Paper Menu >>
Journal Menu >>
 Applied Mathematics, 2010, 1, 137-145 doi:10.4236/am.2010.13018 Published Online September 2010 (http://www.SciRP.org/journal/am) Copyright © 2010 SciRes. AM Some Models of Reproducing Graphs: I Pure Reproduction Richard Southwell, Chris Cannings School of Mathematics and Statistics, University of Sheffield, Sheffield , UK E-mail: bugzsouthwell@yahoo.com Received June 26, 2010; revised August 10, 2010; accepted August 15, 2010 Abstract Many real world networks change over time. This may arise due to individuals joining or leaving the net- work or due to links forming or being broken. These events may arise because of interactions between the vertices which occasion payoffs which subsequently determine the fate of the nodes, due to ageing or crowding, or perhaps due to isolation. Such phenomena result in a dynamical system which may lead to complex behaviours, to self-replication, to chaotic or regular patterns, to emergent phenomena from local interactions. They give insight to the nature of the real-world phenomena which the network, and its dynam- ics, may approximate. To a large extent the models considered here are motivated by biological and social phenomena, where the vertices may be genes, proteins, genomes or organisms, and the links interactions of various kinds. In this, the first paper of a series, we consider the dynamics of pure reproduction models where networks grow relentlessly in a deterministic way. Keywords: Reproduction, Graph, Network, Adaptive, Evolution 1. Introduction There has been much recent interest in the way in which networks such as the World Wide Web grow, and the structures which result from various rules by which new vertices are added and link to the existing vertices. One of the most studied is the so called Preferential Attach- ment model whereby a new node is added at each time tN (we use N and N for the non-negative inte- gers and positive integers respectively) and links to some set of existing vertices with probabilities which depend on the degrees of the latter. In the simplest case the pro- babilities are simply proportional to the degree, a model introduced by Yule [1], again by Simon [2], and then more recently by Barábasi and Albert [3]. The outcome of this process (see [4,5]) is a network in which the de- gree of a randomly selected node follows a power law distribution (i.e., if X is that degree then the probability Pr(=)= b X kak ), and the network is scale-free in the sense that (=)/ (=)=(=)/ (=) P r XlcPr XcPr XldPr Xd for all l, c and d. On the other hand there has been relatively little atten- tion paid to the growth of networks through the repro- duction of existing vertices and the generation of links between these new vertices and the old vertices, although, of course, the preferential attachment model where a new vertex is linked to an existing vertex could be regarded as the production of an offspring by the latter. This is clearly a situation which arises in a biological population which reproduces itself and in which we track related- ness. In a population which reproduces asexually, if we join each individual to its parent, then we simply produce a tree for each clone. More interestingly, if in a sexually reproducing population we join each individual to their two parents we obtain a genealogy (see [6] for alternate ways of representing this network). A further biological example happens when a genome duplication occurs [7]. The genes in the genome each code for some specific protein. If one considers the set of proteins of some organism as vertices in some network and joins any pair of vertices if the corresponding pair of proteins can bind then one obtains the protein-protein- interaction network. In a genome duplication every gene is essentially duplicated, so that there are now two copies producing the protein previously produced by one copy (we assume for simplicity that there is a simple one-one mapping of proteins to genes, ignore post-translational modification and other interactions, and splicing varia- tion). If we then distinguish between the two copies of the genes and the proteins produced by those two copies 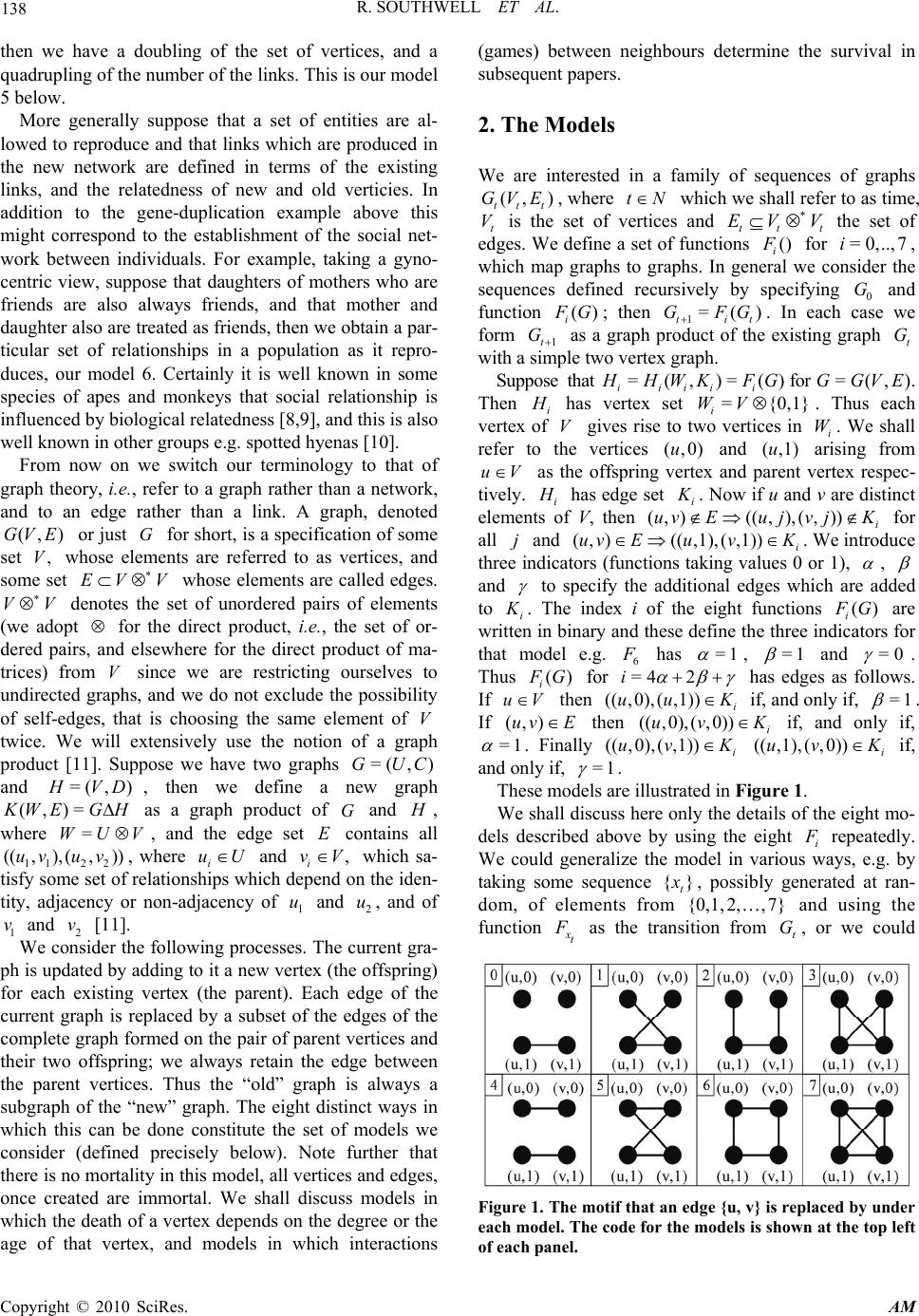 R. SOUTHWELL ET AL. Copyright © 2010 SciRes. AM 138 then we have a doubling of the set of vertices, and a quadrupling of the number of the links. This is our model 5 