Paper Menu >>
Journal Menu >>
 Journal of Signal and Information Processing, 2012, 3, 469-480 http://dx.doi.org/10.4236/jsip.2012.34060 Published Online November 2012 (http://www.SciRP.org/journal/jsip) Analysis of fMRI Single Subject Data in the Fourier Domain Acquired Using a Multiple Input Stimulus Experimental Design Daniel Rio1, Robert Rawlings2*, Lawrence Woltz3, Jodi Gilman4, Daniel Hommer1 1Section of Brain Electrophysiology and Imaging, LCTS, National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health, Bethesda, USA; 2National Institute on Alcohol Abuse and Alcoholism, Columbia, USA; 3Synergy Research Inc., Monrovia, USA; 4Laboratory of Neuroimaging and Genetics, Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, USA. Email: drio@nih.gov Received July 28th, 2012; revised August 27th, 2012; accepted September 12th, 2012 ABSTRACT Analysis of functional MRI (fMRI) blood oxygenation level dependent (BOLD) data is typically carried out in the time domain where the data has a high temporal correlation. These analyses usually employ parametric models of the hemo- dynamic response function (HRF) where either pre-whitening of the data is attempted or autoregressive (AR) models are employed to model the noise. Statistical analysis then proceeds via regression of the convolution of the HRF with the input stimuli. This approach has limitations when considering that the time series collected are embedded in a brain image in which the AR model order may vary and pre-whitening techniques may be insufficient for handling faster sampling times. However fMRI data can be analyzed in the Fourier domain where the assumptions made as to the structure of the noise can be less restrictive and hypothesis tests are straightforward for single subject analysis, espe- cially useful in a clinical setting. This allows for experiments that can have both fast temporal sampling and event-related designs where stimuli can be closely spaced in time. Equally important, statistical analysis in the Fourier domain focuses on hypothesis tests based on nonparametric estimates of the hemodynamic transfer function (HRF in the frequency domain). This is especially important for experimental designs involving multiple states (drug or stimulus induced) that may alter the form of the response function. In this context a univariate general linear model in the Fourier domain has been applied to analyze BOLD data sampled at a rate of 400 ms from an experiment that used a two-way ANOVA design for the deterministic stimulus inputs with inter-stimulus time intervals chosen from Poisson distribu- tions of equal intensity. Keywords: fMRI (BOLD); Time Series Analysis; Single Subject Analysis; Fourier Domain; Statistical Analysis; Complex General Linear Model; Alcoholism Research 1. Introduction Functional magnetic resonance imaging (fMRI) has be- come a standard technique for investigating changes in brain physiology over time. The most important tech- nique developed for this purpose measures the blood oxygen level dependent (BOLD) response to given stim- uli. The BOLD signal arises from localized variations in magnetic susceptibility caused by changes in blood oxy- genation levels instigated by the underlying stimu- lus-induced neuronal activation [1]. These changes can be seen in T2* weighted MRI time series data [2,3] that have a low signal-to-noise (SNR) and temporal SNR [4] as measured in fMRI time series that also have high temporal correlation [5]. Essentially from the very first attempts to analyze fMRI BOLD time series data analyses have been carried out in the time domain [6-8], most often in the context of analyses using a general linear model applied to groups of subjects [7,9-12]. The majority of these methodological approaches require a preliminary attempt to model fMRI time series data from single subjects with a few papers looking at this problem more directly [13]. However there are a number of concerns to deal with in analyses of fMRI time series data carried out in the temporal domain. Foremost among these is the problem associated with temporal correlation as seen in fMRI BOLD time series data [5] where a number of methods to control for this problem [14,15] have been developed. These methods *Retired. Copyright © 2012 SciRes. JSIP  Analysis of fMRI Single Subject Data in the Fourier Domain Acquired Using a Multiple Input Stimulus Experimental Design 470 attempt to remove the autocorrelation in single subject data (a form of signal estimation) in order to ultimately perform statistical inference or signal detection. One of the most common approaches is pre-whitening of the data [15]. Another common approach is to represent the correlation in the data with a fixed order AR model [16] that is often embedded in a restricted maximum likely- hood (REML) estimation formulation [17]. However pre-whitening of the fMRI BOLD data has been shown to have limitations [15]. In particular it may be insuffi- cient to remove the correlation across the temporal do- main for rapidly sampled fMRI data or experimental de- signs that include short inter-stimulus intervals. As well, the application of fixed order AR models also have a number of problems. Primarily, that in practice the cho- sen order of the model is typically not verified for time series at every spatial position or voxel acquired within the brain. In fact the AR model order is typically not even verified for a new experimental design and it would be remarkable if such a constrained model were univer- sally suitable. However if a variable order AR model were fitted to every time series collected at every voxel investigated then the modeled covariance structure would change across these voxels and any resultant statistic would be difficult to interpret across regions of the brain. The use of parametric models for the hemodynamic response function (HRF), somewhat separate from the statistical analysis of fMRI data is another limitation in temporal-based analysis of fMRI data [18,19]. Since the general shape of the HRF is usually assumed before the fMRI BOLD data is analyzed there is some concern with experimental designs in which the shape is state-de- pendent. This can for instance happen with the introduc- tion of a vaso-active drug, such as alcohol, to a subject during part of an experimental procedure [20]. Addition- ally many additional parametric forms along with regres- sion coefficients are typically included to correct for