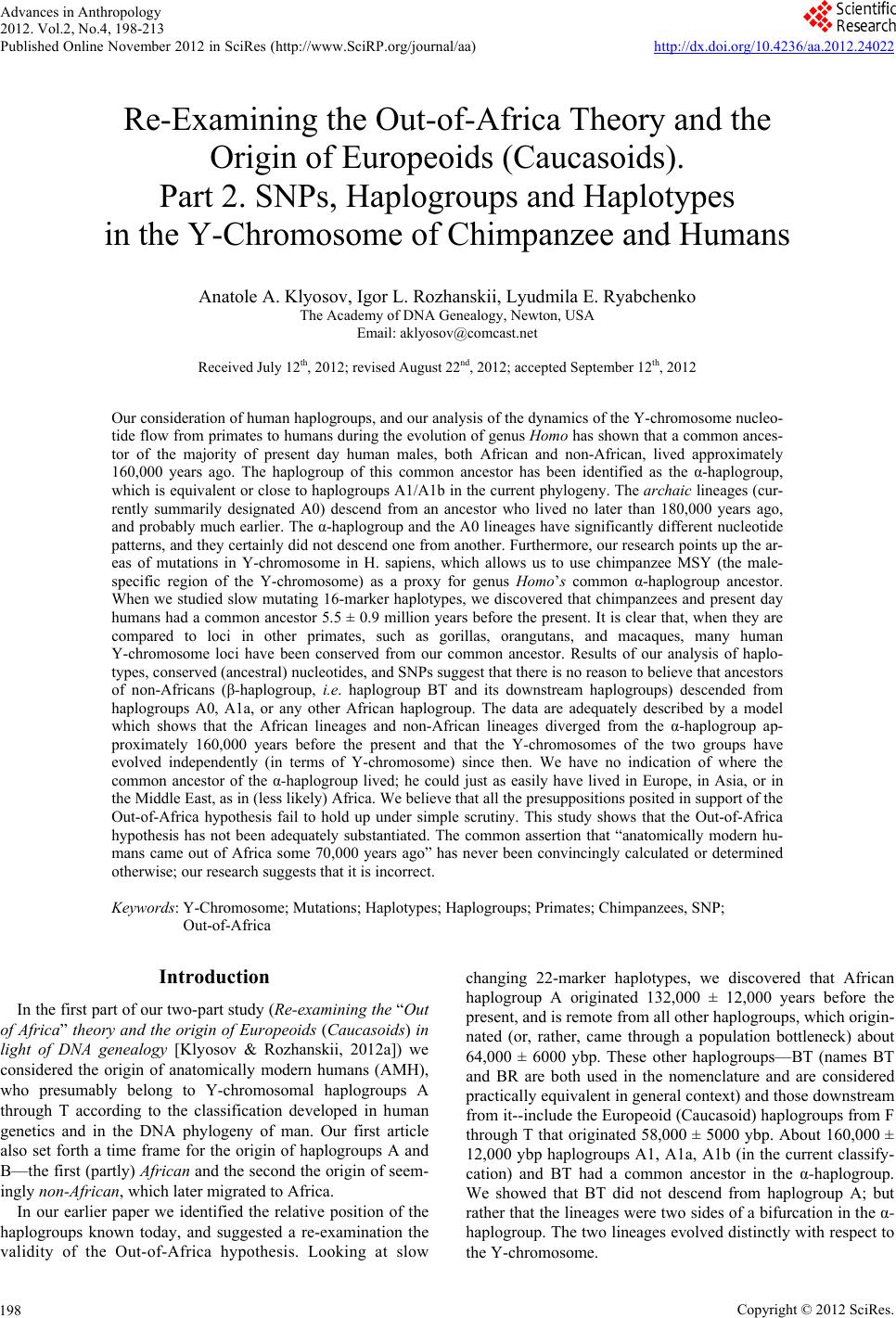
 Advances in Anthropology 2012. Vol.2, No.4, 198-213 Published Online November 2012 in SciRes (http://www.SciRP.org/journal/aa) http://dx.doi.org/10.4236/aa.2012.24022 Copyright © 2012 SciRes. 198 Re-Examining the Out-of-Africa Theory and the Origin of Europeoids (Caucasoids). Part 2. SNPs, Haplogroups and Haplotypes in the Y-Chromosome of Chimpanzee and Humans Anatole A. Klyosov, Igor L. Rozhanskii, Lyudmila E. Ryabchenko The Academy of DNA Genealogy, Newton, USA Email: aklyosov@comcast.net Received July 12th, 2012; revised August 22nd, 2012; accepted September 12th, 2012 Our consideration of human haplogroups, and our analysis of the dynamics of the Y-chromosome nucleo- tide flow from primates to humans during the evolution of genus Homo has shown that a common ances- tor of the majority of present day human males, both African and non-African, lived approximately 160,000 years ago. The haplogroup of this common ancestor has been identified as the α-haplogroup, which is equivalent or close to haplogroups A1/A1b in the current phylogeny. The archaic lineages (cur- rently summarily designated A0) descend from an ancestor who lived no later than 180,000 years ago, and probably much earlier. The α-haplogroup and the A0 lineages have significantly different nucleotide patterns, and they certainly did not descend one from another. Furthermore, our research points up the ar- eas of mutations in Y-chromosome in H. sapiens, which allows us to use chimpanzee MSY (the male- specific region of the Y-chromosome) as a proxy for genus Homo’s common α-haplogroup ancestor. When we studied slow mutating 16-marker haplotypes, we discovered that chimpanzees and present day humans had a common ancestor 5.5 ± 0.9 million years before the present. It is clear that, when they are compared to loci in other primates, such as gorillas, orangutans, and macaques, many human Y-chromosome loci have been conserved from our common ancestor. Results of our analysis of haplo- types, conserved (ancestral) nucleotides, and SNPs suggest that there is no reason to believe that ancestors of non-Africans (β-haplogroup, i.e. haplogroup BT and its downstream haplogroups) descended from haplogroups A0, A1a, or any other African haplogroup. The data are adequately described by a model which shows that the African lineages and non-African lineages diverged from the α-haplogroup ap- proximately 160,000 years before the present and that the Y-chromosomes of the two groups have evolved independently (in terms of Y-chromosome) since then. We have no indication of where the common ancestor of the α-haplogroup lived; he could just as easily have lived in Europe, in Asia, or in the Middle East, as in (less likely) Africa. We believe that all the presuppositions posited in support of the Out-of-Africa hypothesis fail to hold up under simple scrutiny. This study shows that the Out-of-Africa hypothesis has not been adequately substantiated. The common assertion that “anatomically modern hu- mans came out of Africa some 70,000 years ago” has never been convincingly calculated or determined otherwise; our research suggests that it is incorrect. Keywords: Y-Chromosome; Mutations; Haplotypes; Haplogroups; Primates; Chimpanzees, SNP; Out-of-Africa Introduction In the first part of our two-part study (Re-examining the “Out of Africa” theory and the origin of Europeoids (Caucasoids) in light of DNA genealogy [Klyosov & Rozhanskii, 2012a]) we considered the origin of anatomically modern humans (AMH), who presumably belong to Y-chromosomal haplogroups A through T according to the classification developed in human genetics and in the DNA phylogeny of man. Our first article also set forth a time frame for the origin of haplogroups A and B—the first (partly) African and the second the origin of seem- ingly non-African, which later migrated to Africa. In our earlier paper we identified the relative position of the haplogroups known today, and suggested a re-examination the validity of the Out-of-Africa hypothesis. Looking at slow changing 22-marker haplotypes, we discovered that African haplogroup A originated 132,000 ± 12,000 years before the present, and is remote from all other haplogroups, which origin- nated (or, rather, came through a population bottleneck) about 64,000 ± 6000 ybp. These other haplogroups—BT (names BT and BR are both used in the nomenclature and are considered practically equivalent in general context) and those downstream from it--include the Europeoid (Caucasoid) haplogroups from F through T that originated 58,000 ± 5000 ybp. About 160,000 ± 12,000 ybp haplogroups A1, A1a, A1b (in the current classify- cation) and BT had a common ancestor in the α-haplogroup. We showed that BT did not descend from haplogroup A; but rather that the lineages were two sides of a bifurcation in the α- haplogroup. The two lineages evolved distinctly with respect to the Y-chromosome.  A. A. KLYOSOV ET AL. The Essence of the Out-of-Africa Hypothesis The Out-of-Africa hypothesis arose in the middle of the 1980s, under heavy influence of DNA-related interpretations. One of the first articles (Stringer & Andrews, 1988), which initiated the idea, stated that “Genetic data on present human population relationships and data from the Pleistocene fossil hominid record are used to compare two contrasting models for the origin of modern humans”. We now know that early genetic data were inaccurate and that the hominid fossil record was (and still is) controversial. In the same year, by studying 42 world populations Cavalli-Sforza et al. (1988) found that the phylogenetic tree separates Africans from non-Africans. They took this as an indication that “the origin of modern humans was in Africa”, even though the opposite, that is “the origin of modern humans was not in Africa” would have been equally valid. Later, the Out-of-Africa hypothesis acquired five essen- tial presuppositions: 1) All haplogroups, from B to T, descended from haplogroup A. 2) The first anatomically modern human (AMH) came Out- of-Africa around 70,000 years ago. 3) Africa has the highest genetic diversity. Therefore, the AMH descended from African populations. 