below. More generally suppose that a set of entities are al- lowed to reproduce and that links which are produced in the new network are defined in terms of the existing links, and the relatedness of new and old verticies. In addition to the gene-duplication example above this might correspond to the establishment of the social net- work between individuals. For example, taking a gyno- centric view, suppose that daughters of mothers who are friends are also always friends, and that mother and daughter also are treated as friends, then we obtain a par- ticular set of relationships in a population as it repro- duces, our model 6. Certainly it is well known in some species of apes and monkeys that social relationship is influenced by biological relatedness [8,9], and this is also well known in other groups e.g. spotted hyenas [10]. From now on we switch our terminology to that of graph theory, i.e., refer to a graph rather than a network, and to an edge rather than a link. A graph, denoted (, )GV E or just G for short, is a specification of some set ,V whose elements are referred to as vertices, and some set EV V whose elements are called edges. VV denotes the set of unordered pairs of elements (we adopt for the direct product, i.e., the set of or- dered pairs, and elsewhere for the direct product of ma- trices) from V since we are restricting ourselves to undirected graphs, and we do not exclude the possibility of self-edges, that is choosing the same element of V twice. We will extensively use the notion of a graph product [11]. Suppose we have two graphs =( , )GUC and =( ,) H VD , then we define a new graph (,)=KW EGH as a graph product of G and H , where =WUV, and the edge set E contains all 112 2 (( ,),(,))uvuv, where i uU and , i vV which sa- tisfy some set of relationships which depend on the iden- tity, adjacency or non-adjacency of 1 u and 2 u, and of 1 v and 2 v [11]. We consider the following processes. The current gra- ph is updated by adding to it a new vertex (the offspring) for each existing vertex (the parent). Each edge of the current graph is replaced by a subset of the edges of the complete graph formed on the pair of parent vertices and their two offspring; we always retain the edge between the parent vertices. Thus the “old” graph is always a subgraph of the “new” graph. The eight distinct ways in which this can be done constitute the set of models we consider (defined precisely below). Note further that there is no mortality in this model, all vertices and edges, once created are immortal. We shall discuss models in which the death of a vertex depends on the degree or the age of that vertex, and models in which interactions (games) between neighbours determine the survival in subsequent papers. 2. The Models We are interested in a family of sequences of graphs (, ) tt t GVE , where tN which we shall refer to as time, t V is the set of vertices and tt t EV V the set of edges. We define a set of functions () i F for =0,..,7i, which map graphs to graphs. In general we consider the sequences defined recursively by specifying 0 G and function () i F G; then 1=() tit GFG . In each case we form 1t G as a graph product of the existing graph t G with a simple two vertex graph. Suppose that =(,)=() iiiii H HWK FG for =(,)GGVE. Then i H has vertex set ={0,1} i WV. Thus each vertex of V gives rise to two vertices in i W. We shall refer to the vertices (,0)u and (,1)u arising from uV as the offspring vertex and parent vertex respec- tively. i H has edge set i K. Now if u and v are distinct elements of V, then (,)((,),(,)) i uvEu jv jK for all j and (,)((,1),(,1)) i uv EuvK . We introduce three indicators (functions taking values 0 or 1), , and to specify the additional edges which are added to i K. The index i of the eight functions () i F G are written in binary and these define the three indicators for that model e.g. 6 F has =1 , =1 and =0 . Thus () i F G for =4 2i has edges as follows. If uV then (( ,0),( ,1))i uu K if, and only if, =1 . If (,)uv E then ((,0),(,0))i uv K if, and only if, =1 . Finally ((,0),(,1))i uv K ((,1),( ,0))i uv K if, and only if, =1 . These models are illustrated in Figure 1. We shall discuss here only the details of the eight mo- dels described above by using the eight i F repeatedly. We could generalize the model in various ways, e.g. by taking some sequence {} t x , possibly generated at ran- dom, of elements from {0,1, 2,,7} and using the function x t F as the transition from t G, or we could Figure 1. The motif that an edge {u, v} is replaced by under each model. The code for the models is shown at the top left of each panel.  