Health
Vol.5 No.10(2013), Article ID:38248,11 pages DOI:10.4236/health.2013.510232
GroESL protects superoxide dismutase (SOD)— Deficient cells against oxidative stress and is a chaperone for SOD
![]()
Department of Physiology and Biochemistry, University of Malta, Msida, Malta; *Corresponding Author: therese.hunter@um.edu.mt
Copyright © 2013 Gary J. Hunter, Thérèse Hunter. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 19 July 2013; revised 19 August 2013; accepted 19 September 2013
Keywords: Superoxide Dismutase; Chaperone; Heat Shock; Oxidative Stress
ABSTRACT
Superoxide dismutase (SOD)-deficient Escherichia coli OX326A cells are protected against chemically-induced oxidative stress by expression of the chaperonin GroESL. This protection is equivalent to expression of superoxide dismutase even though GroESL has no inherent SOD activity. Co-overexpression of GroESL and SOD in the same cells results in higher protein yields of SOD and greater metallation of SOD when compared with expression of SOD alone. Greater metallation results in the higher specific activity of SOD that is observed in heat shock, and is not due to increased synthesis of SOD mRNA or protein.
1. INTRODUCTION
Escherichia coli has two cytoplasmic superoxide dismutases (EC 1.15.1.1, SOD): an iron-containing SOD (FeSOD) encoded by the sodB gene [1-3] and a manganese-containing SOD (MnSOD) encoded by the sodA gene [4]. These two antioxidant enzymes have similar catalytic activity and protect the cell by removing superoxide anions 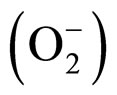 which are the toxic and highly reactive products of the univalent reduction of dioxygen [5]. The dismutation reaction involves an oxidation of superoxide to produce molecular oxygen and a reduction of superoxide to form hydrogen peroxide as a product. The enzyme’s active-site metal is consequently redox cycled between oxidation states during the dismutation. Despite their high degree of sequence (45% identity [6-8]) and structural (91% identity [8]) homology, FeSOD and MnSOD are active only when folded around their respective metal ion cofactor. This is in spite of the fact that both Fe and Mn ions may compete in vivo for the active site of either SOD [9,10]. The activity of SOD is therefore dependent upon the metal ion content of the cell. We show that for human MnSOD expressed in E. coli, activity increases with increasing manganese concentrations in the growth medium.
which are the toxic and highly reactive products of the univalent reduction of dioxygen [5]. The dismutation reaction involves an oxidation of superoxide to produce molecular oxygen and a reduction of superoxide to form hydrogen peroxide as a product. The enzyme’s active-site metal is consequently redox cycled between oxidation states during the dismutation. Despite their high degree of sequence (45% identity [6-8]) and structural (91% identity [8]) homology, FeSOD and MnSOD are active only when folded around their respective metal ion cofactor. This is in spite of the fact that both Fe and Mn ions may compete in vivo for the active site of either SOD [9,10]. The activity of SOD is therefore dependent upon the metal ion content of the cell. We show that for human MnSOD expressed in E. coli, activity increases with increasing manganese concentrations in the growth medium.
MnSOD associates with the E. coli nucleoid and preferentially protects DNA while FeSOD appears to protect cytoplasmic components [11,12]. The biosynthesis of MnSOD is affected by various stimuli [13-15] and the transcription of sodA is regulated by the protein products of six regulons [16]. Folding of nascent polypeptides and refolding of denatured proteins in the cytosol are facilitated by a group of specialised proteins, molecular chaperones [17,18]. In E. coli, GroESL is the only chaperone system essential for cell growth [19]. This chaperonin complex mediates the folding of a variety of protein substrates without being part of the final structure [20]. In vivo GroESL preferentially assists the folding of only approximately 10% of newly translated polypeptides. The typical GroESL substrate generally has a Mr of 20 kDa to 60 kDa and possesses several ab domains with buried hydrophobic residues [21]. In this present study, direct co-overexpression of GroESL with each of FeSOD and MnSOD has revealed differential effects on the in vivo folding of these homologous proteins.
Evidence suggests a relationship between the heat shock response and oxidative stress response. Elevated temperatures elicit oxidative stress and induce a heat shock response [22]. In E. coli, MnSOD provides protection against the superoxide radicals that are produced during aerobic heat shock [23]. However, there are conflicting reports on whether heat shock does induce the synthesis of MnSOD in E. coli. Privalle and Fridovich (1987) [24] previously showed that in E. coli a heat shock at 48˚C increased the SOD activity by a factor of two. Furthermore, these authors identified MnSOD as the responsible agent. Contrary to these results, Hassan and Lee (1989) [25] showed that the synthesis of FeSOD is increased and MnSOD decreased by temperature shift from sub-optimal (22˚C) to elevated (42˚C) temperatures. Although similar environmental stimuli may result in both a heat shock and an oxidative stress response, these two systems are in fact regulated independently. In E. coli there are two oxidative stress stimulons: the H2O2 induced regulon controlled by the oxyR gene product [15, 26] and the superoxide-inducible regulon controlled by the soxRS gene products [27-29]. The heat shock regulon is under the control of the rpoH gene product [30,31] while the synthesis of FeSOD reported at elevated temperatures was shown to be independent of the rpoH gene product [25]. The production of heat shock proteins GroEL and GroES does appear to be indirectly influenced by oxidative stress. Evidence shows that GroEL and GroES are synthesised in response to oxidative stress caused by the recycling redox agent paraquat and H2O2 [15,29,32]. We present evidence that GroESL can in itself protect SOD-deficient cells against oxidative stress even though it does not exhibit SOD activity. We also show that GroESL, when co-expressed with SOD, acts to increase the specific activity of the SOD through an increase in SOD metallation. Furthermore, during heat shock, the observed increase in MnSOD activity in E. coli is demonstrated to be a post-translational phenomenon and is not due to increased transcription or protein synthesis.
2. MATERIALS AND METHODS
2.1. Bacterial Strains and Vectors
E. coli K12 strain TG1 [sup E, hsd D5, thi, D (lac− proAB), F’(tra D36 pro A+B+lac Iq lac ZDM15)], was from Amersham Biosciences. E. coli OX326A (ΔsodA ΔsodB) and its parent strain MG1665 were kindly supplied by Prof. H. Steinman, Albert Einstein College of Medicine, New York, USA. The pREP4-groESL vector [33] encoding the GroEL and GroES operon was a gift from Dr. Stieger, Hoffman La Roche Ltd.
Oligonucleotides required for the modification of the vectors and the PCR of the superoxide dismutase genes were synthesized on an Applied Biosystems model 394 DNA synthesizer using cyanoethyl phosphoramidite chemistry and purified by preparative gel electrophoresis in 20% polyacrylamide gel containing 7 M urea. Oligonucleotides for Real Time PCR were purchased from MWG-Biotech (Table 1).
A tetracycline-resistant derivative of pREP4GroESL was prepared for this work. The tetracycline resistance gene was amplified by PCR of the plasmid pBR322

Table 1. Sequences of oligonucleotides synthesised and used in this study.
