Advances in Microbiology
Vol.2 No.4(2012), Article ID:25956,6 pages DOI:10.4236/aim.2012.24076
In-Situ Biostimulatory Effect of Selected Organic Wastes on Bacterial Atrazine Biodegradation
1Biological Sciences Programme, Abubakar Tafawa Balewa University, Bauchi, Nigeria
2Queens University Environmental and Genomic Research (QUESTOR) Center, Queens University of Belfast, Belfast, Northern Ireland, UK
3Microbiology Department, Usmanu Dan Fodio University, Sokoto, Nigeria
Email: *likitacool@yahoo.co.uk
Received September 18, 2012; revised October 16, 2012; accepted October 26, 2012
Keywords: Biostimulation; Biodegradation; Bioremediation; Mineralization; Atrazine
ABSTRACT
The biostimulatory effect of selected organic wastes on bacterial biodegradation of atrazine (2-chloro-4-ethylamino-6- isopropylamino-1,3,5-triazine) in three agricultural soils in Bauchi state, Nigeria, was carried out. The soil physicochemical characteristics were investigated to further understand the environmental conditions of the sampling sites. Enrichment technique was used to isolate the atrazine-degrading strains. Mineralization studies were carried out to determine atrazine biodegradation potentials of strains. Polymerase Chain Reaction (PCR) amplification of total nucleic acid of strains revealed several bacterial species based on nucleotide sequence analyses. Biostimulatory effect of selected organic wastes carried out showed minimal to average extent of biodegradation. The highest mean values, in CFU/mL, increase in biomass was recorded in Pseudomonas sp for both Cow dung 16.76 (42.03%) and Chicken droppings 12.32 (38.46%). However, biostimulatory effect using consortia provided more promising results, with 41.51% and 42.08% in Cow dung and Chicken droppings, biomass increase, respectively, in studies conducted. This proves that competition, survival of inoculums, bioavailability of organic amendments and nature of chemical are important factors affecting bioremediation.
1. Introduction
Soil contamination and its adverse effect on the overall ecosystem is one of the major problems we are facing today. Agrochemicals needed to boost agricultural activities have adverse effect on soil microflora, especially soil-acting herbicides. Many are deadly substances to human and soil microcosms [1]. Degradation of these chemical pollutants by indigenous microbes in soil has shown to be limited by sub-optimal growth conditions. Nutrient supplementation has been used to stimulate biodegradation on land [2].
Among the available technologies for soil restoration, bioremediation has been recognized as one of the promising alternative. Bioremediation is a natural process which naturally occurring microorganisms metabolize the pollutants to harmless end products [3]. The microbial processes that will employ in the clean-up dictate what nutritional supplements the bioremediation system must supply. The byproduct of microbial processes will provide indicators that bioremediation is successful [4].
Organic wastes contribute to the fertility of the soil by adding organic matter and nutrients, such as Nitrogen, Phosphorus, Sulphur, etc. [5]. Clean up of herbicides contaminated soil has been a dire environmental concern since the advent of industrial era. Although microorganisms are excellent degraders of herbicides in the soil, some reparation may need to be brought about in order to stimulate them to degrade pollutants. However, effective use of biostimulants requires thorough understanding of amendment/herbicide and microbial interaction in the soil. This work investigates the prospects of using selected organic wastes (Cow dung and Chicken droppings) in biostimulation of atrazine biodegradation in agricultural soils.
2. Materials and Methods
2.1. Study Sites and Sampling
Soils were sampled from two agricultural lands in Dass Local Government Area, Bauchi State, Northeastern Nigeria. The two sampling sites are maize (Zea mays) producing land, with history of atrazine usage (contaminated soil, ALA and non-contaminated soil, ALB). A third site, an organic farm (ALC), was also sampled. Soil samples, top 10 cm, were collected, approximately 100 g each, from centre and perimeter of agricultural land [3]. Five different samples from each field were collected on weekly basis for 5 weeks starting from April, 2011. Samples collected were properly mixed, made in composite and transported in ice-pack cooling system. The soil samples were characterized to understand their nature.