R. SOUTHWELL ET AL. Copyright © 2010 SciRes. AM 139 take the , and themselves to be probabilities that the corresponding edges are included at each stage. We shall derive results for our models for the number of vertices with degree d, the total number of edges, the chromatic number, diameter and average distance, and comment briefly on the automorphisms. A further paper will explore additional graph entities such as cliques, and cycles, the number of polygons of given size arising from a given polygon at the previous time step, and add further information regarding some of the entities ex- plored here. As stated above the operations introduced here are all equivalent to taking a graph product of T G with some simple graph Z. Table 1 specifies the products for each of the models. 3. Binary String Representation As mentioned above, a useful, alternative way to define the rules is in terms of the notion of parent and offspring vertices, and binary strings. If we begin with a graph 00 0 (, )GV E, then if 0 uV, this vertex gives rise to (,1)u and (,0)u in 1 G, which we write as u0 and u1, and to (( ,1),1)u, ((,1),0)u ((,0),1)u and (( ,0),0)u in 2 G, which are written in the obvious way as u11, u10, u01 and u00, and so on. In t G there will be 2t verti- ces arising from u. We denote these vertices as strings of length (1)t written u where runs over the bin- ary strings of length t. We refer to these representations as vertex strings. The eight models, all of which at time t have 0 ||2 t V vertex strings, give rise to distinct edge sets. We now specify precisely the edge set for each model at time t. Consider two vertices t ux and t vy (possibly identical) at time t, so t x and t y are two binary strings of length t, whose I’th elements are denoted by i t x and i t y. Now we define a third string t z, where the i'th element of t z, i t z , is determined by the pair (, ) ii tt x y. The purpose of t z is to specify the sequence of edges which need to be added to u and v in order to reach t ux and t vy . In specifying the models earlier we introduced , and , as indicators for the three distinct types of new edge. Here we identify the elements of t z with , , and two new terms * and . Thus if we have (, )=(0,0) ii tt xy , indicating that an edge must be placed between the offspring of the individuals 1t ux and 1t vy , then we record = i t z . Similarly we track the other edges, as is detailed below. Note for ease we in- troduce a corresponding to the choice (, )=(1,1) ii tt xy , and differentiate between (, )=(0,1) ii tt xy and (, ) ii tt x y =(1,0) by using and * respectively. When we use the t z’s to specify which edges exist in t G, we shall in fact take =1 always, and * = . Table 1. The products are denoted by a single letter K = Kronecker, C = Cartesian, H = Comb, S = Strong = AND, N = non-standard. Model α β γ Product Edges of Z 0 0 0 0 K {(1,1)} 1 0 0 1 K {(0,1), (1,1)} 2 0 1 0 H {(0,1)} 3 0 1 1 N N 4 1 0 0 K {(0,0), (1,1),} 5 1 0 1 K {(0,0), (0,1), (1,1)} 6 1 1 0 C {(0,1)} 7 1 1 1 S {(0,1)} If 1) (, )=(0,0) ii tt xy then = i t z , 2) (, )=(1,1) ii tt xy then = i t z , 3) (, )=(0,1) ii tt xy or (, )=(1,0) ii tt xy and 11 = tt ux vy then = i t z , 4) (, )=(0,1) ii tt xy and 11tt ux vy then = i t z , 5) (, )=(1,0) ii tt xy and 11tt ux vy then * = i t z . The string (,)t uvz specifies the start and the se- quence of operations which need to take place to pro- gress from (,)uv to (,) tt ux vy. As examples consider (A) vertices ( 0010101010)u and ( 0011001110)u, then * 10 ( , )=(( ,)),uvz uu and (B) ( 0011001011)u and (0011001011)v gives rise to ((, ),).uv Further note that t z can contain at most one , and then only if =uv . Now we assert that t ux and t vy are linked for a specific model if, and only if, each of the entries in t z (such as , , etc.) is equal to 1. If we start with =uv then we obtain sequences of the form *1 =((,)<, ><,,, >), ktk t zuu where the powers of the sets <> are the direct products. Note here that ’s in front of the must be taken as having value 1 in every model since they relate to the same ver- tex. If we start with uv then we obtain sequences * =((,)<,,,>) t