DNA (Roche Applied Science) with TET5’ and TET3’ primers (Table 1). In addition to the standard components as described in the Takara PCR kit (Takara Bio Inc, Japan), the reaction mixture (50 μl) contained 1 ng template and 130 ng of each primer. This was subjected to 98˚C for 20 s, 60˚C for 1 min and 68˚C for 2 min, for 30 cycles. Post-cycling addition of Klenow enzyme directly to the PCR tubes in situ and incubation for 30 min at 30˚C ensured completion of the reaction products and improved blunt-end cloning efficiency [34]. The DNA fragments were gel-purified from an agarose gel using microcon-100 centrifugal concentrators (Amicon). This PCR product (1391 bp) incorporating both the promoter and the stop codon of the tetracycline resistance gene was cloned into pREP4-groESL (SmaI). This vector was designated pTETGroESL (Figure 1). A derivative of this vector lacking the GroESL operon was constructed by digestion with NsiI, gel purification of the 5227 bp linear fragment and religation. This vector was designated pTET-1.
The His6-tag derivative pTH-1 (Figure 1), of the expression vector pTrc99A (Amersham Biosciences) was constructed for this work. Linker oligonucleotides (1 μg of each His6-tag A and His6-tag B, Table 1) were annealed by incubation at 85˚C for 10 min followed by slow cooling to 21˚C and then ligated with pTrc99A (BamHI/NcoI digested).

Figure 1. Plasmid maps of expression constructs used. (A) DNA and protein-coding sequences of the pTH-1 expression vector constructed in our laboratory showing the His6 tag immediately followed by a Factor Xa cleavage site. (B) The StuI site of pTH-1 was utilised to construct expression plasmids for E. coli FeSOD (pTH-FeSOD) or MnSOD (pTH-MnSOD) and human MnSOD (pTH-hMnSOD). (C) Plasmid map of the pTET-GroESL expression plasmid used to express GroES and GroEL.
2.2. Cloning and Protein Expression
SodA and SodB genes were isolated from E. coli genomic DNA by the polymerase chain reaction (PCR). PCR was performed following the protocol provided with the GeneAmp PCR reagent kit using AmpliTaq DNA polymerase (Perkin Elmer). In addition to standard components, each 20 μl reaction contained 100 ng of template, 100 ng of each primer (ECM-P5 and ECM-P3 for sodA, or ECF-5’ and ECF-3’ for sodB, Table 1) and 1.5 mM MgCl2.
Thermal cycling conditions were 94˚C for 3 min once followed by 94˚C for 30 s and 64˚C (for sodA or 56˚C for sodB ) for 30 s and 72˚C for 1 min applied for 30 cycles. The reactions were held at 4˚C until 1U Klenow polymerase was added and the reactions incubated at 30˚C for 30 min. The reaction mixtures were purified using microcon-100 centrifugal concentrators (Amicon), washed with water and ligated to StuI digested pTH-1 vector in the presence of 8U T4 DNA ligase. Positive clones in the correct orientation were designated pTHsodA and pTH-sodB (Figure 1). Mature human MnSOD (hMnSOD) was subcloned similarly using PCR and primers HMN-P3 and HMN-P5 (Table 1) with the same cycling conditions and the plasmid phmnsod4 (obtained from ATCC) as the template DNA. The human MnSOD expression plasmid was designated pTH-hMnSOD (Figure 1). Double-stranded DNA was sequenced in both directions utilizing dye terminator cycle sequencing chemistry and Thermus aquaticus DNA polymerase and an ABI 373A DNA sequencer (Applied Biosystems). The kanamycin-resistant OX326A competent cells were either transformed with pTH-sodA, pTH-sodB or pTHhMnSOD singly or co-transformed together with pTETGroESL DNA using the calcium chloride method [35]. Transformants were grown at 37˚C in 2xTY media (50 ml, 16 g/liter tryptone, 10 g/liter yeast extract and 5 g/liter NaCl), supplemented with the appropriate antibiotics, 50 μM MnSO4 and 50 μM FeSO4. When the cells reached an OD600 of 0.4, expression was induced by 1 mM IPTG. Growth was continued for 6 h when cells were harvested by centrifugation, resuspended in KP buffer (50 mM potassium phosphate, 1 mM EDTA, pH 7.8) and phenylmethylsulfonylfloride (PMSF, 0.1 mM), and lysed by passage through a French pressure cell (Amicon) at 16000 p.s.i. Insoluble material was removed by centrifugation at 10000 g for 10 min. Protein concentration was measured by the method of Bradford [36]. SDS-polyacrylamide gel electrophoresis (15%) was carried out after reduction with dithiothreitol according to the method of Laemmli [37].
2.3. Metal Chelation Affinity Chromatography (MCAC)
His6-FeSOD, His6-MnSOD and His6-hMnSOD were purified via metal chelate chromatography using HiTrap columns (Amersham Biosciences). Samples lysed using a French pressure cell (as above) in PBS buffer (30 ml, 20 mM sodium phosphate buffer pH 7.2 and 150 mM sodium chloride) containing 0.03% SDS and 1% Triton X-100, were centrifuged for 20 min at 10000 g and the supernatant then filtered through a 0.45 micron Nalgene syringe filter. The final sodium chloride concentration of the samples was adjusted to 0.5 M. Each Hi-Trap column was washed with 20 ml deionised water, charged with 10 ml 100 mM nickel sulphate and then washed with 10 ml Start buffer (20 mM potassium phosphate pH 7.8, 0.5 M sodium chloride). The protein samples (30 ml) were loaded onto the columns, which were then washed with 30 ml Start buffer followed by 15 ml 100 mM imidazole (in Start buffer). His6-SOD was eluted with Elution buffer (20 mM potassium phosphate pH 6, 1 M imidazole, 0.5 M sodium chloride). The collected samples (3 ml) were subjected to buffer exchange (potassium phosphate, 50 mM, pH 7.8) and concentration by centricon 10 (Amicon) centrifugal concentrators.
2.4. Measurements of Superoxide Dismutase Activity
Superoxide dismutase assays were carried out essentially as described by McCord and Fridovich [38] and Ysebaert-Vanneste and Vanneste [39] using cytochrome c as detector and xanthine-xanthine oxidase as superoxide generator. Native PAGE (8%) gels were stained for superoxide dismutase using nitro blue tetrazolium [40].
2.5. Estimation of Metal Content
Concentrations of iron and manganese in the superoxide dismutases were measured with a Hitachi Z-9000 atomic spectrophotometer (Showa Woman’s University, Japan) by Prof. F. Yamakura (Juntendo University School of Medicine, Japan) and Prof. T. Matsumoto (Showa Woman’s University, Japan) or using ICP-MS by ALS Scandanavia, AB, Sweden.
2.6. Paraquat-Induced Stress in Non-Transformed E. coli Cells
E. coli cells TGI (sod+) and OX326A (sod−) were grown at 37˚C (500 ml) to an OD600 of 0.6. Aliquots of each culture (50 ml in 250 ml flasks) were then incubated for a further two hours at 37˚C, 43˚C and 48˚C respectively. Similarly 50 ml cultures of each strain were grown in the presence of the redox cycling compound paraquat (0 µM, 250 µM and 500 µM). These cultures were grown solely at 37˚C or at 37˚C followed by a two-hour heat shock at 43˚C. Cells were harvested, resuspended in 3 ml PBS buffer and lysed by sonication.