2.2. Isolation of Atrazine Degrading Bacterial Strains
Atrazine degrading bacterial strains was isolated using several enrichment cycles. A modified Mendalbaum et al. [6] and Udikovic et al. [7] methods were employed to isolate degraders, using atrazine, in separate units as sole carbon and nitrogen sources, respectively. All enrichment flasks were kept in shaker incubator at 30˚C at 150 rev·min–1 in the dark to preclude photolysis, for a period of two to four weeks. 0.1 mL was plated on M9-ATZ and CIT-ATZ, with control plates, in triplicates. Positive plates were screened and sub cultured for mineralization studies.
2.3. Genetic Characterization of Isolates
The fully screened isolates capable of atrazine degradation were grown on LB media and single colonies were subjected to DNA extraction using modified Griffith et al. [8] method and GenElute bacterial genomic DNA kit. Genomic DNA isolated was used for PCR analysis. 16S rRNA amplification was carried out using forward primer/ 63 F and reverse primer/1387 R. The PCR reaction on DNA samples was carried out using 18.1 uL of distilled water, 2.5 uL of buffer, 1 uL DNTPs and 1 uL DNA template per reaction, and 0.4 µL Taq polymerase in a 0.2 mL (25 µL) PCR tube loaded in an ESCO-MAXI swift thermo cycler machine. The program used was 1 cycle of 95˚C for 5 mins. 32 cycles of 95˚C for 45 secs, and 55˚C for 45 secs, 72˚C for 2 mins, followed by final extension of 72˚C for 10 mins. Visualized products, after separation in a 1% agarose gel electrophoresis. Gel was purified using GenElute Gel Purification Kit. Excised gel band was trimmed and procedural steps followed to purify DNA. The purified DNA was standardized according to concentration, mostly 15 µL purified product and 15 µL molecular grade water were mixed in Eppendorf tubes. 2 µL of forward/reverse 16S rRNA primers are added to the mixture and sent for DNA sequencing at Dundee Sequencing and Services, MCR/PPU, University of Dundee, Scotland.
2.4. Characterization of Soil Samples and Organic Wastes
Characterization of cow dung and chicken droppings was carried out using various analytical processes. Soil particle size was determined using Hydrometer method, as described by Davidson, [9]. Determination of Phosphorus and Nitrogen were carried out by Bray No.1 [10] and Macro-Kjedahl methods [11], respectively. Organic carbon content was determined using Wakley and Black method, [12].
2.5. Biostimulatory Studies
Characterized organic wastes were sterilized in hot air oven at 160˚C for 1 hour and subjected to biostimulatory study using a modified method of Couto et al. [13] and Moorman et al. [14]. Soil samples and organic wastes, at 80%/20% were used to set up the biodegradation slurries, in 1 liter cylindrical propylene container. Sterile deionised water was added just enough to submerge content. Equal concentration of atrazine and aliquot of 20 mL of strains and consortia culture in LB media harvested at late exponential phase, washed and re suspended in deionised water to yield OD600 of 1.0 was inoculated into biodegradation medium. All suspensions were thoroughly stirred and kept static incubation at 30˚C for 3 days. Control experiments were only set with only deionised water and atrazine in biodegradation media. 0.1 mL of 10–4 of slurries was plated on R2A media to determine increase in biomass of bacterial strains, before and after the study.
3. Results and Discussion
The research work investigates the biostimulatory effect of selected organic wastes on bacterial atrazine biodegradation in Agricultural soils. Soil organic matter is considered one of the most important factors responsible for controlling fate of pesticides in soil environment. It can restrict contaminant availability to microorganisms leading to the evolution of microbial population that specialize in accessing bound contaminants [15]. To fully understand the intricacies of the environment studied, soils collected were analyzed accordingly (Table 1). Cropping and herbicide history for at least the past two years before soil sample collection was obtained. Several researches have established relationship between soil characteristics and atrazine degradation parameters [16]. The correlation of more rapid atrazine mineralization in atrazine contaminated soil (ALA) is consistent with observation of others [17,18]. The bioavailability of most herbicides for microbial biodegradation is limited by sorption to organic matter [19]. In addition, Organic carbon substrates may affect microbial community structure and potential for degradation of herbicides, such as atrazine [20]. High percentage of aerobic heterothrophic bacterial count was observed in ALC (data not shown), compared to ALB and ALA (P < 0.05). Houot et al. [21]
observed that soil pH was a great contributor to atrazine fate in soil, but this can hardly be buttressed as soils studied have closely related values of 5.2 to 7.1. Other relationship on moisture and temperature can be subjective, as changes occur through seasonal influences and organic amendment supplementation. The characterization of the properties of the soils studied is summarized in Table 1.