t zuv . There is one additional complication in the case where we have self-edges in 0 G. We need to consider the am- biguities which may arise if =1 and =1 , since the former acting on a vertex, and the latter acting on a self edge at that vertex will result in the same edge. We can deal with this case efficiently by ensuring that any vertex with a self-edge at any time t is only subjected to one of the operators. For our examples above we have that (A) requires ===1 (recall =1 and * = always) so  R. SOUTHWELL ET AL. Copyright © 2010 SciRes. AM 140 there is an edge only in model 7, while (B) requires only that =1 and uv (note that in the absence of a , =uv would only lead to two copies of the same vertex) so there is an edge in models 4, 5, 6 and 7. 4. Homogeneity Definition. Merger of Two Graphs Given two graphs (,)GU E and (,) H VF then we define the merger of G and H as the graph (, ) J UVEF, and denote this by GH Theorem For each of the models specified above given some 00 0 (, )GV E then with 1=() tt GFG and writing in the obvious way 0 =() t t GFG we have that, 0 ,, t tuv E GFLuv where (,)Luv is the graph with vertices u and v, and one edge (,)uv . Proof This follows immediately from the definition of the functions () i F G. It is clear that for any (, )GV E we have ,, iii F GLuvF GFLuv for each of the possible cases 1) uV, vE and (,)uv E , 2) ,uV vV and (,)uvE, 3) uV and vV, and 4) uV and vV . Then by induction the result follows. In view of the theorem above much of the information is captured by considering the case where 0 G consists of a single edge. We consider how a single edge (and sometimes other equally simple structures) evolve under our models. 4.1. Models 0, 1, 4 & 5; Kronecker Products As Table 1 shows, four of the models use the Kronecker product, in fact those for which =0 . For such products, which we denote by , we have ()=AG H ()() A GAH, where () A G denotes the adjacency ma- trix of G, and also denotes the Kronecker product when applied to matrices. The adjacency matrices for the Z’s of models are 0, 1, 4 and 5, respectively, 00 01 , 01 11 , 10 01 and 11 11 . The t th Kronecker powers of these, 22 tt matrices, are easy to obtain. That for model 0 has a single 1 in the (2,2) tt position, and zeroes elsewhere, model 4 gives the identity matrix, model 5 gives a matrix of 1’s. Only model 1 has an interesting pattern, which is essentially the bitwise AND pattern exhibited in [12], and which is discussed below in the Section 8. 4.2. Model 2 Model 2 is particularly simple as we have a tree structure; we simply add a new branch at each vertex. Here we can capture all the structure by starting with 0 G as a single isolated vertex. There are various ways to describe the resulting tree, and these will be explored in more detail in a subsequent paper. For the moment we give only one such description. Starting from a single vertex labeled u, we obtain vertices u1 and u0 which are linked, then ver- tices u11, u10, u01 and u00. After t steps we have a tree with 21 t edges on the vertices of the cube of dimen- sion t. This tree is necessarily a spanning tree. As we make an extra step we take the current cube, with its spanning tree, and make a copy of the cube, join vertices of the 1t dimensional cube to the matching vertex in the copy. An alternate way of expressing this is to con- sider a t-dimensional cube with all edges present. Choose a particular coordinate and remove all the edges from the cube for which this coordinate is 0, then move to the cube which has this coordinate equal to 1, and within this cube repeat the process. At each stage one removes all the edges of a cube, whose dimension is one smaller than at the previous step. 4.3. Models 3, 6 and 7 Model 3 is by far the most complex and interesting of the models. We shall discuss several aspects of this model, along with the other models, but shall postpone a fuller discussion to subsequent papers. In model 6 the graph arising from a single edge after t steps is the (1)t -dimensional cube. In model 7 the graph arising from a single edge after t steps is the complete graph on 1 2t vertices. 