2.7. Immunoblot Analysis
Total cellular protein (40 µg) was resolved by 15% SDS-PAGE. Cells were lysed by treatment with B-PER Reagent (Pierce) followed by brief sonication. Proteins were electroblotted onto Hybond C Extra nitrocellulose membrane (Amersham Biosciences) under semi-dry conditions using a LKB Multiphor II with a continuous transfer buffer system (Tris 48 mM/glycine 39 mM/SDS 0.0375% w/v/methanol 20% v/v) at 0.8 A/cm2 for 70 min. After transfer, the membrane was blocked by overnight room temperature incubation in 5% skimmed milk/PBST (80 mM Na2HPO4, 20 mM NaH2PO4, 100 mM NaCl and 0.1% Tween 20). Levels of GroEL were detected with an anti-GroEL antiserum (1:10,000 dilution) (kindly donated by Prof. B. Bukau, Universitat Heidelberg, Germany). This was bound to donkey anti-rabbit immunoglobulins G conjugated with horseradish peroxidase (Amersham Biosciences) (1:1000 dilution). Proteins were detected by enhanced chemiluminescence (ECL Western blotting system, Amersham Biosciences). Antibodies to FeSOD and MnSOD were kindly provided by Prof H. Steinman and Dr C. Privalle.
2.8. Paraquat-Induced Oxidative Stress in Transformed E. coli Cells
Overnight cultures (10 ml) of E. coli OX326A transformed with the appropriate plasmid were diluted 1:100 in 2TY medium to a final volume of 10 ml, grown with shaking for 2.5 hr and used as the starter culture to inoculate 50 ml of 2TY media to an OD600 of 0.03. This media was supplemented with the appropriate antibiotics, 500 μM paraquat, 0.1 mM IPTG, 50 µM FeSO4 and 50 µM MnSO4. Cultures were grown aerobically for 6 hr in 250 ml flasks at 37˚C. Aliquots (1 ml) were removed regularly to measure optical density (OD600).
2.9. Heat Shock
An E. coli MG1665 (sod+ parent of OX326A) overnight culture served as inoculant (1:100) for a 500 ml culture (2xTY). This was incubated at 37˚C until an OD600 of 0.4 was reached and then was divided into 50 ml aliquots in 250 ml flasks. Four 50 ml cultures were grown a further two hours under different conditions: 37˚C, 46˚C, 37˚C supplemented with chloramphenicol (150 µg/ml) and 46˚C with chloramphenicol. Cells were harvested by centrifugation and resuspended in DEPCtreated water to an OD600 of 1 and stored in aliquots at −80˚C.
2.10. Quantitation of Transcripts by Real-Time PCR
Total cellular RNA was isolated using the Ambion Totally RNATM kit. All samples were standardised before incorporation into RT-PCR experiments by both cell number and rRNA concentration. Quantitative RT-PCR was performed in a two-step reaction using the Eurogentec Reverse Transcription Core kit with random nonomers. These reactions were then performed on a Smart Cycler (Cepheid) using the qPCR Core Kit for Sybr green I (Eurogentec). A typical amplification protocol involved two stages, an initial 10 min denaturation step at 95˚C proceeded by 45 two-step cycles (95˚C for 15 s and 60˚C for 60 s with a 0.2˚/s ramp rate). The second step annealing/extension temperature was set at 60˚C for the sodB and GroEL template amplifications and 62˚C for sodA. Sybr green I fluorescence was measured during the extension phase of each cycle and analysed during the log-linear phase of the reaction. Primers for the amplification of cDNA were sodA-P and sodA-M (for sodA); sodB-P and sodB-M (for sodB); and GroEL-P and GroEL-M (for GroEL) (Table 1).
Amplification of a dilution series (100 pg, 50 pg, 20 pg, 1 pg, 0.1 pg) of the appropriate synthetic clones (pTH-FeSOD, pTH-MnSOD, pTETGroEL) generated the standard curves.
3. RESULTS
3.1. Co-Overexpression of GroESL Increases the Yield of Recombinant FeSOD and MnSOD
In order to observe any in vivo effect, the chaperone GroEL and its cofactor GroES may have on the solubility and specific activity of FeSOD and MnSOD, kanamycinresistant OX326A cells (ΔsodA ΔsodB) were co-transformed with pTET-GroESL and pTH-FeSOD or pTHMnSOD respectively. pTH-1 is a derivative of pTrc99A which is an expression vector incorporating a tac promoter. Our modification involved the insertion of a DNA sequence encoding a hexahistidine tag in-frame with a Factor Xa coding region and incorporating two cloning sites, BbrPI and StuI (Figure 1). The former may be used to bluntend clone protein-encoding sequences to which a simple hexahistidine tag will be attached. The latter site can be used to similarly clone protein-encoding sequences should it be required to remove the hexahistidine tag from the expressed protein. A recognition sequence for the restriction protease Factor Xa has been incorporated so that cleavage occurs directly before the first amino acid of the cloned protein. In each case, translation begins in-frame at the very first nucleotide of the cloned sequence. The product of Factor Xa-cleavage will produce an authentic protein product with no spurious amino acids at the N-terminus due to cloning. As both pTET-GroESL and pTH-1 plasmids have compatible origins of replication and carry different antibiotic resistance genes, they were maintained stably in the co-transformed OX323A cells.
His6-FeSOD and His6-MnSOD proteins were expressed to high levels in SOD-deficient cells with or without co-expression of GroES and GroEL (Figure 2A). The SODs were purified utilising the hexahistidne tags encoded at their N-termini by metal chelation chromatography (Figure 2B). Overall yields were approx. 5 mg protein from a 50 ml culture. High levels of overexpressed recombinant proteins have a tendency of forming inclusion bodies or aggregates in the cytosol [41]. Consistently in our studies we found the overall yield of each SOD to be increased by at least 30% when overexpressed in the presence of GroESL (Figure 2B). This is presumed to be due to an increase in solubility for each SOD.