other perceived confounds including those associated with signal drift and head motion [21,22]. Furthermore other procedures are used to correct e.g. for cardiac arti- facts [23] and time shifts errors seen in multi-slice acqui- sition of fMRI data [24]. Therefore the current standard analysis of fMRI BOLD data in the temporal domain necessitates the use of a number of empirically based ad hoc analytical and/or statistical techniques. However Fou- rier-domain-based analysis can in many instances provide a more general mathematically suitable methodology. The methodology in this paper focuses on the analysis of fMRI time series data in the Fourier or frequency do- main. While Fourier domain approaches have been ex- tensively applied to time series in the general field of signal processing they have had limited use for this ap- plication. One of the earliest attempts at a Fourier-based analysis of fMRI was that by Lange, et al. [25] that fo- cused on the analysis of data obtained from a block ex- perimental design. This approach used a parametric form for the HRF and spatially averaged over adjacent voxels. Another early paper that analyzed fMRI data in the fre- quency domain was by Marchini and Ripley [26], how- ever it was restricted to periodic stimuli. Neither of the methodologies presented in these papers were developed to carry out multivariate hypothesis testing for compli- cated experimental designs. As such they were not based on the statistical theory in the Fourier domain as devel- oped by Brillinger [27,28] and first applied to investigate fMRI time series data by Rio [29]. A more recent paper, also based on the work by Brill- inger, is that by Bai et al. [30]. It focused on obtaining unbiased estimates of the HRF using stochastic rather than deterministic input stimuli (the usual design for fMRI experiments). It uses a weighted estimate of the transfer function and appropriate chi-square statistics to analyze sample data from an fMRI experiment with a “simple” design. In contrast our paper has deterministic inputs or stimuli and an unweighted estimate of the transfer function, an approach that provides estimates with minimum mean square error. The development in our paper therefore is toward a full “multivariate” ap- proach for hypothesis testing to perform signal detection in the spectral domain using an extension of the general linear model methodology in the complex domain. Here the use of stochastic inputs to model the stimuli and weighted transfer function estimations would be inap- propriate for our clinical studies. Thus the paper by Bai is attempting to find the best estimate to the transfer function, but not necessarily carrying out multivariate statistical hy- potheses testing. Thus while these two papers are starting at the same place they are going in different directions. Focusing on the approach taken in this paper we pre- sent a methodological approach developed for the Fou- rier domain that allows us to analyze event-evoked time series data similar to that collected in many fMRI ex- periments. In this approach the noise model can be made less restrictive and allows more direct analysis of fMRI time series data for single subject [29,31,32]. In particu- lar to demonstrate this methodology, fMRI BOLD data [33] are taken from an experimental design that would not typically be suited for some of the noise assumptions made in analyzing these data in the time domain for sin- gle subjects. Here the sampling rate for the experiment was 400 ms and the experiment used a two-way ANOVA design for the stimulus input. In addition, stimulus types were constructed from Poisson processes of equal inten- sity resulting in some inter-stimulus intervals (ISI)s being very short. Thus the data for single subjects generated from this experiment is well suited to analysis using the Copyright © 2012 SciRes. JSIP  Analysis of fMRI Single Subject Data in the Fourier Domain Acquired Using a Multiple Input Stimulus Experimental Design 471 Fourier-based methodology that will be presented in this paper, in that the fast sampling rate and experimental design are easily handled by this technique as compared to temporal-based approaches. 2. Complex General Linear Model 2.1. The Model The model in the time domain takes the usual form for fMRI BOLD data [29,31] s tatrtt (1) where the fMRI signal s t is considered to be a uni- variate time series collected at discrete time points t and at each spatial position ,, x xyz or voxel (implicit). Here the fixed deterministic input stimuli rt are fil- tered by the hemodynamic response function (HRF), , where the convolution operation is symbolically represented by . Multiple (R) input stimuli require that represents a 1 × R matrix and correspondingly the response function represents a R × 1 matrix. Thus each single-input stimulus time series has a correspond- ing single response function represented by a corre- sponding matrix entry in the response function at rt at at. The only assumption on the noise structure, represented by the univariate time series t , is that it be stationary with zero mean. Therefore the fMRI time series observed at each spatial position is assumed to be described by a linear time-invariant model, where the BOLD signal, s t t , is determined by a constant (with respect to time), , plus a linear filter of several fixed determinis- tic inputs and an additive error or noise term, at rt . The analysis is then carried out in the Fourier do- main, where the complex Fourier transform of Equation 1 can be written as kkk sar k (2) where k2πkT , T represents the number of time points and k represents the wave or frequency number. k a , the response function’s representation in the Fourier domain, is henceforth referred to as the hemody- namic transfer function or HTF. Periodograms are then constructed from the Fourier transform of the input function, k r , and the output function, k s , as follows 1 kk 2πT IH k (3) where and superscript H refers to the Hermitian transpose. Estimates of cross-spectral density functions are then constructed from these periodograms. Thus corresponding to each of the periodogram functions we construct a corresponding cross-spectral function [27] that is ,,rs 1m k km ˆ2m 1 fI (4) These estimates are constructed over disjoint fre- quency bands, size k = −m to m with center frequency and provide stable estimates of the cross-spectral functions. Also, since these spectral functions will be