5. Numbers of Vertices and Edges For a general 00 0 (, )GV E we have immediately that the number of vertices at time t is 0 ||*2 t V for all models. The number of edges on the other hand depends on the particular model, and can be relatively easily deduced from the t z possibilities and the etc. appropriate for each model. For example, for model 3, we have * ====1 , so for uv we have= t z(( , )uv * <,, >) t so there are clearly 3t edges for each (,)uv. We have *1 =,<,> <,,> ktk t z uu and this results in (32 ) tt edges for each u. The com- plete set of formulae are given in Table 2. 6. Chromatic Number A vertex colouring of a graph G is the assignment of a colour to each vertex in such a way that no adjacent ver- tices in G have the same colour. The minimal number of colours required to achieve this is the chromatic number,  R. SOUTHWELL ET AL. Copyright © 2010 SciRes. AM 141 Table 2. Formulae describing the number of edges after t time steps within the different models. Model Number of Edges 0 0 ||E 1 0 3|| tE 2 00 (21) |||| tVE 3 00 (32) ||3|| tt t VE 4 0 2| | tE 5 0 4|| tE 6 1 00 *2||2 || tt tV E 7 11 00 (2 * 42)||4|| tt t VE which we denote by ()G . A colouring which achieves this minimum will be refered to as a minimal colouring. With the exception of model 7, the chromatic number of the growing graphs are easy to obtain. 6.1. Models 0, 1, 4 and 5 Suppose that we have a minimal colouring for 0 G with 0 ()G colours. For models 0, 1, 4 and 5, each offspring can be given the same colour as its parent without vio- lating the condition that we have a colouring, and so the chromatic number remains equal to 0 ()G . 6.2. Models 2 and 6 For models 2 and 6, suppose we have a minimal colour- ing of 0 G using the set 01 ()1 0 {,, ,} G cc c of 0 ()G colours. Now suppose that the offspring of a vertex col- oured with i c is coloured (1)( ) 0 imodG c . Then, provided 0 ()>1G this will constitute a minimal colouring for 0 () i RG for =2i and for =6i. Thus 0 =2, t Gmax G . 6.3. Model 3 In this model 1 ()=()1 tt GG . This is because in any minimal, proper colouring of t G there will be an indi- vidual with ()1 t G distinct colours amongst its neighbours (actually there will be at least () t G such individuals, and since this individual's offspring is joined both to the individual and all its neighbours, this off- spring must be given a new colour. This colour can then be given to every offspring. 6.4. Model 7 This is by far the most difficult case, and will be treated more fully elsewhere. We observe only that ()1 ()2() ttt GGG . The first inequality follows since model 3 produces a subgraph of model 7 when they act on the same G. The latter inequality is evident since giving a minimal, proper colouring of t G using colours 01 ()1 0 {,, ,} G cc c we can colour the offspring of any vertex coloured i c with some i c , from a set ** * 01 ()1 0 {, ,,} G cc c of completely new colours. It is clear that if t G is a clique, or bipartite, then the chromatic number doubles, but this doubling is not general. For example the chromatic number of any polygonal graph Q of odd degree > 3 is 3, while 7 (())=5FQ . 7. The Distance Structure We now turn to the details of the distances between ver- tices. The distance between vertices u and v is denoted by (,)duv, the diameter of a graph g by ()G . For a graph t G we denote the numbers of pairs of vertices with distance x as () t x . For each models we shall de- rive the recursions for the distances through time. Mod- els 0 and 4 are excluded since they lead to disconnected components for which the notion of distance is inappro- priate. We also suppose that our initial graph is con- nected so that all subsequent graphs are. 7.1. Model 2 We begin with model 2 since this will allow an easy de- monstration of our methods. It is clear that 1 ()=( )2 tt GG . As in every model if t uV then (0,1)=1du u, while if (,) t uv E with (,)=duv d then (1,1)=du vd, (1, 0)=(1, 0)=1du vdu vd and (0,0)= 2du vd . We can then write 1(0) = 2(0) tt , 1(1)=(0)(1) ttt , 1(2) = 2(1)(2) ttt and 1()=(2)2 (1)() ttt t kkkk if 3k. This enables us to derive closed form expressions for the () ti , for example 0 (0) = 2(0) t t , 00 (1) =(1)(21)(0) t t , and 00 0 (2) =(2)2(1)2(21)(0) t ttt but the forms rapidly become somewhat unmanageable.  