3.2. Co-Overexpression of GroESL Increases the Specific Activity of SOD
The activity of the purified His6-SODs was examined by staining native-PAGE gels for superoxide dismutase activity (Figure 2C). We found that the activity of the
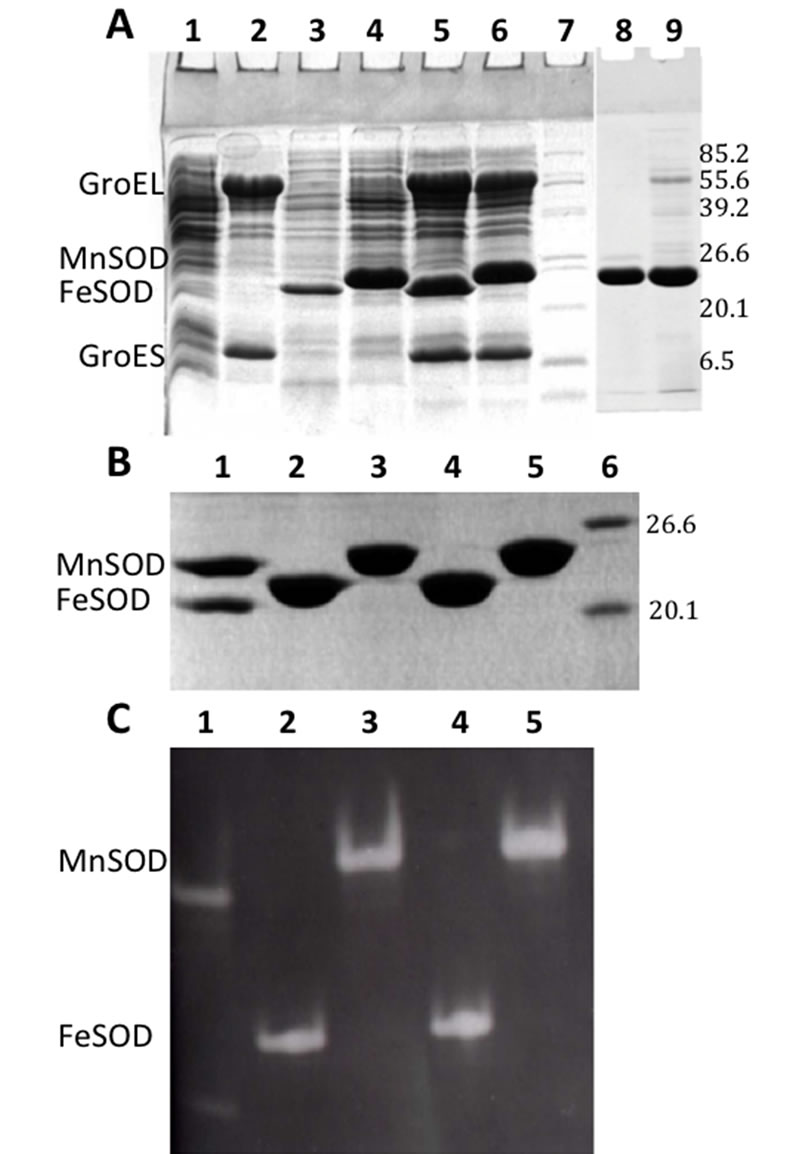
Figure 2. Polyacrylamide gel electrophoresis of proteins. (A) SDS-PAGE (15%) of expressed proteins in cultures of E. coli OX326A cells (20 µg each). Cells harbouring pTET-GroESL, pTH-FeSOD or pTHMnSOD plasmids overexpressed proteins with the expected molecular weights for GroEL and GroES (57.3 and 10.4 KDa respectively, lane 2), His6-FeSOD (22.6 KDa, lane 3) and His6-MnSOD (24.5 KDa, lane 4) respectively. The positions of bands corresponding to GroEL, GroES, FeSOD and MnSOD are shown to the left of the figure. Coexpression of GroEL and GroES together with either His6-FeSOD (lane 5) or His6- MnSOD (lane 6) similarly produced proteins of the expected size. Molecular weight markers (2 µg each) were loaded in lane 7 and their sizes are shown (kDa) to the right of the figure. Control cell lysate harbouring no plasmid was run in lane 1. His6- hMnSOD purified from expression plasmid pTH-hMnSOD without and with co-expression with GroESL are shown in lanes 8 and 9 respectively (taken from a separate gel). (B) SDS_PAGE (15%) of proteins purified by metal chelate chromatography (20 µg each). (His)6FeSOD was purified after expression alone (lane 2) or with GroESL (lane 4). His6-MnSOD was purified after expression alone (lane 3) or with GroESL (lane 5). FeSOD and MnSOD proteins purchased from Sigma were loaded in lane 1 (5 µg each) with positions indicated to the left of the figure, and molecular weight size markers (2 µg each) in lane 6 with sizes indicated to the right of the figure. (C) Native PAGE (8%) stained for SOD activity. The lanes are labelled similarly to (B) and contained 5 µg protein each.
His6-MnSOD purified from cultures in which GroESL had also been induced, was consistently higher than that from cultures in which His6-MnSOD had been expressed alone. Laser densitometry demonstrated an increase in the achromatic band of approx. 60% (results not shown).
In contrast, however, we observed no equivalent difference in the activity of His6-FeSOD. This result was confirmed by spectrophotometric SOD assay (Table 2). Although the specific activity of His6-FeSOD remained unaffected, His6-MnSOD specific activity increased by up to 80%. Metal content analysis showed that this increase in specific activity observed for His6-MnSOD was accompanied by an equivalent increase in manganese content of the enzyme (Table 2).
We conclude therefore that when metal content is less than optimal in MnSOD, the presence of GroESL during expression increases the efficiency of metal capture by the folding protein.
3.3. GroESL Overexpression Protects SOD-Deficient Cells from Oxidative Stress
Under conditions of paraquat-induced oxidative stress, SOD-deficient cells grow poorly, at a reduced rate (Figure 3). This reduced rate of growth can be overcome by the presence of SOD. We examined the effect of expressing His6-FeSOD and His6-MnSOD in the presence or absence of excess GroESL. Both His6-FeSOD and His6-MnSOD increased the growth rates of cells under these conditions, as expected (Figure 3). Consistently, however, His6-MnSOD expression under the conditions used, failed to exhibit any lag phase prior to exponential growth. Cells overexpressing His6-MnSOD therefore grew much faster at an earlier stage than His6-FeSODcontaining cells, and cell densities achieved after 8 hours were significantly higher. Surprisingly, the presence of GroESL overexpressed simultaneously with His6- MnSOD, introduced a lag phase, though cell densities achieved were comparable with His6-MnSOD expressed alone. Rates of growth under these conditions were the highest we observed. Also remarkable is the effect of GroESL co-expression on His6-FeSOD-expressing cells. Growth rates increased during the latter stages and resulted in these cells, too, reaching higher densities when compared to cells expressing the His6-FeSOD alone (Figure 3). This is probably due to the increased amounts of soluble SOD in these cells. Unexpectedly, we found that the expression of GroESL alone could protect oxidatively-stressed cells to an extent comparable with His6- FeSOD expression alone. That this was not due to the presence of the vector in the GroESL plasmid used was demonstrated in cells transformed with pTET-1 (Figure 3). Cell growth in this case was seen to be equivalent to the SOD-deficient cells without plasmid.