used in constructing hypothesis tests a Daniell or equal- weighted averaging window is used to provide estimates for these quantities. These cross-spectral functions take a slightly different form [27] for the band centered at zero frequency, however in this application to fMRI data this band will be discarded due to artifacts (for example low frequency motion drift) and we will not address it in this paper. Band size is picked based on statistical power considerations or on information as to the input power spectrum. If however specific information is known as to the spectral distribution of the input power (e.g. the power of the input stimuli is concentrated in one or more narrow bands) then smaller band sizes should be chosen. Unbiased spectral-banded estimates of the power associ- ated with the input stimuli are given by the cross-spectral function ˆrr f and for the power of the output func- tion or fMRI BOLD signal by ˆss f. Furthermore, estimates can also be constructed for the error spectrum ˆ f represented by g (Equation 10 to be presented in Section 2.3) and provide a measure of the extent to which the output signal can be determined from the input function for the given model. 2.2. Unbiased Non-Parametric Estimates of the HTF In Equation 2 we see that the HTF k a essentially represents regression coefficients relating the stimulus input function k r to the output fMRI BOLD signal k s in the Fourier domain. This represents a form of the general linear model in the complex domain for mul- tiple input and univariate output. Furthermore since the noise k in the Fourier domain is asymptotically independent [27] it is possible to use the method of maximum likelihood estimation as extended to the com- plex domain to construct unbiased estimate of the HTF [27,34] at the center frequencies using the cross-spectral estimates as defined in Equation 4. That is 1 ˆˆ ˆsr rr Af f (5) a R × 1 matrix (one entry for each stimulus input) at every spatial position or voxel. This estimate of the HTF, calculated at the center frequency for each band, is the crucial quantity for which hypothesis tests are made in this implementation of the complex linear model for multiple inputs in the Fourier domain. Furthermore since this estimated HTF is so important to our methodology, Copyright © 2012 SciRes. JSIP  Analysis of fMRI Single Subject Data in the Fourier Domain Acquired Using a Multiple Input Stimulus Experimental Design 472 some additional non-statistical quantitative functions (as a prelude to inference tests) are constructed as follows to investigate the general spatial pattern of the hemody- namic response induced by the stimuli. Using the esti- mated transfer function in the Fourier domain, the power per band can be simply calculated by ˆˆ diag H pAA (6) and the total power (sum over all bands) for the rth transfer function is given by rrr Pp (7) where r = 1 to R. Finally we can construct the total power for all transfer functions as tot trPp (8) where tr refers to the trace (sum of diagonal entries). Thus these functions give us a general sense of the hemody- namic activity at every sampled voxel within the brain in response to each individual input stimulus, , or for all stimuli taken together, . r P tot P 2.3. Hypothesis Testing Consider the test of the hypothesis o H: 0AB where B is the contrast matrix for multiple input and has size R × b, where b = 1 to R depending on the particular test being used. The contrast matrix B allows for testing any individual or combination (contrast, marginal or in- teraction tests) of the R stimulus input (functions). The tests take the form of the following F-distribution [27,31,32] at each spatial position and band (represented by its center frequency ) 2b;2 2m1R 1 1H TT F ˆ ˆ 2m 1 b rr ABBf BBA g ˆ (9) where 1 2m 1ˆˆˆˆ 2m 1 Rsssr rrrs g ffff (10) estimates the spectral error function, ˆ f, a measure of the model’s fit to the data in the frequency domain. A special case of general hypothesis test presented is when the contrast matrix B is equal to the identity matrix. Then the test is for the hypothesis o H: 0A and Equation 9 takes the following form H 2R;2 2m1R ˆ ˆˆ 2m 1 FR rr Af A g (11) This statistic, which we will henceforth refer to as the omnibus test, is related to the squared sample complex multiple correlation coefficient or simply the 2 Rsr statistic in the Fourier domain [27,35]. This statistic takes the following form in our case 1 2ˆˆˆ R1 ˆˆ sr rrrs sr ss ss ff fg ff (12) ranges between 0 and 1, and measures in general how well our statistical model fits the data. Essentially, either spectral statistic conveys information as to how well the fMRI signal is predicted by the stimulus inputs since small values for the error term g lead to large F values for the omnibus test or equivalently 2 sr R values close to 1, in each associated band rep- resented by the center frequency . 3. Application 3.1. Experimental Design Selected data were taken from the following more exten- sive experiment [33], previously analyzed using a stan- dard temporal fMRI group based analysis as imple- mented in AFNI [8]. Four types of visual inputs were presented to control and alcoholic subjects using a split screen format. They consisted of the following paired presentation in a 2-way ANOVA experimental design: 1) Alcoholic beverage and positive image; 2) Non-alcoholic beverage and positive image; 3) Alcoholic beverage and negative image; 4) Non-alcoholic beverage and negative image. Positive and negative visual images were taken from the International Affective Picture System (IAPS) [36] and paired (simultaneously, side-by-side) with al- coholic or non-alcoholic beverages (i.e. milk, orange juice). There were 55 presentations for each type of input stimulus combination. The duration of a single stimulus image presentation was 800 ms and the ISIs between the paired inputs were sampled from an exponential distribu- tion. The average ISI in seconds for each input stimulus presented individually was: Positive and alcohol—8.58 seconds, positive and non-alcohol—9.61 seconds, nega- tive and alcohol—9.24 seconds and negative and non- alcohol—9.77 seconds. The average ISI for all stimulus presentations (without differentiation as to the type of stimulus) was 3.15 seconds. Control and alcoholic sub- jects viewed these images or a neutral background and the entire input stimuli time sequence consisted of 1400 time points and all subjects received the