R. SOUTHWELL ET AL. Copyright © 2010 SciRes. AM 142 We can specify the total number of distances, for all models, 2 00 =(4||2 ||)/2 tt t LV V being simply the number of pairs of vertices plus the number of vertices, and being the same for all the models. The total of the distances ** 111 2 *21 121 00 0 =443 0 =420220 . ttt t tt tt LL L Lt The average distance * =/ ttt dLL and for t large this gives t dct where *2 00 0 =(2(0)) / ((0))cL . The average distance thus increases by 1 at each stage. We can derive this average result in another, more di- rect way. Suppose at some stage we have n vertices and average distance t . Now we add the extra offspring. The average distance is just the distance between a ran- domly selected pair of vertices. We consider the possible pairs in the new graph. There are (2 1)nn such pairs, n are of type (0,1)uu and contribute a distance 1. Of the remaining 2 2,n 1/4 are of type (1,1)uv , 1/2 are of type (1,0)uv , and 1/4 are of type (0,0)uv , which con- tribute (1,1),duv (1,1) 1du v and (1,1) 2du v re- spectively. The average over these 2 2n latter will thus be 1 t and overall we will therefore have 2 1 21 ==1221 21 t tt nn nn nn . A similar argument can be used to obtain an expres- sion for the variance of the distances which asymptoti- cally increases by 1/2 per time step. 7.2. Model 1 In order to derive the appropriate expressions for model 1 we need to track not only the distances but also the number of edges which belong to triangles. Accordingly we define set ()G of the edges which belong to a tri- angle (an edge (,)uvE belongs to a triangle if there exists some k such that (,)ik E and (, ) j kE). De- fine (1)=|()| tG and *(1)=(1)() ttt t . The necessity of considering triangles arises because the off- spring of two linked parents will be distance 3 apart if the parents’ link is not part of a triangle, but only 2 apart if there is a triangle since they are linked through the common neighbour of their parents. In detail we have For all uV we have (0,1)=2duu, and for (,)uvE, if (,)>1duv we have (0, 0)=(0,1)=(1,0)=(1,1)=(, )duvduvdu vdu vduv, if (,)( )uv G then (0,0)=2,du v(0,1)=du v (1, 0)=1du v, (0,1) ()uv G and (1, 0)()uv G , and if (,)\( )uv VG then (0,0)=3du v, (0,1)=du v (1, 0)=1du v, (0,1)()uv G and (1,0)()uv G . We have immediately that 1 ()= ((),3)= ((),3) tt Gmax Gmax G . From these expressions we obtain recursions for the ’s, as follows, 1 (0) =2(0) tt , 1 (1) = 3(1) tt ** 1 (1) = 3(1) tt 11 (2) =(0)(1)4(2) tt tt * 11 (3) =(1)4(3) tt t 1 ()=4 () tt kk if >3k. From the above recursions we can find simple closed form expressions for the ’s, as follows, 0 (0) =2(0) t t , 0 (1) = 3(1) t t ** 0 (1) = 3(1) t t 000 (2)=(42)(0)/ 2(43)(1)4(2) ttttt t * 00 (3) =(43)(1)4(3) tt t t 1 ()=4 () tt kk if >3k. The total distance ** * 000 0 = 4(43 )(2(1)(1))(42)(0) ttttt t LL , so that the average distance converges to a constant. The variance of the distances also converges to a constant. 7.3. Model 3 We obtain, in a straightforward manner that 1 (0) =2(0) tt , 11 (1) =(0)3(1) tt t , 11 (2) =(1)4(2) ttt , 1 ()=4 () tt kk for >2k. We have 1 ()= ((),2)=((),2) tt GmaxG maxG , and the total distance ** 00 00 0 =4((0) (1))3((0) (1)) tt t LL so that the average distance converges to a constant. The vari- ance of the distances also converges to a constant. 7.4. Model 5 We obtain 1 (0) =2(0) tt , 1 (1) =4(1) tt , 11 (2) =(0)4(2) ttt , 1 ()=4 () tt kk for >2k. We have ()=() t GG , and the total distance ** 00 0 =4((0)) 2(0) tt t LL so that the average dis- tance converges to a constant. The variance of the dis- tances also converges to a constant.  R. SOUTHWELL ET AL. Copyright © 2010 SciRes. AM 143 7.5. Model 6 We obtain 1 (0) = 2(0) tt , 11 (1) =(0)2(1) ttt , 11 ()=2(1) 2() tt t kk k for 2k. We have 1 ()=( )1 tt GG , and ** 111 =4 2(0) tt tt LL L , from which we can easily demonstrate that the average distance */ tt L L increases by 1/2 per time step for large t. The variance of the dis- tances increases by 1/4 per time step for large t. 7.6. Model 7 We obtain 1 (0) =2(0) tt , 11 (1) =(0)4(1) ttt , 1 ()=4 () tt kk for 2k. From this we have that the diameter is constant, and ** 00 =4(42)(0)/ 2 tt tt LL , so the average distance converges to a constant. The variance of the distances also converges to a constant. 8. Automorphism In models 0, 1, 4 and 5 we have Kronecker products and this allows us to determine the automorphisms on verti- ces in t G. In these models for a pair of vertices ur and vs , where u and v belong to 0 G, and r and s are binary strings, to be automorphic in t G, they must have u and v automorphic in 0 G and r and s to be automorphic in the Kronecker product of the appropriate . Z This is straightforward in each of the models. The most inter- esting case is model 1. For this the adjacency matrix is 01 =. 