4) Genetic diversity decreases as the distance from Africa increases. Therefore, Africa is the homeland of mankind. 5) The earliest AMH was found in Africa. He presumably is the direct ancestor of all the human beings living today. We concluded in the first article of this set (Klyosov & Roz- hanskii, 2012a) that the Out-of-Africa hypothesis needed re-examination. The mutations in the Y-chromosome haplo- types of Africans and non-Africans clearly indicate that 1) non-Africans could not have descend from Africans; 2) haplogroup B which is about 120,000 years remote from haplogroup A, is not-initially-an African haplogroup; 3) African haplogroup A and all non-African haplogroups (BT) diverged from a common α-haplogroup ancestor who lived 160,000 ± 12,000 ybp. There is no indication that non-African haplogroups resided in Africa. Furthermore, it was recently recognized that there is no such thing as a single haplogroup A. Instead, haplogroup A is an umbrella for a variety of haplogroups, some of which originated more than 160,000 ybp. The Five Presuppositions of the Out-of-Africa Hypothesis Let us briefly consider the five presuppositions listed above, which are the underpinnings of the Out-of-Africa hypothesis. Presupposition 1 was maintained for many years by the In- ternational Society of Genetic Genealogy (ISOGG) with the following description: “The BR haplogroup split off from hap- logroup A 55,000 years before present (bp). It probably ap- peared in North East Africa” Similarly, the introduction to the FTDNA haplogroup A Project (see “Materials and Methods”) begins “Haplogroup A is unique in that all other human hap- logroups spring from its primal branches”. Another similar quotation—“Haplogroup A, first appearing 55,000 years ago, is the oldest of all Y haplogroups and is considered a direct ge- netic link to early man” (Chromosomal Laboratories, 2005). Since we know that the common ancestor of haplogroup A and all other haplogroups belonged initially to the α-haplogroup (which arose 160,000 ybp), and haplogroup B is remote from haplogroup A by at least 120,000 years, it is clear that hap- logroup BT (which arose 64,000 ybp) could not possibly have “split off from haplogroup A 55,000 ybp”. This chronological construction provides inadequate support to the Out-of-Africa hypothesis. Even if some researchers did not consider haplogroup A-M91 to be ancestral to the BR haplogroup (e.g. Cruciani et al., 2002), the literature abounds in interpretations which create an impression that African haplogroups were ancestral to non-African haplogroups. For example, Chiaroni, Underhill and Cavalli-Sforza (2009) state: “haplogroups A and B are the deepest branches in the phylogeny and are essentially restricted to Africa, bolstering the evidence that modern humans first arose there”. Actually, haplogroup B is not among the deepest branches, (cf. Figure 1). Second, Chiaroni, et al., do not indi- cate from which particular haplogroup “modern humans first arose [in Africa]”. From haplogroup A? No, haplogroup A (see Figure 1) is distinct from BT, as Cruciani, et al., pointed out in 2002. From haplogroup B? No, BT was initially one hap- logroup and haplogroup B split from it. From some chromoso- mal Adam 160,000 years ago evolving directly into non-Afri- cans? This cannot be serious. It seems that such Out-of-Africa statements have not been thought through in terms of Y-chro- mosome phylogeny. It is interesting to note that SNP M91 long considered an Af- rican SNP, has recently been re-assigned by ISOGG to non-African haplogroups (ISOGG-2012) (cf. Cruciani et al., 2002; ISOGG, 2006-2011). This move may have been an at- tempt to keep the nomenclature in line with the Out-of-Africa hypothesis. However, the respective nucleotide sequences in chimpanzee MSY and the BT haplogroup are identical (9T), while the African subclade A1a has 8T; therefore, M91 should belong to the African lineages. On the other hand, M91 is 8T in A1b, which is a part of the α-haplogroup. This means, that after its transition from the α-haplogroup to the β-haplogroup (BT) 8T9T, the sequence returned to its ancestral state, and the SNP in BT should be designated differently than M91. All of these data indicate that BT haplogroups did not descend from the “African haplogroup A”; additionally, the slow 22-marker haplotypes quite convincingly show this (Klyosov & Roz- hanskii, 2012a). More genome-based data will be provided below in this paper. Presupposition 2 became a cliché in almost every article in which the Out-of-Africa hypothesis has been mentioned. A few examples, specifying when AMH allegedly came Out-of-Africa, are: “50 thousand years ago” (Jobling & Tyler-Smith, 2003) “50 - 60 thousand years ago” (Shi et al., 2010; Mellars, 2011) “50 - 70 thousand years ago” (Hydjasov et al., 2007; Stoneking & Delfin, 2010) “60 thousand years ago” (Li & Durbin, 2011; Henn et al., 2011) “60 - 70 thousand years” (Ottoni et al., 2010) “60 thousand years ago” (Stewart & Stringer, 2012) “45 - 50 thousand years ago” (Fernandes et al., 2012) “50 - 65 thousand years before present” (Behar et al., 2008) “60 thousand years ago” (Chiaroni et al., 2009) “50 - 75 thousand years ago” (Patin et al., 2009) “50 thousand years ago” (Edmonds et al., 2004) “45 thousand years ago” (Moorjani et al., 2011) “50 - 70 thousand years ago” (Xue et al., 2005) “70 - 80 thousand years ago” (Majumder, 2010) Copyright © 2012 SciRes. 