Atrazine-degrading strains were isolated by enrich-
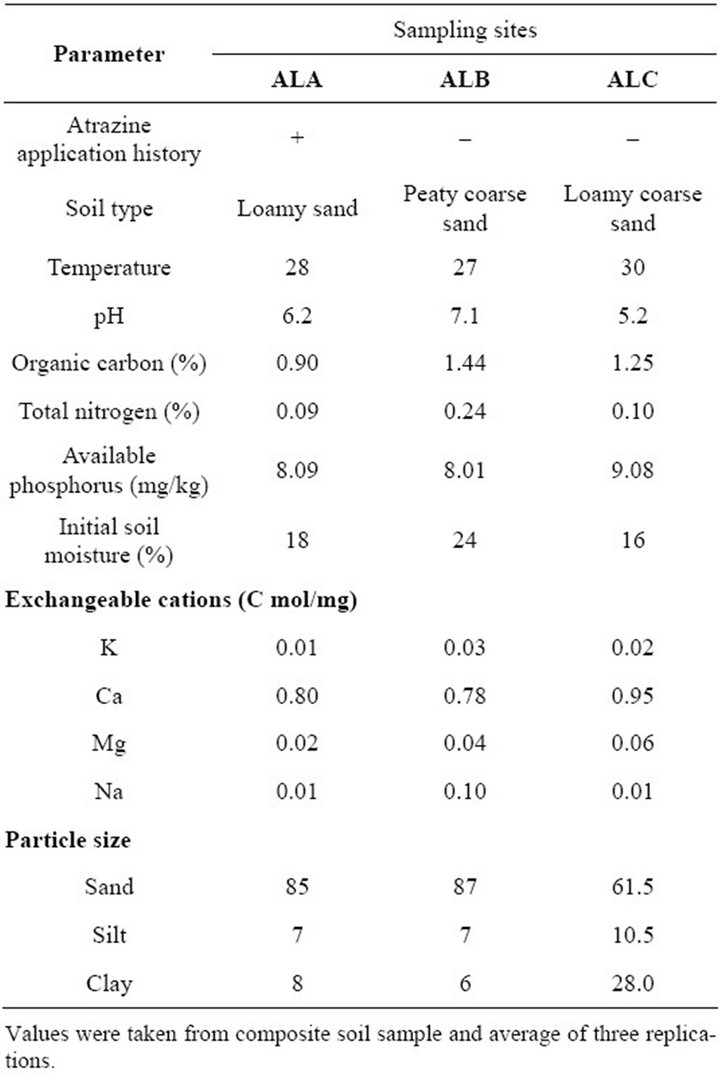
Table 1. Physical and chemical properties of soil in the three sampling sites.
ment techniques using both atrazine as nitrogen and carbon sources with fruitful outcomes. The bacterial populations isolated were analyzed using 16S rRNA-PCR. Searches for nucleotide sequence similarities were carried out using the BLAST program. The reference genomic sequence was compared with nucleotide collection with NCBI data base (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Table 2).
Figures 1 and 2 show the agarose gel electrophoresis of both PCR analysis of chromosomal DNA in strain isolated and subsequently purified products used for sequencing analysis.
Species of Pseudomonas, Rhodococcus and Agrobacterium are among most extensively studied bacteria for bioremediation capabilities [22,23]. Evaluation of proximate composition of selected organic wastes, Table 3, below showed varying concentration of contents, in cow dung and chicken droppings. The combine effect of soil type, organic amendment and atrazine degraders will

Figure 1. PCR analysis of 16S rRNA of putative atrazine degrading bacterial strains isolated.
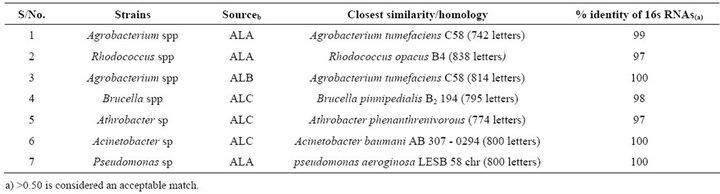
Table 2. 16s rRNA comparison of atrazine-degrading strains with NCBI database.
Figure 2. Purified 16s rRNA-DNA from bacterial strains used for sequencing analysis.