same sequence of input stimuli. Copyright © 2012 SciRes. JSIP 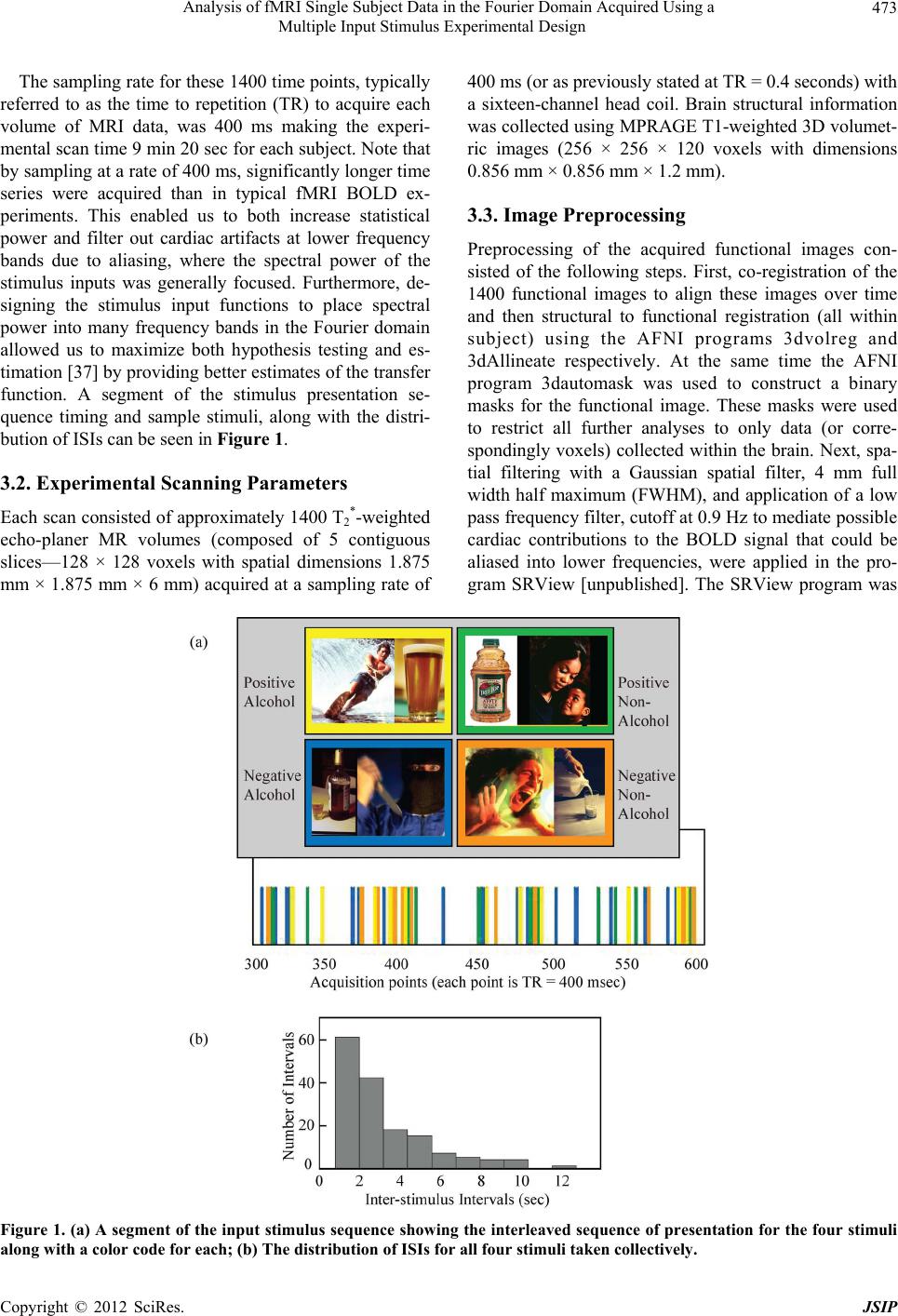 Analysis of fMRI Single Subject Data in the Fourier Domain Acquired Using a Multiple Input Stimulus Experimental Design Copyright © 2012 SciRes. JSIP 473 The sampling rate for these 1400 time points, typically referred to as the time to repetition (TR) to acquire each volume of MRI data, was 400 ms making the experi- mental scan time 9 min 20 sec for each subject. Note that by sampling at a rate of 400 ms, significantly longer time series were acquired than in typical fMRI BOLD ex- periments. This enabled us to both increase statistical power and filter out cardiac artifacts at lower frequency bands due to aliasing, where the spectral power of the stimulus inputs was generally focused. Furthermore, de- signing the stimulus input functions to place spectral power into many frequency bands in the Fourier domain allowed us to maximize both hypothesis testing and es- timation [37] by providing better estimates of the transfer function. A segment of the stimulus presentation se- quence timing and sample stimuli, along with the distri- bution of ISIs can be seen in Figure 1. 3.2. Experimental Scanning Parameters Each scan consisted of approximately 1400 T2 *-weighted echo-planer MR volumes (composed of 5 contiguous slices—128 × 128 voxels with spatial dimensions 1.875 mm × 1.875 mm × 6 mm) acquired at a sampling rate of 400 ms (or as previously stated at TR = 0.4 seconds) with a sixteen-channel head coil. Brain structural information was collected using MPRAGE T1-weighted 3D volumet- ric images (256 × 256 × 120 voxels with dimensions 0.856 mm × 0.856 mm × 1.2 mm). 3.3. Image Preprocessing Preprocessing of the acquired functional images con- sisted of the following steps. First, co-registration of the 1400 functional images to align these images over time and then structural to functional registration (all within subject) using the AFNI programs 3dvolreg and 3dAllineate respectively. At the same time the AFNI program 3dautomask was used to construct a binary masks for the functional image. These masks were used to restrict all further analyses to only data (or corre- spondingly voxels) collected within the brain. Next, spa- tial filtering with a Gaussian spatial filter, 4 mm full width half maximum (FWHM), and application of a low pass frequency filter, cutoff at 0.9 Hz to mediate possible cardiac contributions to the BOLD signal that could be aliased into lower frequencies, were applied in the pro- gram SRView [unpublished]. The SRView program was Figure 1. (a) A segment of the input stimulu s sequence showing the inte rleaved sequence of pr esentation for the four stimuli long with a color code for each; (b) The distribution of ISIs for all four stimuli taken collectively. a 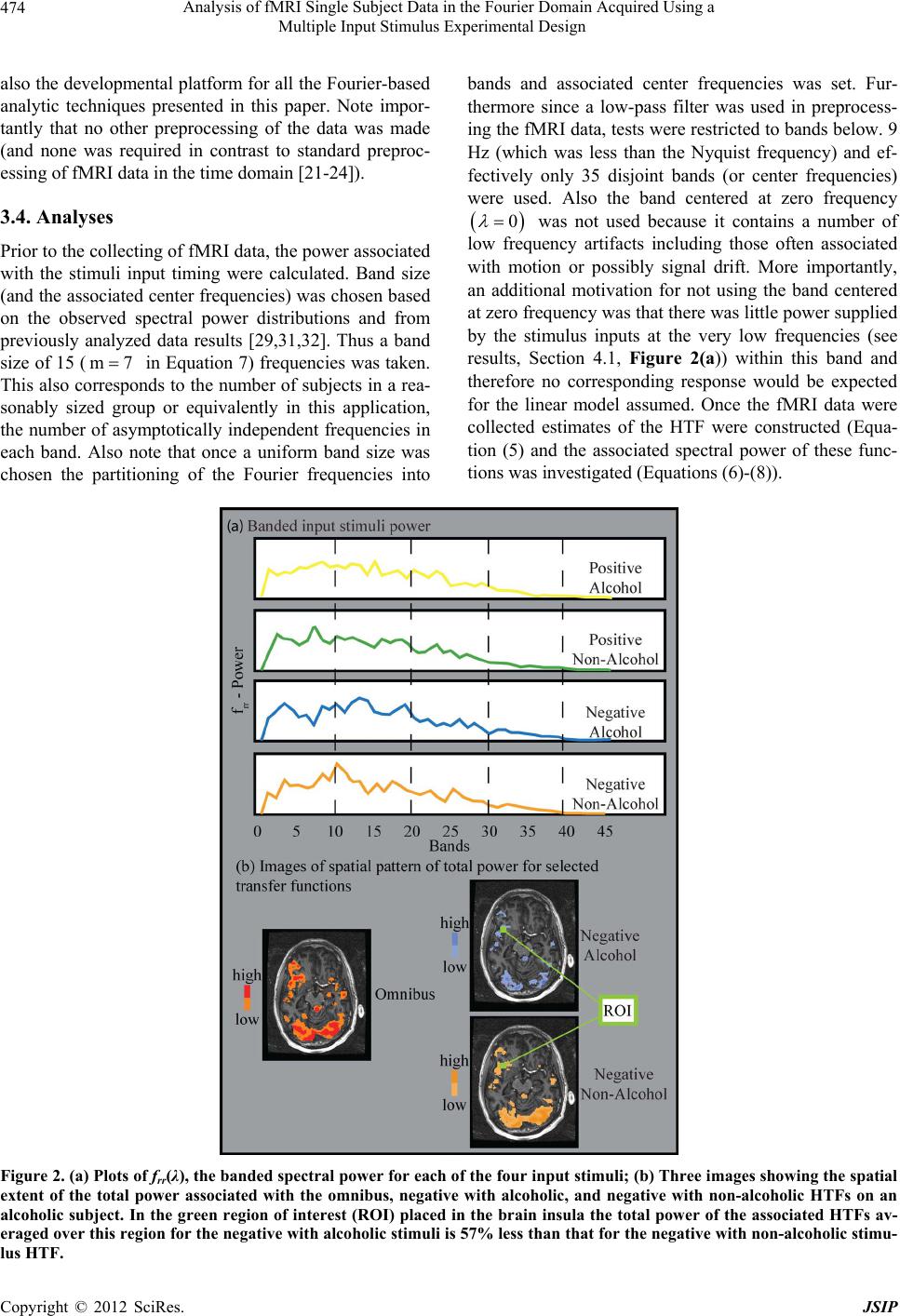 Analysis of fMRI Single Subject Data in the Fourier Domain Acquired Using a Multiple Input Stimulus Experimental Design 474 also the developmental platform for all the Fourier-based analytic techniques presented in this paper. Note impor- tantly that no other preprocessing of the data was made (and none was required in contrast to standard preproc- essing of fMRI data in the time domain [21-24]). 3.4. Analyses Prior to the collecting of fMRI data, the power associated with the stimuli input timing were calculated. Band size (and the associated center frequencies) was chosen based on the observed spectral power distributions and from previously analyzed data results [29,31,32]. Thus a band size of 15 ( in Equation 7) frequencies was taken. This also corresponds to the number of subjects in a rea- sonably sized group or equivalently in this application, the number of asymptotically independent frequencies in each band. Also note that once a uniform band size was chosen the partitioning of the Fourier frequencies into m7 bands and associated center frequencies was set. Fur- thermore since a low-pass filter was used in preprocess- ing the fMRI data, tests were restricted to bands below. 9 Hz (which was less than the Nyquist frequency) and ef- fectively only 35 disjoint bands (or center frequencies) were used. Also the band centered at zero frequency 0 was not used because it contains a number of low frequency artifacts including those often associated with motion or possibly signal drift. More importantly, an additional motivation for not using the band centered at zero frequency was that there was little power supplied by the stimulus inputs at the very low frequencies (see results, Section 4.1, Figure 2(a)) within this band and therefore no corresponding response would be expected for the linear model assumed. Once the fMRI data were collected estimates of the HTF were constructed (Equa- tion (5) and the associated spectral power of these func- tions was investigated (Equations (6)-(8)). Figure 2. (a) Plots of frr(λ), the banded spectral power for each of the four input stimuli; (b) Three images showing the spatial extent of the total power associated with the omnibus, negative with alcoholic, and negative with non-alcoholic HTFs on an alcoholic subject. In the green region of interest (ROI) placed in the brain insula the total power of the associated HTFs av- eraged over this region for the negative with alcoholic stimuli is 57% less than that for the negative with non-alcoholic stimu- us HTF. l Copyright © 2012 SciRes. JSIP  Analysis of fMRI Single Subject Data in the Fourier Domain Acquired Using a Multiple Input Stimulus Experimental Design 475 Model goodness of fit (Equation (12)) was explored using the omnibus hypothesis test o H: 0A using the F-test statistic (Equation (11)). These tests were car- ried out at every voxel with the binary functional brain mask (see Section 3.3) and indicated whether any of the stimuli produced a significant response at any center frequency. In essence the F-test statistic image was con- structed for every band (or center frequency), then a p-value was selected and an associated F-value threshold was used to test for signal response in these images. Al- ternatively, one can consider looking at one spatial posi- tion or voxel, setting a threshold across all bands calcu- lated for that voxel (see Figure 3(a) Section 4.2), and then combining the voxel results at each center frequency to produce spatial images for each band. In either case this produced frequency-specific spatial patterns associ- ated with signal response