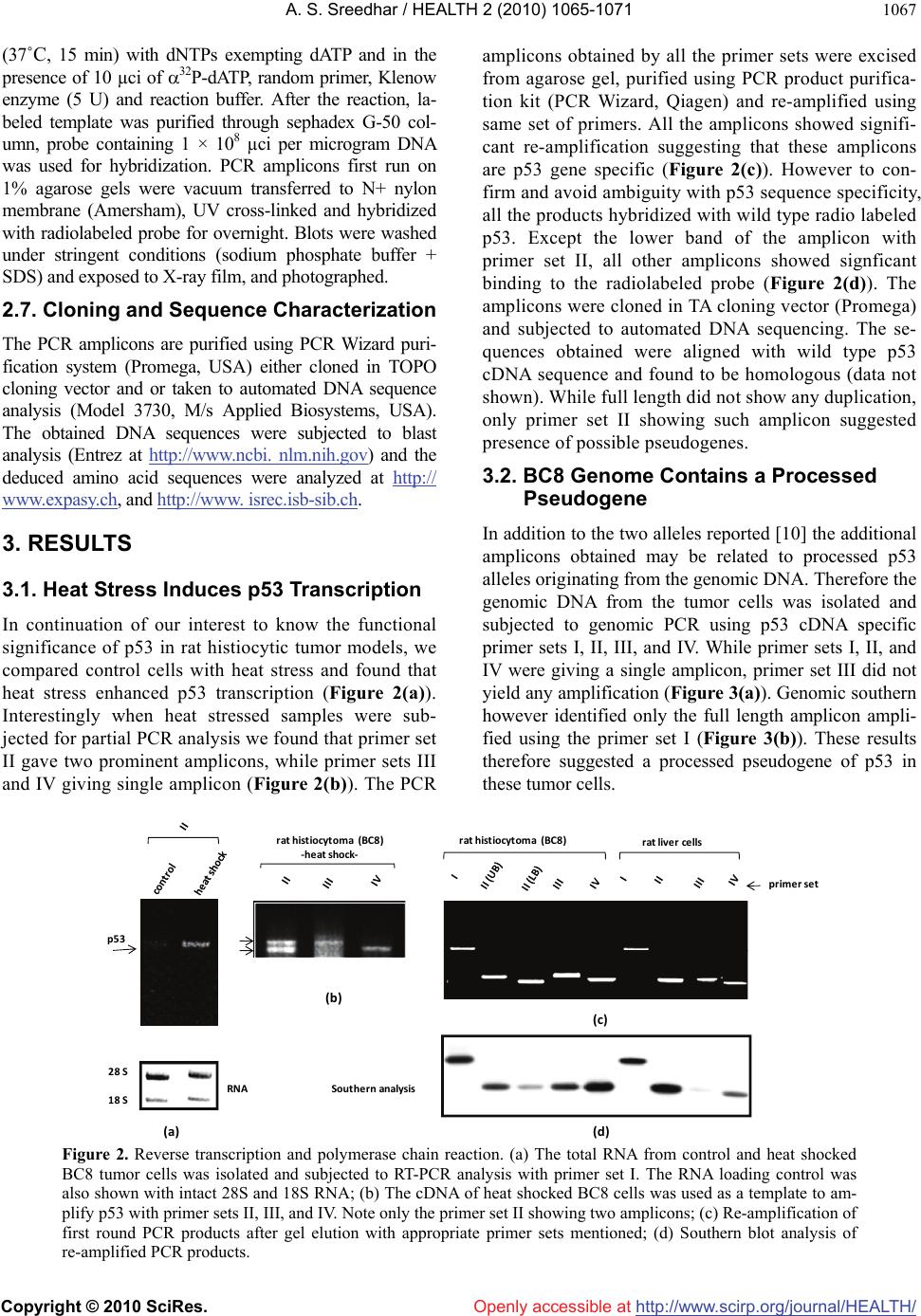 A. S. Sreedhar / HEALTH 2 (2010) 1065-1071 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 1067 (37˚C, 15 min) with dNTPs exempting dATP and in the presence of 10 µci of 32P-dATP, random primer, Klenow enzyme (5 U) and reaction buffer. After the reaction, la- beled template was purified through sephadex G-50 col- umn, probe containing 1 × 108 µci per microgram DNA was used for hybridization. PCR amplicons first run on 1% agarose gels were vacuum transferred to N+ nylon membrane (Amersham), UV cross-linked and hybridized with radiolabeled probe for overnight. Blots were washed under stringent conditions (sodium phosphate buffer + SDS) and exposed to X-ray film, and photographed. 2.7. Cloning and Sequence Characterization The PCR amplicons are purified using PCR Wizard puri- fication system (Promega, USA) either cloned in TOPO cloning vector and or taken to automated DNA sequence analysis (Model 3730, M/s Applied Biosystems, USA). The obtained DNA sequences were subjected to blast analysis (Entrez at http://www.ncbi. nlm.nih.gov) and the deduced amino acid sequences were analyzed at h t tp:// www.expasy.ch, and http://www. isrec.isb-sib.ch. 3. RESULTS 3.1. Heat Stress Induces p53 Transcription In continuation of our interest to know the functional significance of p53 in rat histiocytic tumor models, we compared control cells with heat stress and found that heat stress enhanced p53 transcription (Figure 2(a)). Interestingly when heat stressed samples were sub- jected for partial PCR analysis we found that primer set II gave two prominent amplicons, while primer sets III and IV giving single amplicon (Figure 2(b)). The PCR amplicons obtained by all the primer sets were excised from agarose gel, purified using PCR product purifica- tion kit (PCR Wizard, Qiagen) and re-amplified using same set of primers. All the amplicons showed signifi- cant re-amplification suggesting that these amplicons are p53 gene specific (Figure 2(c)). However to con- firm and avoid ambiguity with p53 sequence specificity, all the products hybridized with wild type radio labeled p53. Except the lower band of the amplicon with primer set II, all other amplicons showed signficant binding to the radiolabeled probe (Figure 2(d)). The amplicons were cloned in TA cloning vector (Promega) and subjected to automated DNA sequencing. The se- quences obtained were aligned with wild type p53 cDNA sequence and found to be homologous (data not shown). While full length did not show any duplication, only primer set II showing such amplicon suggested presence of possible pseudogenes. 