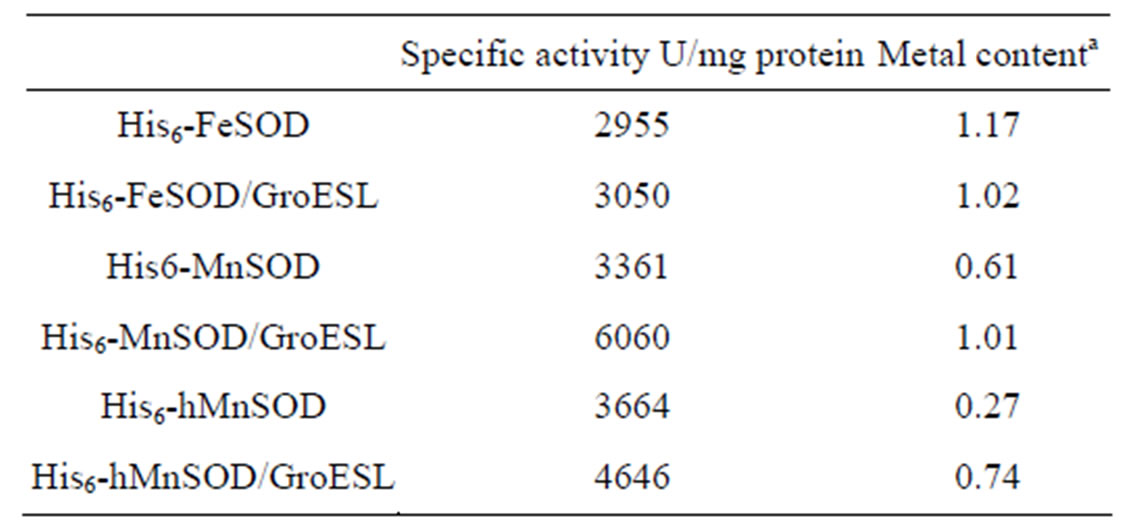
Table 2. Specific activities and metal content of purified SOD proteins.
ag atom/monomer.

Figure 3. Growth curves of E. coli OX326A cells after induction of protein production under conditions of oxidative stress. Paraquat (500 µM) was used to induce oxidative stress and gene expression was initiated using IPTG (0.1 mM). Circles represent growth observed in cultures expressing His6-MnSOD (filled circles coexpressed with GroESL), squares represent growth observed in cultures expressing His6-FeSOD (filled squares coexpressed with GroESL). Open triangles and crosses depict growth of OX326A cells and cells harbouring pTET-1 plasmid respectively. Closed triangles show growth of cells expressing GroESL alone.
3.4. Expression of GroESL Is Increased in SOD-Deficient Cells and during Oxidative Stress
We compared the expression of GroEL in wild-type E. coli (TG1) and in SOD-deficient E. coli cells (OX326A) at 37˚C, 43˚C and 48˚C. The areas of the chemiluminescent signals were scanned by laser densitometry. GroEL was observed to be present in SOD-deficient cells to a higher level than in wild-type (Figure 4A). In both cell types, GroEL was, as expected, inducible by heat.
In each experiment the amount of GroEL synthesised was greater in the cells deficient of cytoplasmic SOD. The amount of GroEL produced in the SOD-deficient cells is consistently higher even under conditions of heat shock at 43˚C and 48˚C although at these higher temperatures the difference between the amount of GroEL in the TG1 and OX326A cell extracts was smaller. Laser densitometry revealed a 3 fold difference between the two strains at 37˚C, a 1.5 fold difference at 43˚C and at 48˚C. When the cells were subjected to paraquat-induced oxidative stress (Figure 4B), an increase in the production of GroEL was observed with increasing doses of paraquat.
3.5. Heat Shock Affects MnSOD Activity and Stability
Extracts from cells exposed to increased temperatures and/or chloramphenicol were analysed for SOD concentration by immunoblotting a 15% SDS-PAGE gel and for SOD activity on an activity-stained 8% native PAGE gel. Laser densitometry of the immunoblot revealed very small increases (30%) in concentrations of both FeSOD and MnSOD when the temperature was raised from 37˚C to 46˚C (Figure 5A). However, we observed a reduction in the activity of FeSOD (by 30%) during heat shock (46˚C, Figure 5B) whereas the activity of MnSOD increased by approx. 80%. Analysis of the chloramphenical-treated heat-shocked samples demonstrated that there was a large reduction in the FeSOD activity but a substantial increase (63%) in the MnSOD activity (Figure 5B). This was accompanied by a small increase in MnSOD (of 30%) and a substantial reduction in FeSOD (of 66%) concentrations (Figure 5A). The former is presumably due to the increase in solubility because protein synthesis is inhibited by chloramphenicol in these samples. Our results have indicated negligible differences between SOD protein concentrations under heat shock and suggest therefore that GroESL probably plays an important role in maintaining the levels of MnSOD during heat shock, by protecting the protein from degradation and ensuring maximum activity of the existing protein.
3.6. Transcription of SOD Is Unaffected by Heat Shock
We have utilised real-time RT-PCR to examine the transcription of FeSOD, MnSOD and GroEL genes under normal and heat-shock conditions (Figure 6). FeSOD demonstrated a small (14%) increase in mRNA sysnthesis with heat shock, while MnSOD mRNA levels increased by 30%. These differences are supported to some extent by minor changes observed in protein concentrations (Figure 5A). We observed a 17-fold increase for GroEL under the same conditions, as expected (Figure 6). We also show that under heat shock in the presence of chloramphenicol, the levels of transcription of all three genes examined is greatly reduced (Figure 6). GroEL
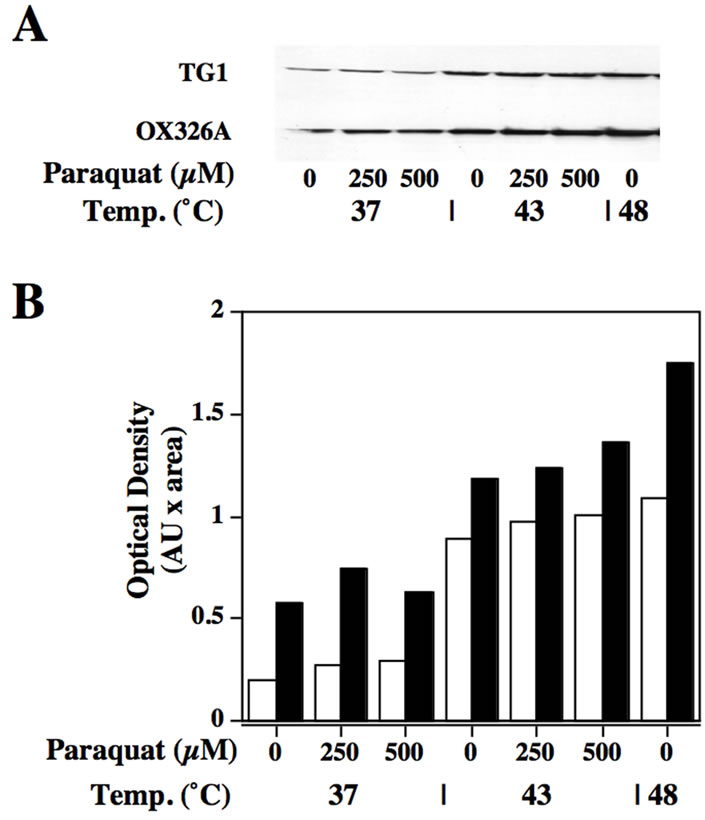
Figure 4. GroEL specific immunoblot of cell samples under heat or oxidative stress. (A) Immunoblot of samples from wild-type (TG1) or SOD deficient (OX326A) cells incubated at 37˚C, 43˚C or 48˚C in the presence or absence of paraquat (PQ) at 250 or 500 µM as indicated in the figure. (B) Histogram of the densities observed using laser densitometry of the immunoblot shown in (A).