11 A The n th Kronecker product, k A , of this matrix have its rows and columns naturally indexed by the n th Kronecker products of vectors (0,1) and (0,1 )T, and will have a zero wherever the row index and column index have a 0 in the same position. It immediately fol- lows that the matrix k A is invariant under any permuta- tion to the elements of the row and column indices. Ac- cordingly the permutations induce an equivalence rela- tion over the binary strings; two strings being automor- phic if they contain the same number of 0’s. 9. Generating All Graphs Now [4] proved that for any graph (,) H UF of order n there exists a set W of size 2/4n such that one can associate distinct subsets i W with the n vertices, such that (, )ij F if, and only if, = ij WW . Consider the case where we evolve a single vertex, linked to itself, for t time steps under model 1. The vertex set of the re- sulting graph will be the set of t length binary strings. Suppose each string t x is associated with a set ()={| =0} i tt Sxi x. Then two vertices, t x and t y , are joined if, and only, () ()= tt Sx Sy . It follows that every graph with n vertices is isomorphic to some sub- graph of the t’th iterate when 2/4tn . Now as pointed out in [4] the bound is exact only when the graph H is bipartite with vertices partitioned equally, when n is even, and differing by 1 when n is odd. Thus we will observe many graphs will appear at earlier stages, for example the graph n K, the complete graph on n vertices, will appear at time =1tn, since we may take ={} i Wi. We shall investigate this phenome- non elsewhere. 10. The Degree Distribution Since we have a deterministic process the degrees of the vertices are well defined. We denote by (,)DGx the number of vertices of degree x in graph G, and refer to the degree of any vertex x as ()deg x. We shall refer to the degree distribution by which we mean the distribu- tion of the degree of a randomly chosen vertex. 10.1. Degree Distribution; Models 0, 1, 4, 5 We begin with the models which are Kronecker products. Given two graphs =( ,) J VE and =( , )KWF with yV with ()deg y and zW with ()deg z, then for (,) y zJK we have ((,))=()*degy zdegy()deg z. It follows that () ,=, , ji D JKx DJjDKij where ()={|,| }ijjNji , | j i having the usual meaning that j divides i. Now in these models we have 0 = tt GG Z, where t Z is the graph obtained by taking the graph Z (as described in Table 1) and taking the Kronecker product of it with itself t times. By knowing the degree distribu- tion of t Z one can easily determine the distribution starting from a generic initial graph. Under model 0, t Z has (2 1) t isolated vertices and a single vertex with degree 1, model 1 has tr C vertices with degree 2r, model 4 has every vertex with degree 1, while model 5 has every vertex with degree 2t. 10.2. Degree Distribution; Model 2 At each stage all the vertices of t G increase their de- 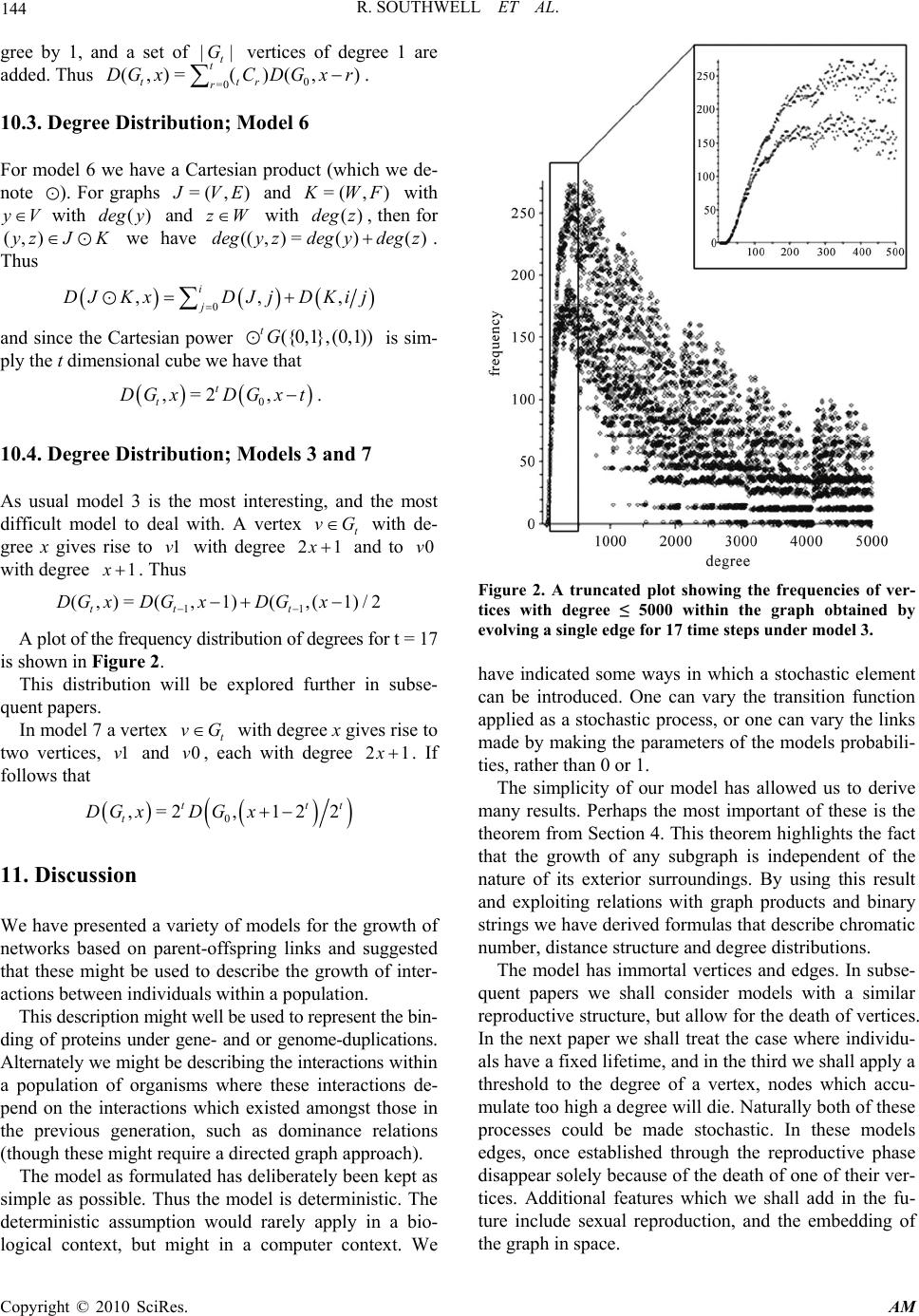 R. SOUTHWELL ET AL. Copyright © 2010 SciRes. AM 144 gree by 1, and a set of || t G vertices of degree 1 are added. Thus 0 =0 (,)=( )(,) t ttr r D GxCDGx r . 10.3. Degree Distribution; Model 6 For model 6 we have a Cartesian product (which we de- note ). For graphs =( ,) J VE and =( , )KWF with yV with ()deg y and zW with ()deg z, then for (,) y zJK we have ((,)=()()degy zdegydegz. Thus 0 ,,, i j DJ KxDJjDKij and since the Cartesian power ({0,1}, (0,1)) tG is sim- ply the t dimensional cube we have that 0 ,=2 , t t D GxDGx t. 10.4. Degree Distribution; Models 3 and 7 As usual model 3 is the most interesting, and the most difficult model to deal with. A vertex t vG with de- gree x gives rise to 1v with degree 21 x and to 0v with degree 1 x . Thus 11 (,)=(,1)(,(1)/2 tt t DG xDGxDGx A plot of the frequency distribution of degrees for t = 17 is shown in Figure 2. This distribution will be explored further in subse- quent papers. In model 7 a vertex t vG with degree x gives rise to two vertices, 1v and 0v, each with degree 21 x . If follows that 0 ,=2, 122 ttt t DG xDGx 11. Discussion We have presented a variety of models for the growth of networks based on parent-offspring links and suggested that these might be used to describe the growth of inter- actions between individuals within a population. This description might well be used to