3.2. BC8 Genome Contains a Processed Pseudogene In addition to the two alleles reported [10] the additional amplicons obtained may be related to processed p53 alleles originating from the genomic DNA. Therefore the genomic DNA from the tumor cells was isolated and subjected to genomic PCR using p53 cDNA specific primer sets I, II, III, and IV. While primer sets I, II, and IV were giving a single amplicon, primer set III did not yield any amplification (Figure 3(a)). Genomic southern however identified only the full length amplicon ampli- fied using the primer set I (Figure 3(b)). These results therefore suggested a processed pseudogene of p53 in these tumor cells. rathistiocytom a(BC8) ratlivercells primerset rathistiocytom a(BC8) ‐heatshock‐ 28S 18S p53 RNA Southernanalysis (a) (b) (c) (d) Figure 2. Reverse transcription and polymerase chain reaction. (a) The total RNA from control and heat shocked BC8 tumor cells was isolated and subjected to RT-PCR analysis with primer set I. The RNA loading control was also shown with intact 28S and 18S RNA; (b) The cDNA of heat shocked BC8 cells was used as a template to am- plify p53 with primer sets II, III, and IV. Note only the primer set II showing two amplicons; (c) Re-amplification of first round PCR products after gel elution with appropriate primer sets mentioned; (d) Southern blot analysis of re-amplified PCR products.  A. S. Sreedhar / HEALTH 2 (2010) 1065-1071 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 1068 rathistiocytoma(BC8) ratascites(AK5)ratfibroblasts(F111) ratliverce lls primerset (a) (b ) (c) (d) Southern analysis Figure 3. Genomic PCR and Southern analysis. (a) BC8 genomic PCR analysis with four primer sets, I, II, III, and IV; (b) Genomic Southern analysis of PCR products obtained from Figure 3(a); (c) Genomic PCR analysis of rat fibroblasts, rat histiocytoma (AK5), and rat liver cell; (d) Genomic Southern analysis of PCR products obtained from Figure 3(c). Processed pseudogenes arise through a mechanism whereby a spliced mRNA is reverse transcribed and sub- sequently inserted into the genome [12]. If pseudo- genes are formed in this way during evolution, these pseudogenes should present in the rat genome and should coexist with all the cell types. To examine this, transformed rat fibroblast cells (F111), parental ascites rat histiocytoma (AK5) were compared with the normal rat genomic DNA. 3.3. Blast and In-Silico Translational Analysis We went ahead of cloning p53 pseudogene and se- quence alaysis of cloned product using automated DNA sequencing. From the sequence analysis we found that there is indeed a processed pseudogene having a potential to provide two gene products with different reading frames. A comparative sequence alig- nment of processed pseudogene with full length RT- PCR product of rat histiocytoma additionally showed high sequence homology. Analysis of pseudogene re- vealed loss of DNA binding region (nt 700-860) and nuclear localization signal (nt 1030-1080) of cDNA (Figure 4). Whole rat genome Blast analysis with cDNA sequence identified its chromosome localization on chromosomes 14, 13, 10, 9 and 2, and the pseu- dogene sequence Blast identified its additional local- ization at chromosome 18 (Table 1). 