Figure 5. Protein concentration and activity of SOD samples from cells (E. coli MG1665) under stress. (A) Immunoblot using two SOD specific antibodies (MnSOD and FeSOD). Positions of the SODs are indicated to the left of the figure. A mixture of FeSOD and MnSOD (0.1 µg each) purchased from Sigma is shown in lane 1, cells grown at 37˚C were used to prepare the sample shown in lane 2, and cells grown at 46˚C were used to prepare the sample in lane 3. Cells under heat stress (46˚C) were also grown in the presence of chloramphenicol (lane 4). (B) A native PAGE stained for SOD activity. Samples were the same as described in (A).
transcription is reduced to a level comparable with that
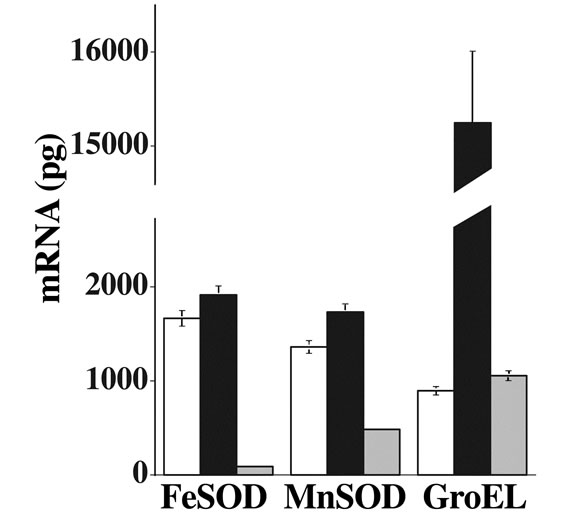
Figure 6. Transcription of mRNA by real-time RT-PCR. Specific primer pairs were used to examine the transcription of FeSOD, MnSOD or GroEL in cultures (E. coli MG1665) grown at 37˚C (open bars), 46˚C (closed bars) or at 46˚C in the presence of chloramphenicol (shaded bars).
observed at 37˚C but the expression levels of the SODs are greatly reduced. FeSOD transcription was reduced to 0.05% of the level observed at 37˚C, and MnSOD transcription reduced to 35%. We measured SOD activity in the same samples used for experiments represented in Figures 5 and 6. At 37˚C, total SOD was found to be 30.6 U/mg. protein, which increased under heat stress to 67.5 U/mg. protein, a 2.2 fold increase. We conclude that this observed increase in activity is not due to transcriptional activation of the SOD genes, nor to an increase in the SOD protein concentration. It is due largely to the increased stability and activity of MnSOD compared with FeSOD under heat shock conditions, or in the presence of GroESL complex.
3.7. Manganese Content of Culture Media Determines the Activity of Human MnSOD Expressed in E. coli
We cultured E. coli OX326A cells harbouring the human MnSOD expression plasmid pTH-hMnSOD with or without pTET-GroESL in minimal M9 media containing various concentrations of MnSO4 from 0 to 10 µM at 30˚C. Protein expression was induced by the addition of 1 mM IPTG when the cell suspension reached an OD600 of 0.3. Levels of protein expression were very high and subsequent purification by MCAC resulted in a single protein band (Figure 2A). After harvesting cells were adjusted for cell number and equal aliquots lysed by sonication and applied to a native PAGE gel stained for SOD activity (Figure 7).
The activity of hMnSOD demonstrated a clear increase with manganese concentration present in the media and was also consistently higher overall when coexpressed with GroESL. Purified protein was analysed for metal content and demonstrated that the higher SOD

Figure 7. SOD activity of his6- hMnSOD from cells cultured in M9 minimal media in the absence (top) and presence (bottom) of co-expressed GroESL, with different amounts of manganese in the growth medium. Lane 1, 0 µM MnSO4, Lane 2, 0.1 µM MnSO4, Lane 3, 1 µM MnSO4, Lane 4, 10 µM MnSO4. Achromatic bands were subjected to densitometry and values normalised to 0 µM MnSO4 (lane 1) and values presented on the figure above each band.
activity from hMnSOD obtained from cells co-expressing GroESL was due to increased metallation of the SOD (Table 2).
4. DISCUSSION
In order to survive, all cells must be capable of responding to changes in their environment. The superoxide dismutases (SODs) are the cell’s first line of defence against the onslaught of reactive oxygen species produced under conditions of oxidative stress. There is evidence that aerobic heat shock also produces conditions of oxidative stress [23] and we have investigated whether SODs are influenced by heat shock. In order to do so, we have utilised real time RT-PCR to examine mRNA synthesis, immunoblotting to reveal changes in protein concentration and examined SOD activity (by both zymography and spectrophotometry) to assess the amount of functionally-folded enzyme within a range of experimental conditions.
Our results unequivocally show that there is only a small induction of expression of FeSOD and MnSOD by heat shock (Figure 6). Furthermore, the concentration of each protein increases by less than 30% during the heat shock response (Figure 5), and actually reflects the small variation observed in expression, in agreement with the observations on protein synthesis during heat shock previously reported [42]. This is in sharp contrast to the two-fold increase in SOD activity observed and previously reported in both E. coli [24,25] and Halobacteriumhalobium [43]. We propose, therefore that the increase in SOD activity during heat shock must be a posttranslational event, and provide evidence that this involves the participation of the chaperonins, GroEL and GroES. The effect of chloramphenicol on SOD activity during heat shock supports this hypothesis (Figures 5, 6), as SOD activity remains high even in the presence of the protein synthesis inhibitor. We also provide evidence that this is due to the activity of MnSOD alone. In support of these findings, we show that co-overexpression of either SOD with the chaperonins affects only MnSOD, which is to a degree comparable to that observed during the heat shock response. Examination of enzyme activity in vitro has revealed that FeSOD is more heat-stable than MnSOD at temperatures close to those required to induce the heat shock response [8,25]. We propose that this situation is, apparently paradoxically, reversed in vivo due to the participation of the chaperonins. The fact that under heat shock and in the absence of protein synthesis, FeSOD activity drops to below detectable levels, while MnSOD activity remains high is strong evidence for a stabilising factor for MnSOD (Figure 5). The presence of GroESL increases the yield of soluble FeSOD and MnSOD by only 30%, which cannot account for our observed and the previously reported increase in activity (Figure 2 and Table 2). Examination of the metal content of purified SODs has also revealed a possible mechanism for the action of GroESL on MnSOD. When GroESL is co-overexpressed with MnSOD, the metal content of the enzyme is increased in direct proportion to the observed activity. Thus we conclude that GroESLchaperonins help MnSOD to fold with optimal manganese content. That this effect was not observed with FeSOD may be accounted for by the fact that FeSOD seemed to contain the maximum amount of metal under all experimental conditions (Table 2). Growth of E. coli is virtually impossible when iron is omitted from the medium, whereas growth continues in the near absence of manganese (results not shown). However, GroESL clearly does not protect FeSOD in vivo under heat shock conditions in the presence of chloramphenicol, in the same way we have observed for MnSOD (above and Figure 5).