represent the bin- ding of proteins under gene- and or genome-duplications. Alternately we might be describing the interactions within a population of organisms where these interactions de- pend on the interactions which existed amongst those in the previous generation, such as dominance relations (though these might require a directed graph approach). The model as formulated has deliberately been kept as simple as possible. Thus the model is deterministic. The deterministic assumption would rarely apply in a bio- logical context, but might in a computer context. We Figure 2. A truncated plot showing the frequencies of ver- tices with degree ≤ 5000 within the graph obtained by evolving a single edge for 17 time steps under model 3. have indicated some ways in which a stochastic element can be introduced. One can vary the transition function applied as a stochastic process, or one can vary the links made by making the parameters of the models probabili- ties, rather than 0 or 1. The simplicity of our model has allowed us to derive many results. Perhaps the most important of these is the theorem from Section 4. This theorem highlights the fact that the growth of any subgraph is independent of the nature of its exterior surroundings. By using this result and exploiting relations with graph products and binary strings we have derived formulas that describe chromatic number, distance structure and degree distributions. The model has immortal vertices and edges. In subse- quent papers we shall consider models with a similar reproductive structure, but allow for the death of vertices. In the next paper we shall treat the case where individu- als have a fixed lifetime, and in the third we shall apply a threshold to the degree of a vertex, nodes which accu- mulate too high a degree will die. Naturally both of these processes could be made stochastic. In these models edges, once established through the reproductive phase disappear solely because of the death of one of their ver- tices. Additional features which we shall add in the fu- ture include sexual reproduction, and the embedding of the graph in space.  R. SOUTHWELL ET AL. Copyright © 2010 SciRes. AM 145 12. Acknowledgements The authors gratefully acknowledge support from the EPSRC (Grant EP/D003105/1) and helpful input from Dr Jonathan Jordan, and others in the Amorph Research Project (www.amorph.group.shef.ac.uk). 13. References [1] G. U. Yule, “A Mathematical Theory of Evolution, Based on the Conclusions of Dr. J. C. Willis, F.R.S.,” Philoso- phical Transactions of the Royal Society of London (Se- ries B), Vol. 213, 1925, pp. 21-87. [2] H. A. Simon, “On a Class of Skew Distribution Func- tions,” Biometrika, Vol. 42, No. 3-4, 1955, pp. 425-440. [3] A. L. Barab´asi and R. Albert, “Emergence of Scaling in Random Networks,” Science, Vol. 286, No. 5439, 1999, pp. 509-512. [4] B. Bollob’as, O. Riordan and G. Spenser, “The Degree Sequence of a Scale-Free Random Graph,” Random Structures and Algorithms, Vol. 18, No. 3, 2001, pp. 279- 290. [5] J. Jordan, “The Degree Sequences and Spectra of Scale-Free Random Graphs,” Random Structures and Algorithms, Vol. 29, No. 2, 2006, pp. 226-242. [6] C. Cannings and A. W. Thomas, “Handbook of Statistical Genetics,” John Wiley and Sons Ltd, New York, 2007. [7] J. S. Taylor and J. Raes, “Duplication and Divergence: The Evolution of New Genes and Old Ideas,” Annual Re- view of Genetics, Vol. 38, No. 1, 2004, pp. 615-643. [8] A. Widdig, P. Nurnberg, M. Krawczak, W. J. Streich and F. B. Bercovitch, “Paternal Relatedness and Age Prox- imity Regulate Social Relationships among Adult Female Rhesus Macaques,” Proceedings of the National Acad- emy of Science USA, Vol. 98, No. 24, 2001, pp. 13769- 13773. [9] G. Hausfater, “Dominance and Reproduction in Baboons (Papio Cynocephalus),” Contributions to Primatology, Vol. 7, 1975, pp. 1-150. [10] L. Frank, K. Holekamp and L. Smale, “Serengeti II: Dy- namics, Management, and Conservation of an Ecosys- tem,” University of Chicago Press, Chicago, 1995. [11] W. Imrich and S. Klavar, “Product Graphs: Structure and Recognition,” John Wiley and Sons Ltd, New York, 2000. [12] S. Wolfram, “A New Kind of Science,” Wolfram Media, Inc., Champaign, 2002. [13] P. Erd¨os, A. W. Goodman and L. Posa, “The Represen- tation of a Graph by Set Intersections,” Canadian Journal of Mathematics, Vol. 18, No. 1, 1966, pp. 106-112. |

