Advances in Microbiology
Vol. 2 No. 4 (2012) , Article ID: 25959 , 5 pages DOI:10.4236/aim.2012.24077
AtzABC Catabolic Gene Probe from Novel Atrazine-Degrading Rhodococcus Strain Isolated from a Nigerian Agricultural Soil
1Biological Sciences Programme, Abubakar Tafawa Balewa University, Bauchi, Nigeria
2Queens University Environmental and Genomic Research (QUESTOR) Center, Queens University of Belfast, Belfast, Northern Ireland, UK
3Microbiology Department, Usmanu Dan Fodio University, Sokoto, Nigeria
Email: *likitacool@yahoo.co.uk
Received September 18, 2012; revised October 16, 2012; accepted October 26, 2012
Keywords: Enrichment; Catabolic Genes; Bioremediation; Atrazine
ABSTRACT
A batch enrichment technique was used to isolate atrazine-degrading Rhodococcus sp strain from an agricultural land with history of atrazine application in Bauchi state, Northeastern Nigeria. The strain was identified on the basis of physiological, biochemical and 16S rRNA gene sequencing. Growth studies and HPLC analysis showed that the strain has potential of atrazine degradation. An investigation into the catabolic genes Atz ABC, which transform atrazine to cyanuric acid, confirms the chromosomal DNA of strain to harbor BC genes, as compared with the positive control, Rhodococcus jostii RHA1. The strain does not possess the Atz A in all catabolic gene probe carried out. The isolation and characterization of the Rhodococcus sp strain showed that catabolic genes may have evolved from a single origin with widespread global distribution, with possible potential in atrazine bioremediation.
1. Introduction
Atrazine, an herbicide used for controlling grassy weeds in maize (Zea mays) cultivation in the world, Nigeria inclusive, is a source of concern due to its hazardous nature. It has been banned in many European countries, but in Nigeria it is still in demand, due to cheap price and availability in all agrochemical dealership all over the country. This has prompted scientist to seek for measures to decontaminate environments polluted with recalcitrant xenobiotics through bioremediation options [1]. Majority of these chemical pollutants can be degraded by microorganisms, often by enzymes that degrade these compounds. The widely-studied Pseudomonas strain ADP has been thoroughly evaluated to contain catabolic genes that encodes degradation of atrazine through Atz ABC [2]. The genes Atz A, B and C, which encodes these enzymes, have been cloned and sequenced. Atrazine chlorohydrolase (Atz A), Hydroxyatrazine ethylaminohydrolase (AtzB) and N-isopropylammelide isopropylaminohydrolase (Atz C) sequencially convert atrazine to cyanuric acid [3]. Further degradation of cyanuric acid have been achieved by many soil bacteria and fungi [4], to carbon dioxide and ammonia [5]. Based on this knowledge, several attempts have been to determine catabolic genes in many atrazine degrading bacteria [6-8]. The pathway for atrazine catabolism to cyanuric acid in Pseudomonas sp strain ADP [3] is as Figure 1.
Comparison of enzyme and gene structure of novel strain in various geographical areas will help in the better understanding of gene transfer and evolution. In this work, a novel atrazine-degrading strain of Rhodococcus sp was isolated from a Nigerian Agricultural soil and subjected to catabolic gene analysis, to further ascertain global gene distribution and conservation.
2. Materials and Methods
Sampling sites: An agricultural soil with history of atrazine application from a Maize (Zea mays) cultivated land in Dass Local Government Area, Bauchi state, Nigeria, was collected. The land has been annually treated with commercial atrazine for more than five years to control grassy weeds. Soil samples were collected on weekly basis for 5 weeks in the first phase and randomly in later stages of investigation. 100 gram each of soil sample was collected from the center and perimeter, randomly in
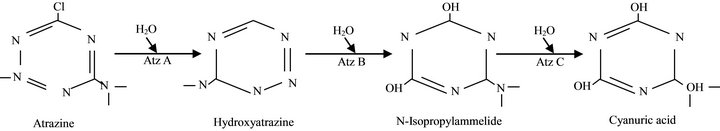
Figure 1. Pathway for Atrazine catabolism to cyanuric acid in Pseudomonas sp strain ADP.