199  A. A. KLYOSOV ET AL. Copyright © 2012 SciRes. 200 Figure 1. Haplogroup tree of the H. sapiens Y-chromosome derived from haplotypes and subclades. This diagram shows the most recent common ancestors (TMRCAs) of H. sapiens as described in this study. To prepare this tree, we analyzed 7415 haplotypes from 46 subclades of 17 major haplogroups. The timescale on the vertical axis shows thousands of years from the common ancestors of the haplogroups and subclades. The tree shows the α-haplogroup, which is ancestral to both the African and non-African haplogroups, and the β-haplogroup, which is equivalent to haplogroup BT in the current classification. The left branch haplogroup A arose ~132,000 ybp. Haplogroups F through R (which includes T) represent Europeoids (Caucasoids) who arose ~58,000 years before the present. The Mongoloid and Austronesian haplogroup C split ~36,000 ybp; the Mid- dle Eastern haplogroups D and E split ~42,000 ybp (Klyosov and Rozhanskii, 2012a). logroup A or B. So, from whom might they have descended in Africa, and when? “40 - 80 thousand years ago” (Campbell and Tishkoff, 2010) Presupposition 3 is phrased variously: Hellenthal et al. (2008) say, “the haplotype diversity is highest in Africans”, (Campbell & Tishkoff, 2010), say that “Africa not only has the highest levels of human genetic variation in the world but also contains a considerable amount of linguistic, environmental and cultural diversity”. In reality, highest diversity does not by any means represent a homeland, unless the homeland has been totally isolated and has not been affected by populations from outside. New York City has a higher diversity than Boston, but that doesn’t mean that New York City is a homeland for Boston. Diversity quite commonly is a result of mixing, and Africa is a home of mixed populations, including quite ancient populations known presently as A0, A1, etc., plus haplogroup B, which did not descend from haplogroup A. When it arrived in Africa, it greatly increased the diversity there. The same is with a popula- tion of R1b-V88, which arrived in Africa on its migratory way along the Mediterranean Sea (Cruciani et al., 2010) and cur- rently resides in Cameroon and Chad, also contributing to di- versity in Africa. As it is shown below, lineages A0 do not only predate the α-haplogroup (A1/A1b), but evolved (in terms of Y-chromosome) independently on the latter. It is likely that A0 represent the African lineages, while the α-haplogroup with its downstream haplogroups represent the non-African ones. “55 - 70 thousand years ago” (Soares et al., 2009) “between 40 and 70 thousand years ago” (Sahoo et al., 2006) “between 35 and 89 thousand years ago” (Underhill et al., 2000) “between 80 and 50 thousand years ago” (Yotova et al., 2011) “between 50 and 100 thousand years ago” (Hublin, 2011) “between 27 - 53 and 58 - 112 thousand years ago” (Carri- gan and Hammer, 2006) “around 70 - 60 thousand years ago” (Curnoe et al., 2012). In fact, however, none of these authors seem actually to have calculated or otherwise obtained the numbers that they cite. Besides, at this point, there is no methodology which can cal- culate those numbers. The numbers are taken from thin air, and cross-cited in dozens of articles. It is astonishing to us that no scholar has ever questioned the figures. The Out-of-Africa hy- pothesis needed the figures, and they seem to have been pro- vided without justification. In fact, neither “haplogroup A” (there are now a number of them), nor any other haplogroup could possibly have come out of Africa 70,000 years ago, since the temporal distance between A and all other haplogroups is at least 120,000 years. Non-African haplogroups clearly did not descend from hap- In short, Africa is home to truly ancient (compared to BT) populations of various haplogroups designated presently as A,  A. A. KLYOSOV ET AL. and other haplogroups which arrived in Africa at different times. So, diversity by itself does not support the idea of a common homeland in this context. Presupposition 4 is stated clearly by Atkinson (2011) in a recent article in which he asserts that “human genetic and phe- notypic diversity declines with distance from Africa ··· under- pinning support for an African origin of modern humans ··· loca l language diversity ··· points to parallel mechanisms shaping genetic and linguistic diversity and supports an African origin of modern human languages”. This effort to shore up the Out-of-Africa hypothesis also fails. Since haplogroups A and BT diverged from the same α-haplogroup (Figure 1), this does not mean that the younger lineage descended from the older. Our research shows that they parallel each other such as two teeth in the proverbial fork. They descend not from each other, but from a common ancestor who was the α-haplogroup. Haplogroup R1a (20,000 ybp) and haplogroup R1b (16,000 ybp) (Klyosov & Rozhanskii, 2012b; Klyosov, 2012), arose in Central Asia (ibid.). Both are younger than haplogroup A (120,000 ybp) but they are not descended from haplogroup A. A boy is not a descendant of his older brother. The fact that the Indo-European (IE) languages are younger than the click lan- guages does not prove that IE languages descended from click languages or that Africa is a homeland of the languages of man. Presupposition 5, that the earliest AMH was found in Africa, is unconvincing for the following reasons: 1) We do not have the DNA of most fossils. Without identi- fying the haplotypes of fossil DNA, any statement about the relationship of fossils to present-day H. sapiens is irrelevant. 