to the input stimuli (see Figure 3(b) Section 4.2). These resultant spatial patterns are presented using multiple p-level mask whose corre- sponding F-statistics with appropriate degrees of freedom (dof) were used as threshold values for the F-test statistic image produced. A color look-up-table (LUT) is used to present these threshold values. The algorithm is: Input: Select multiple p-values p0, ···, pn and associated color values in LUT Calculate corresponding F-value, F(p1), ···,F(pn) such that dof (2b; 2(2 m + 1-R)) Loop over voxels within brain mask Extract F-values at voxel, indexed by band numbers Loop over band numbers Get F for band if (F < F(p 0)) maskPixel = 0 if (F >= F(p 0) and F < F(p1)) maskPixel = 1 ··· if (F >= F(p n – 1) and F < F(pn)) maskPixel = n if (F >= F(p n)) maskPixel = n+1 End loop over band numbers End loop over image voxels Output: Multi-value mask and associated color LUT Next, as a simple method to control for multiple tests (or limit the number of false positives) when looking at every voxel originally sampled within the full brain mask a “voxel limiting” spatial mask was produced as follows. The omnibus F-test images (related to 2 Rsr , a measure of model fit—see Section 2.3) at each center frequency were strictly threshold at p = 0.0001 to pro- duce binary masks for each band. Then the masks were combined to produce one spatial binary mask using a Boolean or operation. This mask enabled us to limit the number of voxels looked at with the specific inference tests for interaction, main and simple effects for the ANOVA design presented. These specific hypothesis tests were selected by using appropriate values for the contrast Figure 3. Representative omnibus multilevel F-test (and associated p-values) results showing some low frequency bands where significant activation occurred in the occipital regions of the brain for four alcoholic subjects. Each row compares two different alcoholic subjects at a particular band or center frequency. matrix B (see Table 1 Section 4.2) in Equation (9). Results for these tests are presented at multiple p-value levels (ranging from 0.001 to 0.01) and overlaid on the regis- tered structural image for the individual subject. These tests were structured by a scheme not typically used in the neuro-imaging field but closer to traditional multi- variate hypothesis testing procedures. First, hypothesis tests for interactions were strictly applied at a p-value threshold of 0.004 and a Boolean OR binary mask pro- duced (similar to that for the omnibus hypothesis tests) that included only those voxels that failed the interaction hypothesis tests (that is an image mask composed of voxels in which no interaction was seen). Only then were main effect hypothesis tests (e.g. contrast of positive versus negative input stimuli images regardless of the beverage currently shown) performed at those voxels for Copyright © 2012 SciRes. JSIP  Analysis of fMRI Single Subject Data in the Fourier Domain Acquired Using a Multiple Input Stimulus Experimental Design 476 which the interaction hypothesis tests were not signifi- cant. On the other hand, in order to better investigate the spatial pattern of the simple effects (e.g. the positive with alcohol combined stimulus image) hypothesis tests asso- ciated with single-input combined stimuli (as seen in Figure 1(a)) were not restricted to voxels in which the interaction hypothesis tests were accepted as would be typically done in rigorous statistically-based multivariate presentations. This may introduce a few possible false positive voxels in the masks for simple effects but was a concession to the importance of presenting their full spa- tial activation pattern (as is typically done in the neuro- imaging field). In fact this is the usual procedure for re- porting fMRI analysis as performed in the temporal do- main. Alternatively it would be completely wrong to in- vestigate voxels with hypothesis tests for main effects where hypothesis test for interactions are affirmative. While the hypothesis-testing logistics seem at first view complicated, Figure 4 (presented in the results Section 4.2) easily clarifies the process. Generally we feel that this type of presentation or a similar approach should be taken when investigating more complicated experimental designs such as presented in this paper, however they often are not [33]. Figure 4. Chart showing available hypothesis tests for in- teraction, marginal and simple effects for one alcoholic subject at one particular slice. 4. Results Only a selected sample of relatively interesting results is presented to demonstrate the utility of analyzing fMRI data in the Fourier domain on single subjects. Note that all functional related results are presented as mask on the subject’s co-registered structural image. 4.1. Spectral Power of the Input Stimuli and Some Selected Examples of the Spatial Patterns of the Power of the HTF and Temporal Shape of the HRF For ideal Poisson processes, the input functions temporal distribution would distribute power equally at all fre- quencies in the power spectrum. Of course in this case, where the stimulus is finite and the sampling is discrete (at a sampling rate of 400 ms), the shape of the power spectra is compromised from this ideal case as can be seen in Figure 2(a) but is much more uniform than for typical fMRI stimulus input designs. While the power spectrum is not uniform from frequency to frequency and decreases in power at higher frequencies, these features would subside for longer-duration experiments and the high frequency fall off would decrease for increasing sampling rates with point stimulations. Focusing further on the specific difference seen in an alcoholic subject we look at the total transfer power for both the omnibus case and the total transfer power asso- ciated for the negative with alcoholic and negative with non-alcoholic image stimuli. As mentioned before, these functions can give us a sense of the general response of the system (in this case the brain) to the input stimuli at every voxel. These results are presented in Figure 2(b). The color scheme in this omnibus mask is unscaled with higher power represented by red to lower power repre- sented by orange. However individual stimuli power masks (again the color scheme simply represents larger power if the color is darker) were normalized between- stimuli by virtue of having the inputs represented by bi- nary sequences. Thus