June, 2011, from the upper 10 cm (top soil) [9]. The samples were made in composite, homogenized and stored at 4˚C until use.
2.1. Enrichment and Isolation of Atrazine-Degrading Rhodococcus sp
Batch enrichment technique was employed in a mineral medium, designated M9-Atz. The medium contains Na2HPO4 (6 g·L–1), KH2PO4 (3 g·L–1), Nacl (0.5 g·L–1), NH4Cl (1 g·L–1), adjusting the pH to 7.4, 1 M MgSO4 (2 mL·L–1), 1 m Cacl2 (1 mL·L–1), distilled H2O (1 L), Agar (15 g·L–1), M9 medium, with atrazine as sole carbon source in concentration 1.25 mg/2.5 mg (M9-Atz medium). A modified Mendalbaum et al. [10] method was adopted to isolate Rhodococcus degrading strain. 1 gram of soil sample was added to M9 medium and vortexed for 1 minute. 1 mL each of slurry was added to 250 mL of Erlenmeyer flask in replicates and supplemented with atrazine (1.25/2.5 mg). Another set-up contains only M9 medium, as control. All enrichment flasks were kept in shaker incubator at 30˚C under shaking at 150 rev·min–1 in the dark to preclude photolysis. After a period of two and four weeks, 0.1 mL were plated on M9-Atz agar and M9-agar, as control. All sets of plates were inoculated in triplicates, after enrichment cycles. A few morphologically distinct colonies on plates were further screened by plating on M9-Atz agar and M9 agar (control). The plates were kept at 25˚C for further analysis.
2.2. Growth and Degradation Studies
Freshly inoculated washed cells in M9-Atz medium were monitored using 6705UV-Vis spectrophotometer to determine cell growth, at interval of 0 hour to 140 hours. All experiment was carried out in triplicate and average (mean) used to plot cell growth versus time (hr) [8]. Heat-killed strain was used as control [10]. HPLC analysis was conducted using water-HPLC autosampler 600 model equipped with 996 photodiode array detector, with a mobile phase of acetonitrile/water (50:50) ratio at a flow rate of 1.0 mL mm, UV 226 nm and standard column. The samples from growth media were centrifuged and ultra filtered, and subjected to HPLC analysis. The parent atrazine was analyzed at every 10 minutes to check for periodical change in chromatographic peak.
2.3. Genetic Characterization of Rhodococcus sp Strain
DNA was extracted using GenElute bacterial genomic DNA kit. Fully grown, discrete colony on Luria-Bertani (LB) agar was picked and inoculated into LB broth. Overnight bacterial culture, with OD600nm:0.5 was used for DNA extraction. PCR amplification of 16S r RNA genes was carried out using 63 F and 1387 R primers. PCR reaction on DNA sample were carried out using a standard set-up of 18.1 µL of distilled water, 2.5 µL of buffer, 1 µL of forward and reverse primers, 0.4 µL Taq polymerase and 1 µL of DNA template per reaction. Each reaction setting was mixed in 0.2ml (25 µL) PCR tube. The amplification was performed with ESCOMAXI swift thermo cycler PCR machine. The program used was 1 cycle of 95˚C for 5 minutes, 32 cycles of 95˚C for 45 sec, 55˚C for 45 sec, and 72˚C for 2 minutes, followed by final extension of 72˚C for 10 minutes. Amplification products were visualized after separation by electrophoresis in a 1% agarose gel and stained with 0.5% ethidium bromide. Results were visualized using UV transilluminator versadoc imager. Purified PCR products were sent for sequencing at Dundee sequencing and services, MRC/PPU, College of life sciences, University of Dundee, Scotland, DD 1 5EU in collaboration with Queens University Belfast, QUESTOR center, Northern Ireland, UK. Sequenced results were compared with NCBI database using BLAST nucleotide search (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.4. Catabolic Gene Analysis
The Atz ABC catabolic genes were determined using primers designed by Sigma-Aldrich, 2011, batch No. 981804499 in the strain DNA (Table 1).