2) The oldest AMH skeletal fragments found in Africa carry many archaic features. They were not exactly AMH, and many clearly do not have modern H. sapiens morphology. Many skeletal fragments are poorly dated. None of the early fossils meet the stringent morphological criteria applicable to living H. sapiens (Tattersall, 2009). A number of authors have argued that the earliest hominids found in Sub-Saharan Africa had African regional morphological features (e.g. Habgood, 1989). Subsequent papers gradually began to claim that sub-Saharan hominids had AMH features. This issue is controversial and complicated; it is the subject of many articles (e.g. Rogers & Jorde, 1995; Hanihara et al., 2003; Tattersall, 2009; Pinhasi et al., 2011; Rightmire, 2009; Prat et al., 2011; Mellars & French, 2011) and by no means solved. 3) The AMH could have migrated to Africa from elsewhere. If our ancestors could have migrated out of Africa, they could equally well have migrated into Africa. 4) Few modern human skulls older than 20,000 years have been discovered in Africa south of Ethiopia. One of the oldest is from Hofmeyr, South Africa, dated to 36 ± 3 thousand years ago. Still, it displays some archaic features (Grine et al., 2007, 2010), although it is the age or even slightly younger than the bones of an AMH of 38 - 45 thousand ybp (e.g. Pinhasi et al., 2011; Prat et al., 2011; Hoffecker, 2011; Benazzi et al., 2011; Higham et al., 2011). Actually, the Hofmeyr skull demonstrates that humans in Africa 36,000 years ago resembled those in Eurasia, which leaves the question of who migrated where unsolved. While the Out-of-Africa hypothesis continues to be presented as truth, it is based on questionable presumptions. It should be noted that the earliest authors were careful in their descriptions of the origin of Africans and non-Africans; they did not claim that one group descended from the other. For example, Nei and Takezaki (1996) wrote: “[Our] results indicate that Africans are the first group of people that split from the rest of human populations”, and “the root of the human population tree still remains unresolved”. The above is only a brief account of the controversies in the anthropology of man and about the alleged African origin of H. sapiens. In this study, we focus mainly on the DNA evidence propounded to support the Out-of-Africa hypothesis. We have found that none of the evidence is convincing. As the principal methodology, we consider base (ancestral) haplotypes of hap- logroups A0, A, and B, determine timespans to their common ancestors as it was initiated in the preceding paper (Klyosov & Rozhanskii, 2012a), compare their haplotypes with that of chimpanzee MSY and show that the methodology of DNA genealogy works even on timespans of millions of years, com- pare ancestral nucleotides of Y-chromosomes of chimpanzee (and the DNA fragments of some other primates) to those in humans, based of that comparison differentiate lineages of haplogroups A0, A/A1b, A1a and their downstream subclades, assign the nucleotides to the α- and β-haplogroups, and finally show that the African and non-African lineages indeed split apart from the α-haplogroup; therefore, non-African lineages did not descend from the African ones. Both African and non-African lineages have evolved-in terms of their Y-chro- mosomes—concurrently (in parallel) from their common an- cestor who lived around 160,000 years before the present. One more set of the African lineages, collectively called A0 hap- logroups, has evolved independently (again, in terms of their Y-chromosomes) on the α- and β-haplogroups as well as on other African lineages. It seems that A0 are the most archaic African lineages, which arose at least 180,000 ybp and possibly much earlier. The Current Structure of Haplogroup A Currently, there is no unified haplogroup A. It is a loose set of lineages in which each has its own set of SNPs and the list of SNPs is growing. Some of them are shown below, thank to the researchers and Institutions named in the “Materials and Meth- ods” section. Figure 2 shows a haplotype tree of haplogroup A. It is based on tests collected by the FTDNA Haplogroup A Project. The tree consists of four principal parts. The most remote branch in the tree is the doublet of two relatives (1 and 2). They do not have SNPs that occur in other haplogroups (ISOGG designated their haplogroup as A0*), and have a base 22-marker haplotype as follows 13 11 12 - 10 11 - 16 - 10 9 14 14 8 8 8 9 12 11 12 8 12 12 11 11 (A0*) This is remote from all known haplotypes in the world; for example, A0* has DYS438 = 16—a slow marker in which a mutation happens about once in 45,000 years. All other hap- logroups have DYS438 = 9-11. In other words, A0* differs from other haplotypes by 5 to 7 mutations. This newly design- nated haplogroup A0* differs from the deduced ancestral hap- lotype of the composite haplogroup A described by Klyosov and Rozhanskii (2012a) by 27 mutations: 12 11 11 - 9 11 - 10 - 10 8 14 15 7 10 8 12 13 11 16 8 13 9 11 12 (A) The differences translate to 250,000 years, and place their common ancestor at 190,000 ybp. With respect to time, this Copyright © 2012 SciRes. 