4. DISCUSSION The p53 gene is frequently lost or rearranged in a large variety of cancers, and most of the alterations in p53 are found in the core domain that interfere with p53 DNA- binding activity [5]. Although p53 has been a wonder molecule and the guardian of genome, mutation of p53 affects its native functions including the antiapoptotic function. Several p53 mutant cells are reported to have lost apoptotic functions but not the cell cycle inhibition [13,14]. While our earlier study suggesting that loss of C-terminal 50 amino acids could have played a role in p53-transcription independent apoptosis via Fas/CD95 translocation from golgi to plasma membrane [11,15], a report from Zhu et al. [16] indicated that the N-terminal 43-63 amino acid are more than sufficient to activate p53 transcription dependent apoptosis. Further, induction of pro-apoptotic factor Bax, a known transcriptional cli- ent for p53 [17], and subsequent activation of intrinsic apoptotic death pathway through mitochondrial dysfunc- tion [18] directed us to look for possible processed genes in the tumor genome. By definition, pseudogenes lack a function. However, the classification of pseudogenes generally relies on computational analysis of genomic sequences using complex algorithms [19]. It has been established that quite a few pseudogenes can go through the process of  A. S. Sreedhar / HEALTH 2 (2010) 1065-1071 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 1069 transcription, either if their own promoter is still intact or in some cases using the promoter of a nearby gene; this expression of pseudogenes also appears to be tis- sue-specific [20]. Pseudogenes are often referred to in the scientific literature as nonfunctional DNA. Failure to observe pseudogenes coding for a product under ex- perimental conditions is no proof that they never do so inside an organism. Homologous recombination between the intact functional p53 gene and the p53 pseudogene is thought to have occurred in such a perturbed intracellu- lar environment with genomic instability, thus inactivat- ing the intact allele of the functional p53, therefore the persistence of pseudogenes is in itself additional evi- dence for their activity. Natural selection would remove p53 cDNAvs. Pseudogene Figure 4. Blast analysis of p53 cDNA and pseudogenes showing the loss of DNA binding domain (DBD) and nuclear localization signal (NLS) in the pseudogene.  A. S. Sreedhar / HEALTH 2 (2010) 1065-1071 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 1070 Table 1. Genome blast analysis showing the chromosome localization of cDNA and pseudogenes. S. No. Accession number chromosome E value % identity cDNA NW 047430.2 NW 001084694.1 14 0.0 84 NW 047390.2 NW 001084680.1 13 0.0 78 NW 047334.2 NW 0010884656.1 10 1e-140 100 NW 047813.2 NW 001084694.1 9 0.0 84 NW 047627.2 NW 001084680.1 2 0.0 87 pseudogene NW 047518.2 NW 001084740.1 18 0.0 99 NW 047430.2 NW 001083694.1 14 0.0 84 NW 047390.2 NW 001084680.1 13 3e-23 76 NW 047334.2 NW 001084656.1 10 1e-42 81 NW 47813.2 NW 001084880.1 9 5e-86 95 this type of DNA if it were useless, since DNA manu- factured by the cell is energetically costly. As the func- tion of more pseudogenes is being