3. Results and Discussion
The identity of the strain was determined by 16S rRNA gene sequencing analysis showed that the strain has maximum homology with Rhodococcus opacus B4 (97%), with both forward and reverse nucleotide sequence analysis. Any > 0.50 is considered an acceptable match [10] (Table 2). The mineralization studies of the Rhodococcus sp showed that the strain grows well in
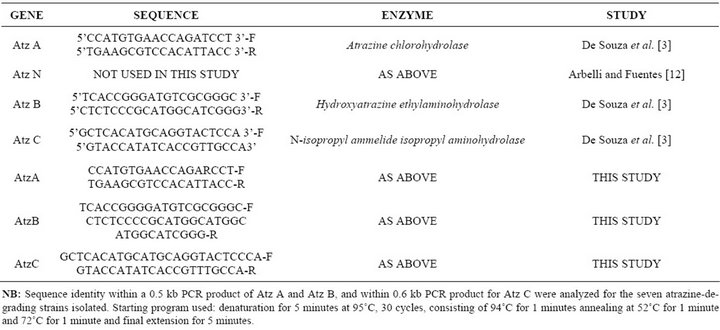
Table 1. Sequence comparison of isofunctional bacterial enzyme that catabolize atrazine.
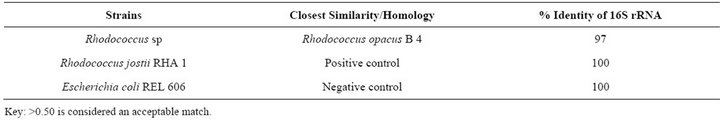
Table 2. 16S RNA comparison of Rhodococcus sp strain with NCBI database.
atrazine media (M9-Atz) (Figure 2). Likewise, HPLC analysis from sample collected at 0 hour time, as compared to 24 hour, showed slight reduction in chromatographic peak in parent compound after 10 minutes retention time. The degradation intermediates of atrazine were not investigated. Uninoculated sample was used as control in HPLC analysis, while heat killed strains were used in the growth studies (Figure 3).
This preliminary knowledge provided the impetus to investigate the presence of homologous gene in Rhodococcus strain isolated (Figures 4 and 5). As a negative control, bacteria known not to degrade atrazine was used, Rhodococcus jostii RHA1 was used as a positive strain. Specific primers Atz ABC designed by Sigma-Aldrich, 2011, were used for catabolic gene probe.
The biodegradation of atrazine generally involves a series of hydrolytic reaction catalyzed by enzymes the amidohydrases super family. Four enzymatic steps, first identified in Pseudomonas sp ADP are dehalogenation, deamination of the two alkylamino side chains and ring cleavage of cyanuric acid, a central metabolite in the degradation of all s-triazine [11,12]. The genes Atz ABC that encodes the first three steps leading to the production of cyanuric acid was investigated in the Rhodococcus sp
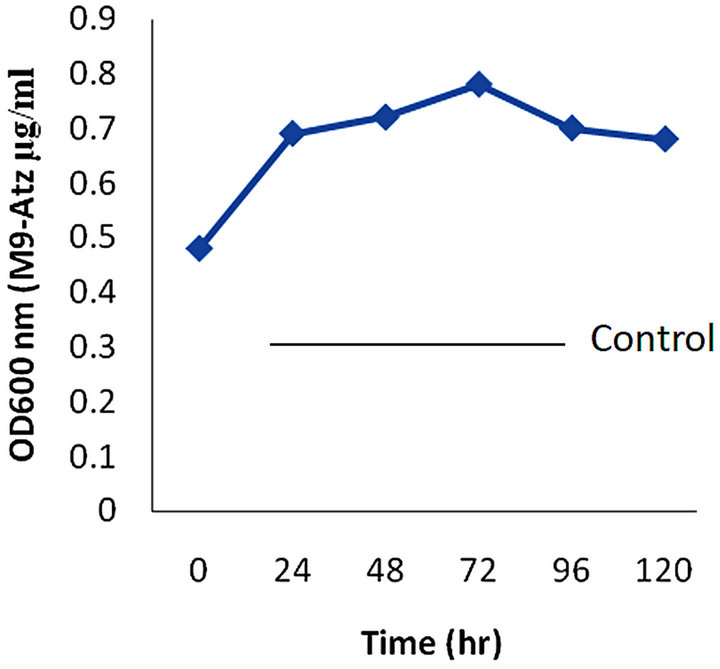
Figure 2. Cell growth determination by Rhodococcus sp strain on M9-Atz media.