201  A. A. KLYOSOV ET AL. Figure 2. A 22 marker tree of 39 haplotypes of haplogroup A. These haplotypes are part of the FTDNA Haplogroup A Project. The upper right branch is haplogroup A1a (presumably M31); it consists of haplotypes from Finland, England, Switzerland, Cape Verde. Number 3—at about 4 o’clock on the figure—is from an African-American. His DNA has close matches in Cameroon and presumably belongs to A0, which is positive at SNP V152 (Simms et al., 2011), Four and 5 (both Afri- can-American) are positive for L896; the former has close matches in Ghana. One and 2 belong to close relatives who have been assigned to haplogroup A0*. The remaining 28 haplotypes belong to haplogroup A1b1b2b-M13/M32. common ancestor is older than the α-haplogroup (160,000 ± 12,000 ybp). As we will see later, A0* has a different pattern of nucleotides than α-haplogroup, which actually might coincide with the A1/A1b haplogroup in the current nomenclature. The A0* haplotype shown above differs from the base hap- logroup B haplotype described in Part 1 of this study (Klyosov & Rozhanskii, 2012a) by 29 mutations: 11 12 11 - 11 11 - 10 - 11 8 16 16 8 10 8 12 10 11 15 8 12 11 12 11 (B) There are 286,000 years between haplogroup A0* and hap- logroup B. Clearly, A0* could not be ancestral to haplogroup B, or to haplogroup BT. The upper right branch in the tree (haplotypes 6 - 11) belongs to haplogroup A1a; it is composed mainly of European haplo- types, and has the following base haplotype: 12 10 11 - 7 13 - 8 - 10 8 15 17 6 10 9 12 13 11 16 8 13 11 11 12 (A1a) It differs from the ancestral base haplotype of haplogroup A by 14 mutations (84,300 years). Using calculations based on 25 and 67 marker haplotypes, we have determined the age of the A1a branch of about 8500 ybp. Since the age of haplogroup A is 132,000 ± 20,000 years, and the A1a base haplotype is 14 mutations apart, their common ancestor lived 122,000 ± 15,000 ybp. This is haplogroup A within the margin of error, hence, A1a is a downstream subclade of haplogroup A. Generally, it is known from the phylogeny, and the haplotypes confirm it. Haplotypes 3, 4 and 5 on Figure 2 belong to three different lineages of A0. They differ by 12, 7 and 11 mutations, respec- tively, from the presumed base haplotype: 12 12 11 - 10 11 - 12 - 11 8 15 15 8 10 8 0 12 11 14 8 13 8 12 12 These differences correspond to conditional generations, or about 52 thousand years before the present. Another way of arriving at the most recent common ancestor (TMRCA) is using the permutation method described in Klyosov (2009). The permutation method does not require the base haplotype, and it is based on the sum of the squared muta- tion differences between each allele in all the loci (22 loci in this particular case) divided by the square of a number of hap- lotypes (3 haplotypes in this case) and by the mutations rate constant (0.00027 mutations per marker per 25 years). Since the number of squared permutations in these three haplotypes was 122, we obtain 122/22/9 = 0.616 mutations per marker, and the common ancestor lived 25 × 0.616/0.00027 = 57,000 years be- fore the present, which is close to the above value. 1685 2167 In the 28 haplotypes which form haplogroup A1b1b2b- M13/M32 on the left side of the tree, there is a young Saudi Arabia branch of four haplotypes with a base haplotype as fol- lows: 12 11 11 - 9 11 - 10 - 10 9 12 12 7 12 8 0 13 11 16 9 14 9 11 11 (A1b1b2b) It differs from the base haplotype of the rest of the hap- logroup by 40 mutations in the 67 marker format, and by 3 mutations in the 22 marker format. This translates to 12,300 years between their common ancestors. Most of the haplotypes of haplogroup A in the dataset are represented by people who report European or Middle Eastern backgrounds. Therefore, the premise that haplogroup A almost exclusively resides in Africa, is not accurate, at least based on data on hand, even if they are clearly not representative. We simply do not know the current distribution of haplogroup A carriers around the world, and we know even less that distribu- tion some hundred thousand years ago. How Old Is Haplogroup A0? Only four A0 haplotypes are currently known in the 22 marker format (numbers 1/2, 3, 4 and 5 on the tree in Figure 2): 13 11 12 - 10 11 - 16 - 10 9 14 14 8 8 8 9 12 11 12 8 12 12 11 11 (haplotype 1/2) 12 12 14 - 11 12 - 14 - 11 8 15 15 7 10 8 0 13 13 14 8 13 8 12 11 (haplotype 3) 12 12 11 - 10 11 - 11 - 11 8 15 15 8 10 8 0 11 13 14 7 13 9 12 13 (haplotype 4) 12 13 10 - 10 11 - 10 - 11 8 15 15 8 9 8 0 10 9 14 8 12 8 11 12 (haplotype 5) Since their ancestral haplotype is unknown, we need again to apply the permutation method (Klyosov, 2009) to determine how deep in time their common ancestor might have lived. The sum of the squared mutation differences between each of them is 310, and we arrive at 310/16/22 = .881 mutations per marker in the four A0 haplotypes. The common ancestor of the five A0 individuals lived 0.881/0.00027 × 25 = 81,550 years before the present. This value makes sense only if all four haplotypes belong to the same haplogroup, but we know that they don’t, therefore, the actual figure is >81,550 ybp. Since the haplotype of the doublet (1/2) is quite distant from the others, we can use another way to estimate a time span between the contemporary haplotype 1/2 and the base haplo- type of 3, 4 and 5. They differ by 23 mutations, which corre- spond to 186,000 years between them. Therefore, the common ancestor for entire A0/A0* lineages lived (186,000 + 52,000)/2 Copyright © 2012 SciRes. 202  A. A. KLYOSOV ET AL. Copyright © 2012 SciRes. 