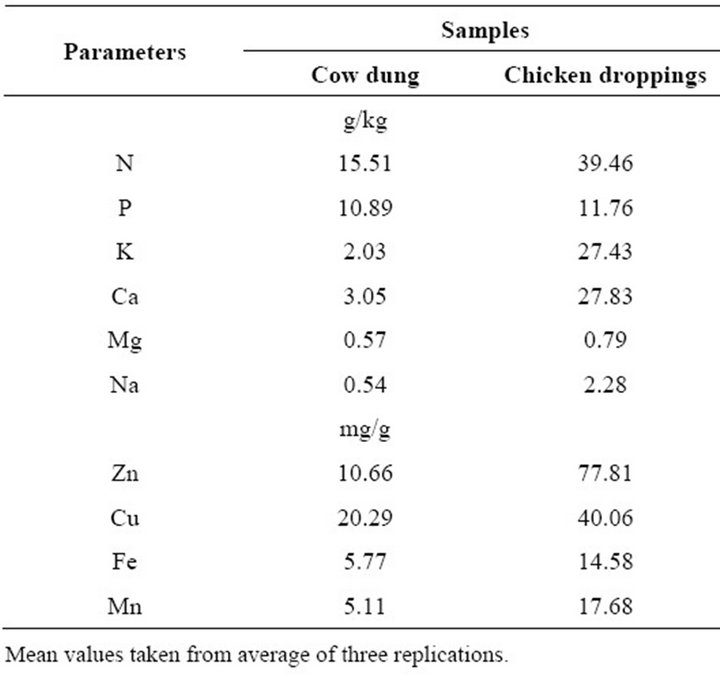
Table 3. Basic proximate mineral element composition of cow dung and chicken droppings used in the study.
translate into knowledge of activity in the environment.
Information obtained before and after stimulatory studies provided basis of understanding changes in soil environment amended with organic wastes. It has been shown that degradation of pollutants by given native microbial population can be favored by presence of required nutrients [24]. Biostimulation accelerate the decontamination rate, as the addition of one or more ratelimiting nutrients to the system improves the degradation potential of inhabiting microbial population [25]. The results showed that the mean comparative biostimulatory effect of bacterial strain, in cfu/10–4 mL, was 10.50 and 06.07; 14.56 and 09.97; 11.06 and 07.45; 12.07 and 08.45; 09.76 and 10.88; 09.88 and 8.88; 16.76 and 12.50 in Agrobacterium sp, Rhodococcus sp Aagrobacterium sp, Brucella sp, Athrobacter sp, Acinetobacter sp and Pseudomonas sp, in cow dung and chicken droppings, respectively (Table 4).
The relative higher value in bacterial biomass in cow dung compared to chicken droppings may be as a result of detrimental effect of nitrogen on atrazine degraders. In all biostimulatory studies, consortia provided a better option when using both cow dung and chicken droppings, with 17.06 (45.51%) and 15.08 (42.08%) during the analysis, with no tangible change in control experiments. These results demonstrated rather a relatively weak option in the use cow dung and chicken droppings for improved metabolism of atrazine bacterial strains studied. As most atrazine use this herbicide as nitrogen source, the presence of preferential nitrogen source in the environment is detrimental to atrazine degradation. Also Moorman et al. [12] showed that poultry litter, commonly applied to soil as manure on atrazine contaminated soil, was two times faster without the litter, nutrients mainly nitrates and phosphate did not play role in the degradation of atrazine. Also, nitrogen amendments were in fact shown to decrease atrazine degradation rates by Pseudomonas sp ADP [26,27]. If atrazine mineraliza-
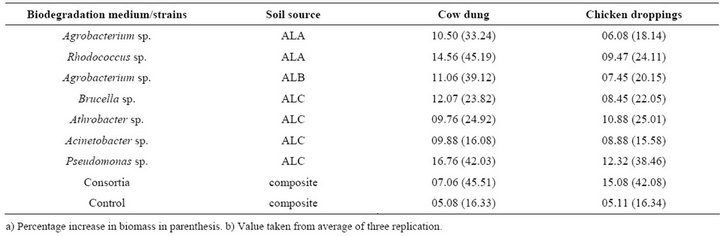
Table 4. Biostimulatory effect of cow dung and chicken droppings during an in-situ incubation using atrazine degrading bacterial strains.
tion is sought, substrates containing high C to N ratio maybe preferable. Citrate or succinate biostimulation markedly increased atrazine mineralization in soil that had potential of atrazine biodegradation.