The mononuclear SODs are two-domain proteins in which the metal ion is secured by two ligands contributed by each domain [6,7]. It may easily be envisaged that for metal release or metal capture, only the domains need to move apart, and an intramolecular movement may be invoked by rotation about any one of several peptide bonds (unpublished observations). If only partially folded, the domain interface may be exposed to solvent. As this interface is composed largely of hydrophobic amino acid residues, it may be proposed that the action of the chaperonins may be affected via this exposed interface, as the GroESL complex has been shown to interact preferentially with hydrophobic regions of misfolded proteins [44].
Interestingly, we have found that expression of GroES and GroEL can complement SOD-deficient cells under conditions of oxidative stress (Figure 3). Surprisingly this observation was supported by the finding that SODdeficient cells were expressing more endogenous GroEL than the equivalent wild-type (Figure 4). The level of superoxide in aerobically grown SOD-deficient cells is far greater than in wild type cells. GroEL expression is induced further by paraquat (used in vivo to produce conditions of oxidative stress) as well as temperature in both cell types (Figure 4). Since the SOD-deficient cells do not readily convert superoxide to hydrogen peroxide, this effect may be due to a superoxide response in the cell. The chaperonin appears to be capable of protecting the cells against superoxide without having inherent superoxide dismutase activity (unpublished results) and without being able to repair oxidatively damaged proteins. GroEL (or its homologues) has been shown to be highly susceptible to damage caused by oxidative stress [45,46]. Susceptibility to and protection from oxidative stress could be part of the same process. In the percolator hypothesis of chaperonin action [47], it is estimated that between 3,000 and 3,500, water molecules could fit within the empty cavity of the chaperonin (the so-called Anfinsen cage). Even in the presence of a target protein, between 500 and 2,000 water molecules may move in and out of the cavity during each chaperonin cycle. It is not difficult to imagine superoxide or other reactive oxygen species being taken up during this process, and once inside, would oxidatively attack the residues within the cavity, producing both the damage reported, and effectively removing the superoxide to protect the cell.
The observed involvement of GroESL with MnSOD activity in E. coli presented here is also of particular relevance to eukaryotic cells, where MnSOD is a protein targeted to mitochondria. Mitochondria obtain their own chaperonins using similar import mechanisms, and contain the hsp60 and hsp10 homologues of GroEL and GroES respectively. During import, MnSOD must presumably be subject to fold (or refolding) inside the mitochondrial matrix, where manganese ions will have been sequestered and then transferred by a manganese carrier protein [48]. The role of chaperonins in the increased metal binding capacity of MnSOD is therefore of interest in this context, and we have evidence which shows that human MnSOD activity is also increased when co-expressed with GroESL in E. coli. Although MnSOD had not previously been identified as a substrate for GroESL [21], we demonstrate that in the presence of GroESL, the metallation, activity and stability of MnSOD increases. Further investigation is currently underway to discover the role and interaction of chaperonins and SODs in mitochondria.
5. ACKNOWLEDGEMENTS
We would like to thank Prof. F. Yamakura and T. Matsumoto for metal analyses, Prof. H. Steinman for the gift of E. coli strains OX326A and MG16655, Dr. C. Privalle for antibodies to SOD, Prof. B. Bukau for antibodies to GroEL and Dr. M. Stieger for the original pREP4- GroESL plasmid. This work was supported by research grants from the University of Malta.
REFERENCES
- Bull, C. and Fee, J.A. (1985) Steady-state kinetic studies of superoxide dismutases: Properties of the iron containing protein from Escherichia coli. Journal of the American Chemical Society, 107, 3295-3304. http://dx.doi.org/10.1021/ja00297a040
- Sakamoto, H. and Touati, D. (1984) Cloning of the iron superoxide dismutase gene (sodB) in Escherichia coli K-12. Journal of Bacteriology, 159, 418-420.
- Carlioz, A., Ludwig, M.L., Stallings, W.C., Fee, J.A., Steinman, H. M. and Touati, D. (1988) Iron Superoxide Dismutase: Nucleotide Sequence of the gene from Escherichia coli K12 and correlations with crystal structures. The Journal of Biological Chemistry, 263, 1555-1562.
- Touati, D. (1983) Cloning and mapping of the manganese superoxide dismutase gene (sodA) of Escherichia coli K-12. Journal of Bacteriology, 155, 1078-1087.
- McCord, J.M. and Fridovich, I. (1969) Superoxide dismutase: An enzymic function for erythrocuprein (Hemocuprein). The Journal of Biological Chemistry, 244, 6049-6055.
- Edwards, R.A., Baker, H.M., Jameson, G.B., Whittaker, M.M., Whittaker, J.W. and Baker, E.N. (1998) Crystal structure of Escherichia coli manganese superoxide dismutase at 2.1Å resolution. Journal of Biological Inorganic Chemistry, 3, 161-171. http://dx.doi.org/10.1007/s007750050217
- Lah, M.S., Dixon, M.M., Pattridge, K.A., Stallings, W.C., Fee, J. A. and Ludwig, M. L. (1995) Structure-function in Escherichia coli iron superoxide dismutase: Comparisons with the manganese enzyme from Thermus thermophilus. Biochemistry, 34, 1646-1660. http://dx.doi.org/10.1021/bi00005a021
- Hunter, T., Bannister, J.V. and Hunter, G.J. (2002) Thermostability of manganeseand iron-superoxide dismutases from Escherichia coli is determined by the characteristic position of a glutamine residue. European Journal of Biochemistry, 269, 5137-5148. http://dx.doi.org/10.1046/j.1432-1033.2002.03200.x
- Ose, D.E. and Fridovich, I. (1979) Manganese-containing superoxide dismutase from Escherichia coli: Reversible resolution and metal replacements. Archives of Biochemistry and Biophysics, 194, 360-364. http://dx.doi.org/10.1016/0003-9861(79)90628-3
- Beyer, W.F. and Fridovich, I. (1991) In vivo competition between iron and manganese for the occupancy of the active site region of the manganese-superoxide dismutase of Escherichia coli. The Journal of Biological Chemistry, 266, 303-308.
- Steinman, H.M., Weinstein, L. and Brenowitz, M. (1994) The manganese superoxide dismutase of Escherichia coli K-12 associates with DNA. The Journal of Biological Chemistry, 269, 28629-28634.
- Hopkin, K.A., Papazian, M.A. and Steinman, H.M. (1992) Functional differences between manganese and iron superoxide dismutases in Escherichia coli K-12. The Journal of Biological Chemistry, 267, 24253-24258.
- Hassan, H.M. and Fridovich, I. (1977) Regulation of the synthesis of superoxide dismutase in Escherichia coli. The Journal of Biological Chemistry, 252, 7667-7672.
- Hassan, H.M. and Fridovich, I. (1977) Enzymatic defenses against the toxicity of oxygen and of streptonigrin. Journal of Bacteriology, 129, 1574.
- Greenberg, J.T. and Demple, B. (1989) A global response induced in Escherichia coli by redox-cycling agents overlaps with that induced by peroxide stress. Journal of Bacteriology, 171, 3933-3939.