203 ≈ 120,000 years ago, again assuming that these haplotypes belong to the same haplogroup. This means that the actual time to their common ancestor is significantly higher than 81,550 ybp. The common ancestor of A0 and the other African haplo- types lived more than 180,000 years before the present (Klyosov & Rozhanskii, 2011). Let us move to the main subject of this study, namely, what the Y-chromosome nucleotides and SNPs show about the con- nection between haplogroup BT and haplogroup A. As we have shown above, haplogroup A cannot be ancestral to haplogroup BT. The two lineages diverged independently from the α-hap- logroup. Additionally, one of several lineages (currently de- signated as A0 and A0*) evolved independently of the hap- logroup A. Nucleotides and SNPs in the Y-Chromosome of H. Sapiens and the Y-Chromosome and/or DNA Fragments of Some Primates As the principal methodology of this study we consider nu- cleotides and SNPs in 1) some living primates of the family Hominidae (e.g. chim- panzee, gorilla, orangutan) and the macaque (of the family Cercopithecidae, tribe Papionini) 2) newly discovered lineages in a variety of haplogroups A, 3) haplogroup BT and its descendant haplogroups R1a and R1b (as specific examples of Europeoids [Caucasoids]). First, we should consider whether or not we are correct when we choose to compare nucleotides and SNPs in living primates and living humans. We can assume that human and chimpanzee genomes diverged 6.5 million years ago (cit. Green et al., 2010) or 6 million years ago (Hughes et al., 2012; Scally et al., 2012), or 5 to 7 million years ago (Prüfer et al., 2012). The tribe Papi- onini emerged 6 - 8 million years ago and the Hominidae fam- ily emerged 13 - 18 million years ago (Perelman et al., 2011). Following Cruciani et al. (2011) and Scally et al. (2012), and references therein, we can take 1.0 × 10−9 single-nucleotide substitutions per base per year as an estimate of the mutation rate in the Y-chromosome. This tells us that in 6.5 million years a nucleotide in the Y-chromosome would mutate 0.013 - 0.015, or between 1.3% and 1.5%. Whatever the exact figure, it is clear that it is safe to use primate nucleotides as proxies for nucleotides in present day humans. We know that chimpanzee Y-chromosomes and human Y-chromosomes are remarkably divergent in structure and gene content (Hughes et al., 2010); however, nucleotides in the Y-chromosome positions we have considered (see below) appear to have retained their structure quite remarkably (see Figure 3). Figure 3 shows a comparison of a 97-nucleotide human DNA fragment with the respective MSY fragment in chimpan- zee and the DNA fragments from the whole genome shotgus sequence of gorilla, pongo, and macaque. Eighty seven percent (84 of the 97 nucleotides) are identical in humans and in these primates (see Figure 3). The fragments contain nucleotide C (position 51) which becomes SNP P82 (mutation CA) con- verting haplogroup A1 into A1a. Other SNPs which mark this conversion are M31, V4, V14, and V25. One can see that the ancestral nucleotides in the DNA of these primates have been stable for 15 million years or so; thus, they can be employed for comparison with the respective nu- cleotides in human Y-chromosomes. Since the chimpanzee genome is by many accounts the closest to humans and the most thoroughly studied, and since the chimpanzee genome sequence is available (see Materials and Methods), it was employed in our further studies. How Long Ago Did the Split between the Common Ancestor of Chimpanzees and Humans Occur? To answer this question we have employed slow markers available for the 22-marker panel (Klyosov & Rozhanskii, 2012a); the respective alleles were determined for chimpanzee and human MSYs (see Materials and Methods). Overall, 16 markers out of the 22 were recovered in the chimpanzee Y-chromosome, as follows: DYS 426, 388, 392, 455, 438, 578, 641, 472, 425, 594, 436, 490, 617, 568, 640, 492. Six other markers in chimpanzee MSY did not have homology with the human MSY as a result of a rearrangements of genome that occurred during the past~6.5 million years. Therefore, the 16- marker haplotype of present-day chimpanzee MSY was ana- lyzed (see Materials and Methods) as follows: 8 15 10 4 5 9 10 5 10 4 4 7 4 4 8 9 In present-day H. sapiens males the respective haplotype is as follows. Only the predominant alleles in all haplogroups, particularly in the oldest ones, were included (see Materials and Methods): 11 12 11 11 10 8 10 8 12 10 12 12 12 11 11 11 The mutation rate constant for those 16 markers equals .00410- Figure 3. Comparison of the MSY fragments (1 - 97) from Humans and Chimpanzee, and the DNA fragments in the genome of Gorilla, Pongo and Macaque (whole genome shotgun sequence), along with a consensus tree fitting mutation patterns (inset).  A. A. KLYOSOV ET AL. AK mutations per haplotype (Klyosov, 2011). The two haplo- types differ by 64 mutations, or, by 4 mutations per marker on average. The correction for back mutations is determined as (1 + e4)/2 = 28 (ibid.). This means that the calculated time to the common ancestor of H. sapiens and Pan troglodytes must be multiplied by 28. The sixty-four mutation difference between the MSY human and chimpanzee haplotypes computes to a common ancestor equal to 64 0.004115,610437,080 conditional generations (the arrow indicates a correction for back muta- tions), or 10.9 ± 1.7 million years distance between the present day Y-chromosomes of chimpanzees and humans. In other words, the split occurred 5.5 ± 0.9 million years ago (the ob- tained timespan should be divided by two, since it separates the present day MSYs). This fits the numbers which are commonly cited by geneticists, e.g. 5.6 - 8.3 million years ago (Green et al., 2010) or 3.9 - 5.9 million years ago (Sun et al., 2011), or the figures of 6 and 5 - 7 million years ago, given in the preceding section of this paper. This result also shows that calculations using slow mutated markers can be employed even for millions of years back. Nucleotides and SNPs in the Y-Chromosomes of Chimpanzee and Humans Let us follow some mutations in the Y-chromosome from chimpanzee to several principal human lineages: A0, A1, A1a, A1b, and the β-haplogroup (which is close to or identical with BT). This might give us a clue to the direction of mutational flow and, therefore, to which lineage was derived from which. Here