strain isolated. Several researches have established that the self transmissible genes are widely distributed in other atrazine-degrading bacteria [3]. Also, it has been confirmed that not all atrazine-degrading bacteria contain full complements of Atz ABC. Some, as seen in this study may contain Atz N, which substitutes of Atz A [13]. The strain contains BC catabolic genes, but not the Atz A gene, at various optimization of annealing temperature.

Figure 3. Atrazine metabolism determination by Rhodococcus sp strain in M9-Atz media by HPLC analysis.
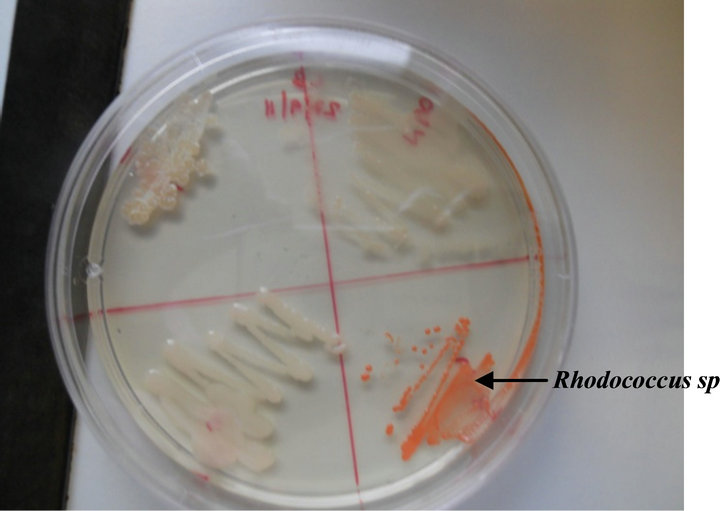
Figure 4. Rhodococcus sp strain growth pattern on LB medium.
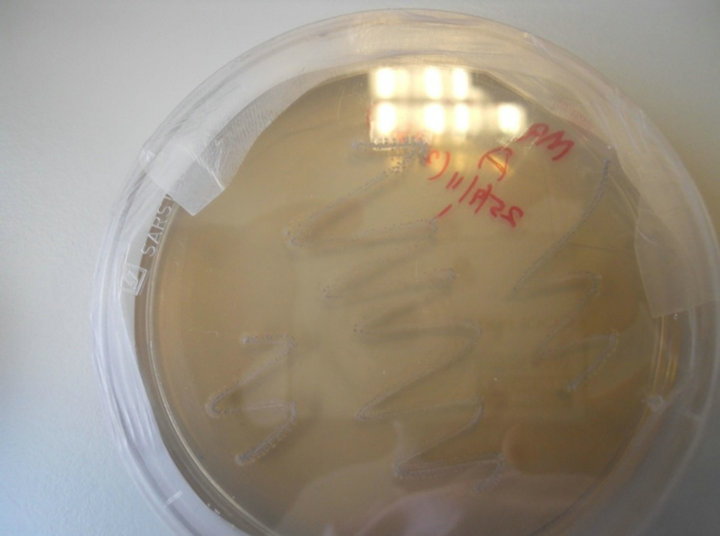
Figure 5. Rhodococcus sp strain growth pattern on M9-Atz medium
The lack of using Atz N catabolic gene probe altered conclusive assumption on the pathway of the atrazinedegrading strain isolated.