Enhanced mineralization of contaminants in natural habitants by nutrient addition will require proper knowledge of natural microbial communities and their growth requirement, and problem of organic matter sorption represses biodegradation [15]. The study showed that biostimulatory effect were better in adapted (ALA) and non-contaminated (ALB) soils, than organic soil (ALC) atrazine degrading strains, in both cow dung and chicken droppings studied.
4. Conclusions
To date, most of soil biostimulation experience have been carried out in the laboratory (controlled conditions), while results from the field experiment are sparse. The paradox of achieving as good or better result in the field as in the laboratory is the greatest challenge to face in implementing a bioremediation strategy.
In this study it was noted that cow dung and chicken droppings can increase the level of organic nutrient needed by plants for growth (soil fertility), but have minimal or average effect on atrazine degraders, thus possibly increasing sorption of atrazine and subsequently its uptake (translocation) by plants. Also, soils especially ALC, composed generally have trapping nature and lack release factor due to high sorption ability.
5. Acknowledgements
This work was fully supported by the Education Trust Fund (ETF) intervention, 2011, for research fellowship of the first author.
REFERENCES
- ATSDR, “Agency for Toxic Substances and Disease Registry: Toxicological Fact for Atrazine,” Mailstop Atlanta, Georgia, 2004, pp. 4-9
- I. E. Adieze, R. N. Nwabueze and G. O. C. Onyeze, “Effect of Poultry Manure on the Microbial Utilization of Hydrocarbon in Oil Polluted Soil,” Nigerian Journal of Microbiology, Vol. 17, No. 1, 2003, pp. 12-16.
- R. D. Fletcher, “Bioremediation of Aviation Oil Spill: An Environmental Alternative,” Journal Industrial Microbial, Vol. 7, No. 1, 2002, pp. 28-111.
- R. M. Atlas and R. Bartha, “Stimulated Biodegradation of Oil Slicks Using Leophillic Fertilizer,” Environmental Science Technology, Vol. 7, 1981, pp. 538-541 doi:10.1021/es60078a005
- G. C. Opkokwasili and W. A. James, “Microbiol Contamination of Kerosene, Gasoline and Crude Oil and Their Spoilage Potential,” Nigerian Journal of Microbilogy, Vol. 29, 1995, pp. 147-156.
- R. T. Mandelbaum, D. L. Allan and L. P. Wackett, “Isolation and Characterization of a Pseudomonas sp. That Mineralizes the S-Triazine Herbicide Atrazine,” Journal Applied and Environmental Microbiology, American Society for Microbiology, Vol. 61, No. 4, 1995, pp. 1451- 1457.
- N. Udikovic, D. Hrsak, G. Mendas and D. Filipcic, “Enrichment and Characterization of Atrazine Degrading Bacterial Communities,” Food Technology and Biotechnology, Vol. 41, No. 3, 2003, pp. 211-217.
- R. I. Griffith, A. S. Whiteley, A. G. O’Donnell and M. J. Bailey, “Rapid Method for Co-Extraction of DNA and RNA from Natural Environments for Analysis of Ribosomal DNA and rRNA-Based Microbial Community Composition,” Applied Environmental Microbiology, American Society of Microbiology, Vol. 66, No. 12, 2000, pp. 5488-5491.
- A. L. Davidson, “Soil Chemical Analysis,” Prentice Hall, New York, 1955, pp. 44-48.
- R. H. Bray, “Determination of Total Organic and Available Forms of Phosphorus in Soil,” Soil Science, Vol. 59, 1965, pp. 39-45. doi:10.1097/00010694-194501000-00006
- C. A. Black, “Methods of Soil Analysis,” American Society of Agronomy, Vol. 9, No. 2, 1965, pp. 12-15.
- A. Wakley and I. A. Black, “An Examination of the Method for Determining Soil Organic Matter and Proposed Modification of the Acid Titration Method,” Journal of Soil Science, Vol. 37, 1965, pp. 29-38.