we have to adhere to one condition: we cannot call anyone of lineages African unless we know that it actually originated in Africa. We know only where the majority of bearers of the subclades presently live. For example, all 11 individuals as- signed to the A0 haplogroup live in the United States, with “unknown origin” marked on the list in the Haplogroup A Pro- ject (June 2012); one is marked as African American. In other words, we simply do not know where their ancestors lived, say, 100,000 years ago; we assume that they lived in Africa, how- ever, it is not enough to determine that “their common ancestor arose in Africa”. Seventeen individuals assigned to the A1a subclade and its four sub-lineages, currently live in England, Switzerland, Finland, Cape Verde, and the US. Most of them marked un- known origin; however, some of them indicated European countries as origin of their recent ancestors. Forty-six bearers of A1b1 and its downstream subclades currently live in England, Ireland, Scotland, Italy, Algeria, Turkey, Kuwait, Saudi Arabia, Tunisia (one), Chad (one), South Africa (one), the US. None of the individuals in the project belongs to upstream A1 or A1b subclades. Therefore, SNPs V168, P108 and V221, which are actually observed in BT (β-haplogroup) and down to hap- logroups R1a and R1b, as well as in parallel downstream sub- clades of A1b, could have been inherited as mutations from a common α-haplogroup ancestor of both sides of the bifurca- tion shown in Figure 1. In the description below, one should take into account that haplogroup A is a collective, a tentative grouping, which in- cludes all that is not in haplogroup BT. Indexes, which are (tentatively) assigned to haplogroup A subclades, change every few months or even weeks. There is nothing African in the indexes of the subclades. We can only assume, for the sake of explanation, that 1) the α-haplogroup in Figure 1 corresponds to A1/A1b haplogroups in the current classification; 2) one side of the bifurcation represented in Figure 1 leads to the β-haplogroup (currently BT or sometimes BR in the classifica- tion); and 3) another leads to the “African” subclades, hap- logroups A1a and downstream subclades of A1b. We can as- sume that the latter are “African”, since their SNP mutations are not observed in the β-haplogroup, including Europeoids. The word “African” is in the quotations marks since we actu- ally do not know where their common ancestors lived some 100,000 ybp or before (or after) that. A0 lineages. There are two principal types of A0 lineages: 1) those which retain the nucleotides found in MSY of living chimpanzee (Table 1), and 2) those which contain specific SNPs compared with the ancestral nucleotide in chimpanzees (Ta- ble 2). These two types make haplogroup A0 very distinct from those (Africans and non-Africans) ancestors of which descended from the α-haplogroup. For the most cases A1 A1bBT bearers, for example, have those 1)—type nucleotides mutated into the respective SNPs; and 2)—type nucleotides retained as ancestral in chimpanzee. It does not mean that the A0 bearers are closer to chimpanzees than to other humans. It means that the Y-chromosome in A0 bearers evolved (mutated) independ- ently compared to other “African” and “non-African” lineages. Besides, as shown above, A0 is significantly older than the α- haplogroup. As a result, H. sapiens living today have three identified thus far principal MSY lineages: 1) “Common African” (part of subclades of haplogroup A, branched from the α-haplogroup and formed the left-hand bifurcation in Figure 1); 2) “non- African” (descendants of the β-haplogroup, branched from the α-haplogroup and formed the right-hand bifurcation in Figure 1); and 3) bearers of A0 lineages. Those three principal MSY lineages are very different in terms of their Y-chromosomes, however, they are brought much closer together by maternal part of their DNA, so their resulting genome is adjusted with every new generation. In addition to those listed in Table 1, the following SNPs have been assigned to the BT haplogroup in the current classi- fication, and may be directly related to the α-haplogroup: L413, L418, L438, L440, L604, L957, L962, L969, L970, L971, L977, L1060, L1061, L1062, SRY10831.1, M42, M94, M139, M299, P97, PK1. The α-haplogroup. Although no individuals bearing A1 or A1b SNPs are listed in the FTDNA Haplogroup A Project or in the Walk Through YDNA Project, it can be safely assumed, based on the SNPs patterns, that the α-haplogroup includes A1 and A1b subclades. Haplogroup A (in its entirety) is a hap- logroup which split from the α-haplogroup some time after 160,000 ± 12,000 ybp; haplogroup A contains many mutated nucleotides (SNPs) which are not mutated in A0 lineages. The mutated SNPs in the α-haplogroup (A1/A1b) are inherited not only by the African subclades but also by the non-African sub- clades. All 77 mutations shown in Table 1 represent SNPs for the α- and β-haplogroups. The SNPs assigned to A1b are P108 and V221; the SNPs assigned to BT are V29, V59, V64, V235 (and those immediately below Table 1), total 27 SNP’s. The re- maining 50 mutations in Table 1 are the SNPs assigned to A1, according to the current classification. SNPs P305 and L986 are mistakenly assigned to the A0 subclades in the current version of ISOGG-2012 (June 2012); mistakenly, because they do not Copyright © 2012 SciRes. 