Several studies have linked the genes to be plasmid encoded [8]. This is to be investigated in further characterization of the strain. The control, Rhodococcus jostii RHA1, provided strong and informative probes of 500 bp and 610 bp for Atz B (Figure 6) and Atz C (Figure 7) respectively in putative strain isolated. The Atz A was absence in both positive control and strains (Table 2) investigated, but maybe likely that they possess Atz N as initially asserted. The presence of Atz BC catabolic gene is a strong indication of versatility, conservativeness and global distribution of atrazine-degrading, even if they belong to novel species. A putative Atrazine degradation pathway can be generated for the Rhodococcus sp strain investigated, as below (Figure 8).
4. Acknowledgements
This work was fully supported by the Education Trust Fund (ETF) intervention, 2011, for research fellowship of the first author. We are grateful to Prof. M.J Larkin for the use of Laboratory facilities at QUESTOR center, Queens University Belfast and for the kind and skillful assistance rendered. We thank Aaron McLaughlin, a PhD Student at Queens University, Belfast, for providing the
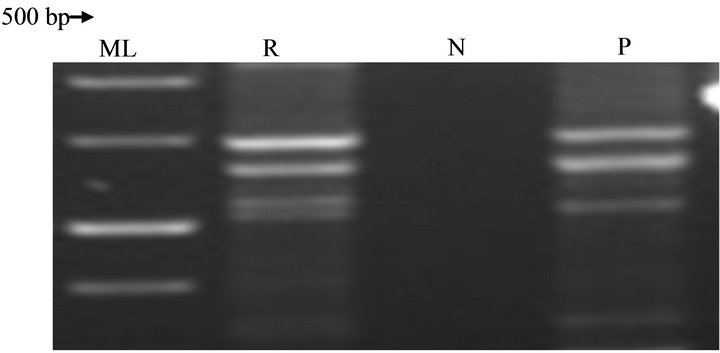
Figure 6. Chromosomal DNA-PCR analysis of Atz B gene in Rhodococcus sp strain at 65˚C.
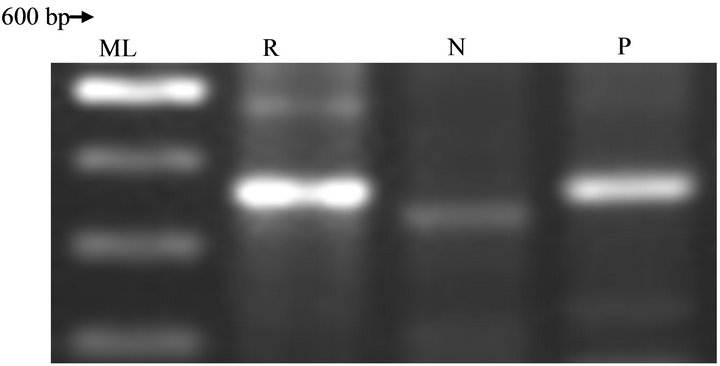
Figure 7. Chromosomal DNA-PCR analysis of Atz C gene in Rhodococcus sp strain at 52˚C.
KEY: ML: Molecular ladder; R: Rhodococcus sp strain; N: Negative control; P: Positive control.
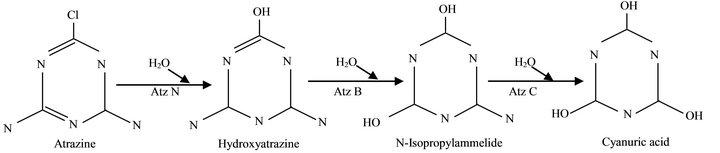
Figure 8. Putative pathway for atrazine-degrading Rhodococcus sp strain isolated in the study.
positive control strain, Rhodococcus jostii RHA1 used in the study.