it is possible to compare values across stimuli where a region of interest (ROI) was sam- pled in the insula region for this alcoholic subject. Here we see a 57% decrease in power between the negative stimuli with a non-alcoholic beverage to that with the alcoholic beverage for the region (green box) shown. This can be compared to the often-reported signal change in the temporal domain for the BOLD response of typically a few percent [38]. Using these results it might be possible to conclude that the BOLD response to a negative image in association with an alcohol image is diminished as compared to the BOLD response seen when a negative images is combined with a non-alcoholic image for this alcoholic subject. Copyright © 2012 SciRes. JSIP  Analysis of fMRI Single Subject Data in the Fourier Domain Acquired Using a Multiple Input Stimulus Experimental Design Copyright © 2012 SciRes. JSIP 477 Looking now at the banded (or frequency) power dis- tribution of the transfer function for selected brain regions in the alcoholic subject (Figure 5) shows an interesting result in that the occipital region of this subject shows dominant low frequency responses while the insula region response is generally at much higher frequencies. A neighborhood of selected voxels where the power has been plotted as a function of bands from these two regions shows this quite dramatically. It generally appears than that the occipital region response is slower and more ex- tended in the temporal domain than that for the insula, which shows a more peaked response due to the higher- frequency components. 4.2. A Sampling of Hypothesis Testing Results A synopsis of all hypothesis tests for this experimental design is presented in Table 1, for which a more com- plete description of these tests follows. Sample images of spatial masks associated with the hypothesis tests are Figure 5. A look at 3 × 3 voxel ROI neighborhoods in the occipital and insula regions of the brain from an alcoholic subject showing the frequency-banded power of the omnibus HTF (Nyquist cut off at band 35 has been applied) along with some slices showing the spatial extent of frequency bands that showed relatively high power (voxels having the maximum value represented in red). Note especially the difference in the transfer function frequency dependence between the occipital and insula regions of the brain. Table 1. Synopsis of hypothesis tests. Test Type Mask Used Contrast Matrix (B) Omnibus whole brain Identity Interaction omnibus mask BT=[1/2 –1/2 –1/2 1/2] Main Effects Emotional Valence non-interaction voxels only BT=[1/2 1/2 –1/2 –1/2] Beverage Type non-interaction voxels only BT=[1/2 –1/2 1/2 –1/2 Simple Effects—Individual Stimuli (Positive, Alcohol) omnibus mask BT = [1 0 0 0] (Positive, NonAlc) omnibus mask BT = [0 1 0 0] (Negative, Alcohol) omnibus mask BT = [0 0 1 0] (Negative, NonAlc) omnibus mask BT = [0 0 0 1] Simple Effects—Contrast (Pos, Alc vs. NonAlc) omnibus mask BT = [1 –1 0 0] (Neg, Alc vs. NonAlc) omnibus mask BT = [0 0 1 –1] (Pos vs. Neg, Alc) omnibus mask BT = [1 0 –1 0] (Pos vs. Neg, NonAlc) omnibus mask BT = [0 1 0 –1]  Analysis of fMRI Single Subject Data in the Fourier Domain Acquired Using a Multiple Input Stimulus Experimental Design 478 presented in Figures 3 and 4 for selected subjects and slices. Testing of the HTF for signal detection o H: 0A was performed using the omnibus F-sta- tistic (Equation (11)) at all center frequencies as previ- ously detailed (see Section 3.4). Examples of the thresh- old being applied at six adjacent voxels in the occipital brain region of the brain for a sample subject are pre- sented in Figure 3(a). In addition, some selected low frequency bands are presented for four alcoholic subjects in Figure 3(b) using multilevel p-values (see Section 3.4). These were chosen to demonstrate the spatial extent of the low-frequency activation (generally restricted to gray matter) among a number of subjects. Note that there is no expectation that the activations will occur in exactly the same bands across every subject since each subject’s transfer function can have a slightly different shape. But it is of interest to note that activation in the occipital re- gions for all four subjects shown have a major low fre- quency response. Also note the red regions (p < 0.0001) that were used to spatially restrict the more detailed hy- pothesis tests that follow (see Section 3.4). Next, hypothesis tests of the form o H: 0AB for interaction, main and simple effects for the experi- mental design presented were systematically performed using Equations (9) and (10) with the appropriate values for the B contrast matrix. A list of all hypothesis test, spatial mask applied and associated B contrast matrix values used for these tests can be seen in Table 1. Sam- ple results for these hypothesis tests can be seen in Fig- ure 4 for a sample slice that included the insula region of the brain. These hypothesis tests were performed in the following systematic order. First, hypothesis tests for interactions between the input stimuli for emotional con- tent and beverage were performed. Results from these tests for interaction (threshold of p < 0.004) were used for voxel selection where a binary mask was created by selecting voxels in which no interactions were seen (light green, p > 0.004). Tests for main effects were then re- stricted to the voxels within this mask. This prevents combining difference in the emotional states with the alcohol beverage input image and those same differences with the non-alcohol beverage input image when they have different slopes. Hypothesis tests for main effects were then applied and the results are presented in the top right hand side and bottom left hand side of Figure 4. Finally hypo thesis tests for simple effects associated with the four stimuli (see Section 3.1) were performed (see Ta b le 1) and the results are also presented in Figure 4 (center of figure along with a color key). Hypothesis tests for simple contrast can be easily found by scanning either vertically or horizontally for the desired test result. Note that color keys were not provided for all hypothesis tests results presented in Figure 4, however all color keys were generally the same (having the same range of p-values, dark 0.001 to light 0.02), similar to that shown for the individual stimuli. There is a main effect of emotional valence (positive versus negative) in the insula for this alcoholic subject (Figure 4, bottom left of figure). We also see a main effect of beverage type (alcohol versus non-alcohol), but it is somewhat muted. This is also clarified by looking at the simple effects hypothesis tests results (Figure 4, four images in the center). Here we see the hypothesis test results mask for the effect of the positive-alcohol stimu- lus is much stronger than the hypothesis test results mask for the negative-alcohol stimulus. This is further con- firmed by looking at the hypothesis test results for the simple contrast of positive versus negative emotion stim- uli with an alcoholic beverage image (presented Figure 4, bottom row, center). Finally it should be noted that in general both the occipital and insula regions of the brain for this alcoholic subject’s chosen brain slice show a reasonable activation (or hemodynamic response) spatial pattern for the hypothesis tests presented [33]. 