uncovered by testable and repeatable science, it is evident that these genetic elements, which are copiously spread in the genomes of different organisms, have been created with purpose. In addition, and in contrast to previously believed in- formation that pseudogenes are non functional copies of genes [21,22], growing evidence suggests that at least some pseudogenes are functional. It has been demon- strated that pseudogenes notably arise from seemingly absent or disabled promoters, premature stop codons, splicing errors, frameshift-causing deletions and inser- tions, etc., and do not necessarily abolish gene expres- sion [23,24]. McCarrey et al. [25] have suggested that pseudogenes can be functional in terms of the regulation of the expression of its paralogous genes, otherwise an- tisense to pseudogenes should not interfere with cellular functions. In support of this earlier we have used N- terminal siRNA to p53 and could inhibit its functions [10]. With respect to the evolution of regulatory func- tions of pseudogenes we must now conclude that tran- scribed pseudogenes are not necessarily without function. Indeed, they would appear to be especially suited to roles involving the antisense regulation of the active genes to which they are related [24]. In summary we report a processed pseudogene and additional transla- tional products for p53 in a rat histiocytoma that differ from the parental tumor and from the rat genome may have function roles upon stress and tumorigenesis. REFERENCES [1] Green, D.R. and Kroemer, G. (2009) Cytoplasmic func- tions of the tumour suppressor p53. Nature, 458(7242), 1127-1130. [2] Darzynkiewicz, Z. (1995) Apoptosis in antitumor strate- gies: modulation of cell cycle or differentiation. Journal of Cellular Biochemistry, 58(2), 151-159. [3] Halevy, O., Michalovitz, D. and Oren, M. (1990) Differ- ent tumor-derived p53 mutants exhibit distinct biological activities. Science, 250(4977), 113-116. [4] Michalovitz, D., Halevy, O. and Oren, M. (1991) P53 mutations—gains or losses. Journal of Cellular Bio- chemistry, 45(1), 22-29. [5] Levine, A.J. (1997) P53, the cellular gatekeeper of growth and division. Cell, 88(3), 323-331. [6] Weghorst, C.M., Buzard, G.S., Calvert, R.J., Hulla, J.E. and Rice, J.M. (1995) Cloning and sequence of a proc- essed p53 pseudogene from rat: A potential source of false ‘mutations’ in PCR fragments of tumor DNA. Gene, 166(2), 317-322. [7] de Fromentel, C.C. and Soussi, T. (1992) TP53 Tumor suppressor gene: A model for investigating human mutagenesis. Genes Chromosomes and Cancer, 4(1), 1- 15. [8] El-Deiry, W.S., Kern, S.E., Pietenpol, J.A., Kinzler, K.W. and Vogelstein, B. (2002) Definition of a consensus binding site for p53. Nature Genetics, 1(1), 45-49.  