REFERENCES
- R. D. Fletcher, “Bioremediation of Aviation Oil Spill: An Environmental Alternative,” Journal of Industrial Microbial, Vol. 7, No. 1, 2002, pp. 28-111.
- J. K. Struthers, K. Jayachandranand and T. B. Moorman, “Biodegradation of Atrazine by Agrobacterium radiobacter J14a and Use of This Strain in Bioremediation of Sediments and Surface Water,” American Chemical Society, Vol. 42, No. 2, 1998, pp. 432-436.
- M. L. De Souza, J. S. Martinez, M. J. B. Sadousky and L. P. Wackett, “The Atrazine Catabolism Genes atz ABC Are Widespread and Highly Conserved,” Journal of Bacteriology, American Society of Microbiology, Vol. 180, No. 7, 1998, pp. 1951-1954.
- P. K. Donelly, J. A. Entry and D. L. Crawford, “Degradation of Atrazine and 2, 4-Dichlorophenoxyacetic Acid by Mycorrhizal Fungi at Three Nitrogen Concentrations in Vitro,” Applied and Environmental Microbiology, Vol. 59, 1993, pp. 2642-2647.
- Y. Zeng, L. S. Collin, S. Stephen and P. Kothar, “Atrazine Degradation Pathway,” 2006. http://umbb.msi.umn.edu./atr/map.html
- P. A. Vaishampayan, P. P. Kanekar and P. K. Dhakephalkar, “Isolation and Characterization of Athrobacter sp Strain MCM B-436, an Atrazine-Degrading Bacterium, from Rhizospheric Soil,” International Biodeterioration, Vol. 60, 2007, pp. 273-278. doi:10.1016/j.ibiod.2007.05.001
- V. Garcia-Gonzalez, F. Gorantes, O. Porrua and E. Santero, “Regulation of the Pseudomonas sp Strain ADP Cyanuric Acid Degradation Operon,” Journal of Bacteriology, Vol. 187, No. 1, 2006, pp. 155-167. doi:10.1128/JB.187.1.155-167.2005
- M. Fazlurrahmam, M. Batra, J. Pandey, C. R. Suri and R. K. Jain, “Isolation and Characterization of an AtrazineDegrading Rhodococcus sp Strain MP-P1 from Contaminated Soil,” Letters in Applied Microbiology, Vol. 49, No. 6, 2009, pp. 721-729.
- E. L. Kruger, J. C. Anhal, D. Sorensen, B. Nelson, A. L. Chouhy, T. A. Anderson and J. R. Coats, “Atrazine Degradation in Pesticide-Contaminated Soils, Phytoremediation,” American Chemical Society of Symposium Series 664,Washington DC, 1997, pp. 54-64.
- R. T. Mandelbaum, D. L. Allan and L. P. Wackett, “Isolation and Characterization of a Pseudomonas sp That Mineralizes the S-Triazine Herbicide Atrazine,” Journal of Applied and Environmental Microbiology, American Society for Microbiology, Vol. 61, No. 4, 1995, pp. 1451- 1457.
- M. Radosevich and O. H. Tuovinen, “Microbial Degradation of Atrazine in Soils, Sediments and Surface Water,” American Chemical Society, Vol. 42, No. 2, 2002, pp. 432-436.
- Z. Arbelli and C. Fuentes, “Prevalence of the Genes trzN and Biogeographic Patterns among Atrazine-Degrading Bacteria Isolated from 13 Colombian Agricultural Soils. Federation of European Microbiological Societies,” Microbial Ecology, Vol. 72, 2010, pp. 611-623.
- M. De Souza, J. Seffernick, B. Martinez, M. J. Sadousky and L. P. Wackett, “The Atrazine Catabolism Genes atz ABC Are Widespread and Highly Conserved,” Journal of Bacteriology, American Society for Microbiology, 1998, pp. 1951-1954.
NOTES
*Corresponding author.