5. Conclusions In conclusion we have presented a number of analytic techniques in the Fourier domain that provides a some- what new and different perspective into the analysis of fMRI BOLD data as compared to what have become the standard temporal-domain-based approaches. The meth- odology has a number of advantages that can be espe- cially useful for single subject fMRI BOLD data analysis obtained from complicated event-driven experimental designs. This has allowed us to analyze fMRI BOLD time series data for single subjects obtained at a sampling rate of 400 ms without resorting to unrealistic restrictions on the structure of the noise and to implement a full set of statistical hypothesis tests based on a complex general linear model framework. This is critically important for those designs that include relatively short inter-stimulus intervals and fast acquisition rates as presented in this paper, but could also be a potential problem in the analy- sis of data obtained from any fMRI experiment. This has enabled us to analyze fMRI time series data with a large number of sampled points (collected in a short time) and thereby increase statistical power for single-subject ana- lyses. Finally we have shown that it is possible to use tradi- tional signal processing and statistical techniques to in- vestigate and detect signal changes in fMRI BOLD time series data acquired from a relatively complicated evoked response experimental design. Toward this end we have implemented a number of investigative and statistical procedures and shown that in particular a Fourier-based Copyright © 2012 SciRes. JSIP  Analysis of fMRI Single Subject Data in the Fourier Domain Acquired Using a Multiple Input Stimulus Experimental Design 479 approach can be implemented for single subjects analysis of fMRI BOLD time series data with fast acquisition times. This gave robust results even for an experimental design that is challenging to analyze in the temporal do- main using a multistage group analysis approach [33]. We have also shown how to look at the analysis of these data in a systematic manner using a complex univariate multiple-regression methodology. Furthermore, this me- thodology approach to fMRI data analysis is still rela- tively novel and a number of extensions are in the proc- ess of being developed. These include techniques to bet- ter handle the multiple comparison problem (beyond what was implemented in the paper to restrict the hypo- thesis testing of main and simple effects by the general omnibus hypothesis test produced mask) and to more fully implement the complex general linear model in the Fourier domain to handle multivariate inputs (needed to analyze multiple fMRI runs data for an individual subject) and analyses for subject groups. REFERENCES [1] S. Ogawa, T. M. Lee, A. S. Nayak and P. Glynn, “Oxy- genation-Sensitive Contrast in Magnetic Resonance Im- age of Rodent Brain at High Magnetic Fields,” Magnetic Resonance in Medicine, Vol. 14, No. 1, 1990, pp. 68-78. doi:10.1002/mrm.1910140108 [2] S. Ogawa, D. W. Tank. R. Menon, J. M. Ellermann, S. G. Kim, H. Merkle and K. Ugurbil, “Intrinsic Signal Changes Accompanying Sensory Stimulation: Functional Brain Mapping with Magnetic Resonance Imaging,” Proceed- ings of the National Academy of Sciences, Vol. 89, No. 13, 1992, pp. 5951-5955. doi:10.1073/pnas.89.13.5951 [3] K. K. Kwong, J. W. Belliveau, D. A. Chesler, I. E. Gold- berg, R. M. Weisskoff, B. P. Poncelet, D. N. Kennedy, B. E. Hoppel, M. S. Cohen and R. Turner, “Dynamic Mag- netic Resonance Imaging of Human Brain Activity during Primary Sensory Stimulation,” Proceedings of the Na- tional Academy of Sciences, Vol. 89, No. 12, 1992, pp. 5675- 5679. doi:10.1073/pnas.89.12.5675 [4] K. Murphy, J. Bodurka and P. A. Bandettini, “How Long to Scan? The Relationship between fMRI Temporal Sig- nal to Noise Ratio and Necessary Scan Duration,” Neuro- Image, Vol. 34, No. 2, 2007, pp. 565-574. doi:10.1016/j.neuroimage.2006.09.032 [5] M. W. Woolrich, B. D. Ripley, J. M. Brady and S. M. Smith, “Temporal Autocorrelation in Univariate Linear Mo- deling of FMRI Data,” NeuroImage, Vol. 14, No. 6, 2001, pp. 1370-1386. doi:10.1006/nimg.2001.0931 [6] K. J. Worsley and K. J. Friston, “Analysis of Time-Series Revisited—Again,” NeuroImage, Vol. 2, No. 3, 1995, pp. 173-181. doi:10.1006/nimg.1995.1023 [7] K. J. Friston, A. P. Holmes, K. J. Worsley, J. P. Poline, C. D. Frith and R. S. J. Frackowiak, “Statistical Parametric Maps in Functional Imaging: A General Linear Approach,” Human B rain Mapping, Vol. 2, No. 4, 1995, pp. 189-210. doi:10.1002/hbm.460020402 [8] R. W. Cox, “AFNI: Software for Analysis and Visualiza- tion of Functional Magnetic Resonance Neuroimages,” Computers and Biomedical Research, Vol. 29, No. 3, 1996, pp. 162-173. doi:10.1006/cbmr.1996.0014 [9] K. J. Friston, W. Penny, C. Phillips, S. Kiebel, G. Hinton and J. Ashburner, “Classical and Bayesian Inference in Neuroimaging: Theory,” NeuroImage, Vol. 16, No. 2, 2002, pp. 465-483. doi:10.1006/nimg.2002.1090 [10] K. J. Worsley, C. H. Liao, J. Aston, V. Petre, G. H. Dun- can, F. Morales and A. C. Evans, “A General Statistical Analysis for fMRI Data,” NeuroImage, Vol. 15, No. 1, 2002, pp. 11-15. doi:10.1006/nimg.2001.0933 [11] C. F. Beckmann, M. Jenkinson and S. M. Smith, “General Multilevel Linear Modeling for Group Analysis in fMRI,” NeuroImage, Vol. 20, No. 2, 2003, pp. 1052-1063. doi:10.1016/S1053-8119(03)00435-X [12] S. M. Smith, M. Jenkinson, M. W. Woolrich, C. F. Beck- mann, T. E. J. Behrens, H. Johansen-Berg, P. R. Ban- nister, M. De