A. S. Sreedhar / HEALTH 2 (2010) 1065-1071 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 1071 [9] Blagosklonny, M.V. and El-Deiry, W.S. (1998) Acute overexpression of wt p53 facilitates anticancer drug- induced death of cancer and normal cells. International Journal of Cancer, 75(6), 933-940. [10] Sreedhar, A.S., Pardhasaradhi, B.V.V., Khar A. and Srini- vas, U.K. (2002) Effect of C-terminal deletion of P53 on heat induced CD95 expression and apoptosis in a rat histiocytoma. Oncogene, 21(25), 4042-4049. [11] Bennett, M., Macdonald, K., Chan, S.W., Luzio, J.P., Simari, R. and Weissberg, P. (1998) Cell surface traf- ficking of Fas: A rapid mechanism of p53-mediated apoptosis. Science, 282(5387), 290-293. [12] Lewin, B. (1990) Structural genes evolve in families. In: Genes IV, Oxford University Press, New York, 497-517. [13] Rowan, S., Ludwig, R.L., Haupt, Y., Bates, S., Lu, X., Oren, M. and Vousden, K.H. (1996) Specific loss of apoptotic but not cell-cycle arrest function in a human tumor derived p53 mutant. EMBO Journal, 15(4), 827- 838. [14] Ryan, K.M. and Vousden, K.H. (1998) Characterization of structural p53 mutants which show selective defects in apoptosis but not cell cycle arrest. Molecular Cell Biology, 18(7), 3692-3698. [15] Sreedhar, A.S., Pardhasaradhi, B.V.V., Khar, A. and Srinivas, U.K. (2000) Heat induced expression of CD95 and its correlation with the activation of apoptosis upon heat shock in rat histiocytic tumor cells. FEBS Letters, 472(2-3), 271-275. [16] Zhu, J., Zhou, W., Jiang, J. and Chen, X. (1998) Identi- fication of a novel p53 functional domain that is necessary for mediating apoptosis. Journal of Cellular Biochemistry, 273(21), 13030-13036. [17] Miyashita, T. and Reed, J.C. (1995) Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell, 80(2), 293-299. [18] Sreedhar, A.S., Pardhasaradhi, B.V.V., Khar, A. and Srinivas, U.K. (2002) A cross talk between cellular sig- nalling and cellular redox state during heat-induced apoptosis in a rat histiocytoma. Free Radical Biology and Medicine, 32(3), 221-227. [19] Harrison, P.M., Zheng, D., Zhang, Z., Carriero, N. and Gerstein, M. (2005) Transcribed processed pseudogenes in the human genome: An intermediate form of ex- pressed retrosequence lacking protein-coding ability. Nucleic Acids Research, 33(8), 2374-2383. [20] Zheng, D., Frankish, A., Baertsch, R., Kapranov, P., Rey- mond, A., Choo, S.W., Lu, Y., Denoeud, F., Antonarakis, S.E., Snyder, M., Ruan, Y., Wei, C.L., Gingeras, T.R., Guigó, R., Harrow, J. and Gerstein, M.B. (2007) Pseu- dogenes in the ENCODE regions: Consensus annotation, analysis of transcription, and evolution. Genome Research, 17(6), 839-8351. [21] Zhang, Z.D., Frankish, A., Hunt, T., Harrow, J. and Ger- stein, M. (2010) Identification and analysis of unitary pseudogenes: Historic and contemporary gene losses in humans and other primates. Genome Biology, 11(3), R26. [22] Torrents, D., Suyama, M., Zdobnov, E. and Bork, P. (2003) A genome-wide survey of human pseudogenes. Genome Research, 13(12), 2559-2567. [23] Woodmorappe, J. (2003) The potential immunological functions of pseudogenes and other ‘junk’ DNA. Journal of Creation, 17(3), 102-108. [24] Woodmorappe, J. (2003) Pseudogene function: Regulation of gene expression. Journal of Creation, 17(1), 47-52. |