- Compan, I. and Touati, D. (1993) Interaction of Six Global Transcriptional Regulators in Expression of Manganese Dismutase in Escherichia coli K-12. Journal of Bacteriology, 175, 1687-1696.
- Ellis, R.J. and Hartl, F.U. (1996) Protein folding in the cell: Competing models of chaperonin function. The FASEB Journal, 10, 20-26.
- Hartl, F. U. (1996) Molecular chaperones in cellular protein folding, Nature, 381, 571-579. http://dx.doi.org/10.1038/381571a0
- Fayet, O., Ziegelhoffer, T. and Georgopoulos, C. (1989) The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. Journal of Bacteriology, 171, 1379-1385.
- Sigler, P.B., Xu, Z., Rye, H.S., Burston, S.G., Fenton, W. A. and Horwich, A. L. (1998) Structure and function in GroEL-mediated protein folding. Annual Review of Biochemistry, 67, 581-608. http://dx.doi.org/10.1146/annurev.biochem.67.1.581
- Houry, W.A., Frishman, D., Eckerskorn, C., Lottspeich, F. and Hartl, F.U. (1999) Identification of in vivo substrates of the chaperonin GroEL. Nature, 402, 147-154. http://dx.doi.org/10.1038/45977
- Lee, P.C., Bochner, B.R. and Ames, B.N. (1983) AppppA, heat-shock stress, and cell oxidation. Proceedings of the National Academy of Sciences USA, 80, 7496-7500. http://dx.doi.org/10.1073/pnas.80.24.7496
- Benov, L. and Fridovich, I. (1995) Superoxide dismutase protects against aerobic heat shock in Escherichia coli, Journal of Bacteriology, 177, 3344-3346.
- Privalle, C.T. and Fridovich, I. (1987) Induction of superoxide dismutase in Escherichia coli by heat shock, Proceedings of the National Academy of Sciences USA, 84, 2723-2726. http://dx.doi.org/10.1073/pnas.84.9.2723
- Hassan, H.M. and Lee, F.J. (1989) Effect of temperature and htpR on the biosynthesis of superoxide dismutase in Escherichia coli. FEMS Microbiology Letters, 58, 133- 137. http://dx.doi.org/10.1111/j.1574-6968.1989.tb03033.x
- Christman, M.F., Morgan, R.W., Jacobson, F.S. and Ames, B.N. (1985) Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell, 41, 753-762. http://dx.doi.org/10.1016/S0092-8674(85)80056-8
- Demple, B. and Amabile-Cuevas, C.F. (1991) Redox redux: The control of oxidative stress responses. Cell, 67, 837-839. http://dx.doi.org/10.1016/0092-8674(91)90355-3
- Tsaneva, I.R. and Weiss, B. (1990) soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. Journal of Bacteriology, 172, 4197-205.
- Walkup, L.K. and Kogoma, T. (1989) Escherichia coli proteins inducible by oxidative stress mediated by the superoxide radical. Journal of Bacteriology, 171, 1476- 1484.
- Yamamori, T. and Yura, T. (1982) Genetic control of heat-shock protein synthesis and its bearing on growth and thermal resistance in Escherichia coli K-12. Proceedings of the National Academy of Sciences USA, 79, 860-864. http://dx.doi.org/10.1073/pnas.79.3.860
- Grossman, A.D., Erickson, J.W. and Gross, C.A. (1984) The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell, 38, 383-390. http://dx.doi.org/10.1016/0092-8674(84)90493-8
- VanBogelen, R.A., Kelley, P.M. and Neidhardt, F.C. (1987) Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. Journal of Bacteriology, 169, 26-32.
- Amrein, K.E., Takacs, B., Stieger, M., Molnos, J., Flint, N. A. and Burn, P. (1995) Purification and characterization of recombinant human p50csk protein-tyrosine kinase from an Escherichia coli expression system overproduceing the bacterial chaperones GroES and GroEL. Proceedings of the National Academy of Sciences USA, 92, 1048-1052. http://dx.doi.org/10.1073/pnas.92.4.1048
- Hunter, T. and Hunter, G.J. (1998) GST fusion protein expression vector for in-frame cloning and sitedirected mutagenesis. BioTechniques, 24, 194-196.
- Sambrook, J., Fritsch, E.F. and Maniatis, T. (1989) Molecular cloning: A laboratory manual. 2nd Edition, Cold Spring Harbor Laboratory, Cold Spring Harbour, NY.
- Bradford, M.M. (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilising the principle of protein binding. Analytical Biochemistry, 72, 248-254. http://dx.doi.org/10.1016/0003-2697(76)90527-3
- Laemmli, U. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680-685. http://dx.doi.org/10.1038/227680a0
- McCord, J.M. and Fridovich, I. (1969) Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). The Journal of Biological Chemistry, 244, 6049-6055.
- Ysebaert-Vanneste, M. and Vanneste, W.H. (1980) Quantitative resolution of Cu, Znand Mn-superoxide dismutase activities. Analytical Biochemistry, 107, 86-95. http://dx.doi.org/10.1016/0003-2697(80)90496-0
- Beauchamp, C. and Fridovich, I. (1971) Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Analytical Biochemistry, 44, 276-287. http://dx.doi.org/10.1016/0003-2697(71)90370-8
- Clark, E.D.B. (1998) Refolding of recombinant proteins. Current Opinion in Biotechnology, 9, 157-163. http://dx.doi.org/10.1016/S0958-1669(98)80109-2
- Farewell, A. and Neidhardt, F.C. (1998) Effect of temperature on in vivo protein synthetic capacity in Escherichia coli. Journal of Bacteriology, 180, 4704-4710.
- Begonia, G.B. and Salin, M.L. (1991) Elevation of superoxide dismutase in Halobacterium halobium by heat shock. Journal of Bacteriology, 173, 5582-5584.
- Lilie, H. and Buchner, J. (1995) Interaction of GroEL with a highly structured folding intermediate: Iterative binding cycles do not involve unfolding. Proceedings of the National Academy of Sciences of the United States of America, 92, 8100-8104. http://dx.doi.org/10.1073/pnas.92.18.8100
- Reverter-Branchat, G., Cabiscol, E., Tamarit, J. and Ros, J. (2004) Oxidative damage to specific proteins in replicative and chronological-aged Saccharomyces cerevisiae: Common targets and prevention by calorie restriction. The Journal of Biological Chemistry, 279, 31983-31989. http://dx.doi.org/10.1074/jbc.M404849200
- Cabiscol, E., Piulats, E., Echave, P., Herrero, E. and Ros, J. (2000) Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. The Journal of Biological Chemistry, 275, 27393-37398.
- Csermely, P. (1999) Chaperone-percolator model: A possible molecular mechanism of Anfinsen-cage-type chaperones. Bioessays, 21, 959-965. http://dx.doi.org/10.1002/(SICI)1521-1878(199911)21:11<959::AID-BIES8>3.0.CO;2-1
- Luk, E., Carroll, M., Baker, M. and Culotta, V. C. (2003) Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a member of the mitochondrial carrier family. Proceedings of the National Academy of Sciences of the United States of America, 100, 10353-10357. http://dx.doi.org/10.1073/pnas.1632471100