Luca, I. Drobnjak, D. E. Flitney, R. K. Ni- azy, J. Saunders, J. Vickers, Y. Zhang, N. De Stefano, J. M. Brady and P. M. Matthews, “Advances in Functional and Structural MR Image Analysis and Implementation as FSL,” NeuroImage, Vol. 23, 2004, pp. 208-219. doi:10.1016/j.neuroimage.2004.07.051 [13] H. Zhang, W. L. Luo and T. E. Nichols, “Diagnosis of Single-Subject and Group fMRI Data with SPMd,” Hu- man Brain Mapping, Vol. 27, No. 5, 2006, pp. 442-451. doi:10.1002/hbm.20253 [14] T. E. Lund, K. H. Madsen, K. Sidaros, W. Luo and T. E. Nichols, “Non-White Noise in fMRI: Does Modelling Have an Impact?” NeuroImage, Vol. 18, No. 7, 2006, pp. 54-66. doi:10.1016/j.neuroimage.2005.07.005 [15] J. L. Marchini and S. M. Smith, “On Bias in the Esti- mation of Autocorrelations for fMRI Voxel Time-Series Analysis,” NeuroImage, Vol. 18, No. 1, 2003, pp. 83-90. doi:10.1006/nimg.2002.1321 [16] B. Lenoski, L. C. Baxter, L. J. Karam, J. Maisog and J. Debbins, “On the Performance of Autocorrelation Esti- mation Algorithms for fMRI Analysis,” IEEE Journal of Selected Topics in Signal Processing, Vol. 2, No. 6, 2008, pp. 828-838. doi:10.1109/JSTSP.2008.2007819 [17] K. J. Friston, K. E. Stephan, T. E. Lund, A. Morcom and S. Kiebela, “Mixed-Effects and fMRI Studies,” Neuro- Image, Vol. 24, No. 1, 2005, pp. 244-252. doi:10.1016/j.neuroimage.2004.08.055 [18] M. A. Lindquist and T. D. Wager, “Validity and Power in Hemodynamic Response Modeling: A Comparison Study and a New Approach,” Human Brain Mapping, Vol. 28, No. 8, 2007, pp. 764-784. doi:10.1002/hbm.20310 [19] M. A. Lindquist, J. M. Loh, L. Y. Atlas and T. D. Wager, “Modeling the Hemodynamic Response Function in fMRI: Efficiency, Bias and Mis-Modeling,” NeuroImage, Vol. 45, 2009, pp. 187-198. doi:10.1016/j.neuroimage.2008.10.065 [20] M. Luchtmann, K. Jachau, C. Tempelmann and J. Ber- narding, “Alcohol Induced Region-Dependent Alterations of Hemodynamic Response: Implications for the Statisti- Copyright © 2012 SciRes. JSIP  Analysis of fMRI Single Subject Data in the Fourier Domain Acquired Using a Multiple Input Stimulus Experimental Design Copyright © 2012 SciRes. JSIP 480 cal Interpretation of Pharmacological fMRI Studies,” Ex- perimental Brain Research, Vol. 204, No. 1, 2010, pp. 1-10. doi:10.1007/s00221-010-2277-4 [21] J. Tanabe, D. Miller, J. Tregellas, R. Freedman and F. G. Meyer, “Comparison of Detrending Methods for Optimal fMRI Preprocessing,” NeuroImage, Vol. 15, No. 4, 2002, pp. 902-907. doi:10.1006/nimg.2002.1053 [22] T. Johnstone, K. S. O. Walsh, L. L. Greischar, A. L. Alex- ander, A. S. Fox, R. J. Davidson and T. R. Oakes, “Mo- tion Correction and the Use of Motion Covariates in Mul- tiple-Subject fMRI Analysis,” Human Brain Mapping, Vol. 27, No. 10, 2006, pp. 779-788. doi:10.1002/hbm.20219 [23] G. H. Glover, T. Li and D. Ress, “Image-Based Method for Retrospective Correction of Physiological Motion Ef- fects in fMRI: Retroicor,” Magnetic Resonance in Medi- cine, Vol. 44, No. 1, 2000, pp. 162-167. doi:10.1002/1522-2594(200007)44:1<162::AID-MRM23 >3.3.CO;2-5 [24] R. Sladky, K. J. Friston, J. Tröstl, R. Cunnington, E. Mo- ser and C. Windischberger, “Slice-Timing Effects and Their Correction in Functional MRI,” NeuroImage, Vol. 58, No. 2, 2011, pp. 588-594. doi:10.1016/j.neuroimage.2011.06.078 [25] N. Lange and S. L. Zeger, “Non-Linear Fourier Time Series Analysis for Human Brain Mapping by Functional Magnetic Resonance Imaging,” Applied Probability & Statistics, Vol. 46, No. 1, 1997, pp. 1-29. doi:10.1111/1467-9876.00046 [26] J. L. Marchini and B. D. Ripley, “A New Statistical Ap- proach to Detecting Significant Activation in Functional MRI,” NeuroImage, Vol. 12, No. 4, 2000, pp. 366-380. doi:10.1006/nimg.2000.0628 [27] D. R. Brillinger, “TIME SERIES Data Analysis and The- ory,” Holden-Day, San Francisco, 1981. doi:10.2307/2530198 [28] D. R. Brillinger, “The General Linear Model in the De- sign and Analysis of Evoked Response Experiments,” Journal of Theoretical Neurobiology, Vol. 1, 1981, pp. 105-119. [29] D. E. Rio, R. R. Rawlings and D. W. Hommer, “Applica- tion of a Linear Time Invariant Model in the Fourier Do- main to Perform Statistical Analysis of Functional Mag- netic Resonance Images,” Proceedings SPIE, Vol. 3978, 2000, pp. 265-275. doi:10.1117/12.383406 [30] P. Bai, Y. Truong and X. Huang, “Nonparametric Esti- mation of Hemodynamic Response Function: A Fre- quency Domain Approach,” IMS Lecture Notes—Mono- graph Series. Optimality: The Third Erich L. Lehmann Symposium, Vol. 57, 2009, pp. 190-215. doi:10.1214/09-LNMS5712 [31] D. E. Rio, R. R. Rawlings, L. A. Woltz, J. B. Salloum and D. W. Hommer, “Single Subject Image Analysis Using the Complex General Linear Model—An Application to Functional Magnetic Resonance Imaging with Multiple Inputs,” Computer Methods and Programs in Biomedi- cine, Vol. 82, No. 1, 2006, pp. 10-19. doi:10.1016/j.cmpb.2005.12.003 [32] D. Rio, R. Rawlings, L. Woltz, J. Gilman and D. Hommer, “An Application of the Complex General Linear Model to Analysis of fMRI Single Subjects Multiple Stimuli Input Data,” Proceedings in Medical Imaging, Biomedical Ap- plications in Molecular, Structural, and Functional Im- aging, Vol. 7262, 2009. doi:10.1117/12.811811 [33] J. M. Gilman and D. W. Hommer, “Modulation of Brain Response to Emotional Images by Alcohol Cues in Al- cohol-Dependent Patients,” Addiction Biology, Vol. 13 No. 3-4, 2008, pp. 423-434. doi:10.1111/j.1369-1600.2008.00111.x [34] R. H. Shumway and D. S. Stoffer, “Time Series Analysis and Its Applications: With R Examples,” Springer, New York, 2006. [35] H. Akaike, “On the Statistical Estimation of the Fre- quency Response Function of a System Having Multiple Input,” Annals of the Institute of Statistical Mathematics, Vol. 17, No. 1, 1965, pp. 185-210. doi:10.1007/BF02868166 [36] P. J. Lang, M. M. Bradley and B. N. Cuthbert, “Interna- tional Affective Picture System (IAPS): Technical Man- ual and Affective Ratings,” The Center for Research in Psychophysiology, University of Florida, Gainsville, 1995. [37] A. M. Dale, “Optimal Experimental Design for Event-Re- lated fMRI,” Human Brain Mapping, Vol. 8, No. 2-3, 1999, pp. 109-114. doi:10.1002/(SICI)1097-0193(1999)8:2/3<109::AID-HB M7>3.0.CO;2-W [38] J. E. Desmond and G. H. Glover, “Estimating Sample Size in Functional MRI (fMRI) Neuroimaging Studies: Statistical Power Analyses,” Journal of Neuroscience Methods, Vol. 118, No. 2, 2002 pp. 115-128. doi:10.1016/S0165-0270(02)00121-8 |