- M. N. P. F. S. Couto, E. Monteiro and M. T. S. D. Vasconceles, “Mesocosm Trials of Bioremediation of Contaminated Soil of a Petroleum Refinery: Comparison of Natural Attenuation, Biostimulation and Bioaugmentation,” Environmental Science and Pollution Research, Vol. 17, No. 7, 2010, pp. 1339-1346. doi:10.1007/s11356-010-0318-y
- T. B. Moorman, J. K. Cowan, E. LArthur and J. R. Coats, “Organic Amendments to Enhance Herbicide Biodegradation in Contaminated Soils,” Biology and Fertility of Soil, Vol. 33, No. 6, 2001, pp. 541-545.
- G. M. Coleres, “Soil Organic Matter-Driven Selection of Atrazine-Degrading Microbial Population,” 2005. http://www.reis.usda.gov./webcrisprojectpages/195956
- R. M. Zablotowicz, M. A. Weaver and M. A. Locke, “Microbial Adaptation for Accelerated Atrazine Mineralization/Degradation in Missisipi Delta Soils,” Weed Science, Vol. 54, 2006, pp. 538-554. doi:10.1614/WS-04-179R3.1
- E. Barriouso and S. Houot, “Influence of Compost Addition to Soil on the Behavior of Herbicides,” Pesticides Science, Vol. 4a, No. 1, 1996, pp. 665-757.
- F. Martin-Laurent, L. Cornet, L. Ranjard, J. C. LopezGutierrez, L. Phillipport, C. Schwartz, C. Cartoux and G. Soulas, “Estimation of Atrazine-Degrading Genetic Potential and Activity in Three French Agricultural Soils,” FEMS Microbiology Ecology, Vol. 48, No. 1, 2004, pp. 5425-5435.
- M. Alexander, “Introduction to Soil Microbiology,” 2nd Edition, John-Wiley and Sons, New York, 1984, pp. 281- 299.
- R. D. Rhin, J. J. Fuhrmann and M. Radosevich, “Microbial Community Response to Atrazine Exposure and Nutrient Availability: Linking Degradation Capacity to Community Structure,” Microbiology Ecology, Vol. 46, 2003, pp. 145-160. doi:10.1007/s00248-002-1048-6
- S. Houo, E. Barriuso and V. Bergheaud, “Modifications Atrazine Degradation Pathways in a Loamy Soil after Addition of Organic Amendments,” Soil Biology Biochemistry, Vol. 30, 14, 2000, pp. 2147-2157.
- M. Farhadian, C. Vachelard, D. Duchez and C. Larroche, “In Situ Bioremediation of Monoaromatic Pollutants in Groundwater: A Review,” Bioresource Technology, Vol. 99, 2008, pp. 5296-5308. doi:10.1016/j.biortech.2007.10.025
- A. K. Haritash and C. P. Kaushik, “Biodegradation Aspects of Polycyclic Aromatic Hydrocarbons (PAHs): A Review,” Journal of Hazardous Materials, Vol. 169, 2009, pp. 1-15. doi:10.1016/j.jhazmat.2009.03.137
- D. Dellile, E. Pelletier, A. Rodriquez-Blanco and J. F. Ghiaglione, “Effects of Nutrient and Temperature on Degradation of Petroleum Hydrocarbon in Sub-Artarctic Coastal Sea Water,” Polar Biology, Vol. 32, 2009, pp. 1521-1528. doi:10.1007/s00300-009-0652-z
- M. Nikolopoulou and N. Kalogeraski, “Biostimulation Strategies for Fresh and Chemically Polluted Marine Environments with Petroleum Hydrocarbon,” Journal of Chemical Technology & Biotechnology, Vol. 84, No. 1, 2009, pp. 802-807.
- G. B. Clausen, L. Larsen, K. Johnson, J. R. Lipthay and J. de Aamand, “Quantification of the Atrazine-Degrading Pseudomonas sp Strain ADP in Aquifer Sediment by Quantitative Competitive Polymerase Chain Reaction,” FEMS Microbiology Ecology, Vol. 41, 2002, pp. 211-229. doi:10.1111/j.1574-6941.2002.tb00983.x
- V. Garcia Gonzalez, F. Gorantes, O. Porrua and E. Santero, “Regulation of the Pseudomonas sp Strain ADP Cyanuric Acid Degradation Operon,” Journal of Bacteriology, Vol. 187, No. 1, 2003, pp. 155-167.
NOTES
*Corresponding author.