204  A. A. KLYOSOV ET AL. Table 1. Ancestral nucleotides in chimpanzee MSY which are retained in A0 lineages, but are mutated in the α-haplogroups (i.e. in A1/A1b/BT). Nucleotides SNP Chimp A0 α-haplogroup C C T P108 A A G P305 A A C L985 G G A L986 T T A L989 A A G L990 A A T L1002 G G del L1003 T T C L1004 A A G L1009 C C A L1053 T T C L1085 G G C L1089 G G C L1090 A A G L1093 C C A L1098 A A G L1099 T T C L1101 C C T L1105 C C A L1013 G G A L1114 A A C L1116 T T C L1118 G G T L1120 C C T L1123 T T C L1124 A A G L1125 A A G L1127 C C T L1128 T T G L1130 A A G L1132 C C A L1135 A A G L1136 C C T L1137 Continued C C T L1142 A A G L1143 C C T L1145 A A G L1150 TAGG TAGG del L1153 G G C L1155 C C T V21 A A G V29 G G A V41 C C T V54 C C T V59 T T A V64 A A G V161 G G A V168 C C G V171 A A G V174 T T G V203 G G T V221 A A G V235 G G T V238 C C T V241 A A G V250 have the respective SNPs in A0, those SNPs belong to the α-haplogroup (see Table 1). Table 2 lists SNPs which belong to the A0 lineages; those nucleotides are retained ancestral in the α-haplogroup. It again shows the principal difference between A0 and BT. In addition to the mutated nucleotides listed in Table 2, the following ancestral nucleotides are mutated in A0 lineages but retained in the α-haplogroup: A—L1086, L1094, L1102, L1109, L1112, L1113, L1117, V79, V81, V141, V223, V242, C—L1088, L1097, L1100, L1103, L1104, L1107, L1108, L1111, V164, G—P114, L1087, L1091, L1096, L1106, L1115, L1119, V139, del—V152. A total 62 SNPs in A0 lineages are listed above plus 30 more (L981, L983, L988, L994, L1007, L1014, L1070, L1072, L1073, L1075, L1076, L1078, L1079, L1080, L1081, L1082, V150, V151, V153, V157, V158, V159, V161.1, V162, V164, V169, V170, V181, V195, V203) which are currently (June 2012) identified and listed in public sources (see Materials and Methods). The difference between the listed 52 mutations in A1/A1b (α-haplogroup) and the 92 mutations in A0 lineages compared to the ancestral nucleotides in chimpanzee MSY Copyright © 2012 SciRes. 205  A. A. KLYOSOV ET AL. Table 2. Ancestral nucleotides in chimpanzees which 1) are retained in the α- haplogroup A1 A1bBT and 2) show a mutation in A0 lineages. Nucleotides SNP Chimp A0 α-haplogroup A G A L529 A T A L896 C A C L982 G T G L984 C A C L991 A C A L993 T C T L995 TTA del TTA L997 T G T L998 C T C L999 C A C L1000 G T G L1001 A G A L1005 A T A L1006 C A C L1008 T C T L1010 C T C L1012 T C T L1016 T C T L1018 G A G V148 T G T V149 A G A V154 G A G V166 G A G V172 T G T V173 A T A V177 G T G V196 A G A V225 G A G V229 C T C V233 G A G V239 shows that A0 lineages and the α-haplogroup are remote from each other; however, these data should not be used for calcula- tions of age, because not all SNPs have been identified or as- signed as yet. A1a subclade. The A1a subclade is downstream from the A1 subclade; thus far, it has five SNPs identified (M31, P82, V4, V14, V25) which were not inherited by the β-haplogroup. It seems that A1a split from the α-haplogroup between 160,000 and 60,000 ybp, and eventually ended up in Africa, Europe, Asia (see above). Most identified bearers of A1a’s currently live in Europe or the US; needless to say, this does not tell us Subclades downstream from haplogrou much about where the subclade originated. p A1b. The SNPs of tly, only one bearer of the A1b1 subclade is listed in th y on nucleotides in ch nstream SNP-mutations occurred in the bo tides and the fo these subclades (e.g. A1b1, A1b1a and A1b1b2-M13) are not observed in the BT haplogroups, therefore, it appears that bear- ers of these subclades ended up in Africa some time after their ancestors split from the α-haplogroup (A1/A1b). However, even this suggestion cannot be certain since many A1b1b2- M13a bearers currently live in England, Ireland, Scotland, Italy, Turkey, Algeria, Kuwait, Saudi Arabia, as well as in Tunisia and Chad (cf. the FTDNA Haplogroup A Project). We do not know when or where the subclades downstream from A1b arose. Curren e FTDNA Haplogroup A Project (as origin unknown). The majority of identified bearers of the haplogroup A subclades belong to A1b1b2-M13 (60%); 23% belong to the A1a sub- clade, 15% belong to the A0 subclades). Once we have established that we can rel impanzee MSY which are either inherited or generally intact in the human Y-chromosome, or which mutate into the respect- tive SNPs in certain lineages of human male haplogroups, we can try to interpret the diagram in Figure 1 in terms of the an- cestral nucleotide and acquired SNPs flow. Figure 1 shows that incoming nucleotides of chimpanzee (from below on the dia- gram) to the α-haplogroup should either flow as unchanged, ancestral, non-mutated, or as mutated into the respective SNPs into the α-haplogroup and then, mutated, to the both sides of the tree. Therefore, those ancestral (from chimpanzee) and the SNP-mutated (in the α-haplogroup) nucleotides are expected to be present in both “common Africans” (not A0 bearers) and all non-Africans. It is exactly what is observed. It does not mean that non-Africans descended from an African common ancestor. Those African and non-African branches of the tree are parallel ones (see Figure 1). Additionally, dow th branches after they split from the α-haplogroup. Those include 1) SNPs of the A1a subclade and its (future) down- stream subclades, as well as downstream subclades of A1b, which are expected to be observed in Africa. However, only few of them have been identified there thus far due to a poor representation of African haplotypes. Those SNPs have not been found in bearers of haplogroups B through T, including most of non-Africans. Those also include 2) relatively recent SNPs in the descendents of the β-haplogroup, which (SNPs) have resulted in the 19 principal haplogroups (B to T) down- stream of the β-haplogroup. Those SNPs, of course, cannot be found among bearers of haplogroups A and A0. What is the pattern of intact (ancestral) nucleo llow-up SNPs? First, we will look at two A0 lineages, actu- ally identified in the WTY Project, and placed on the ISOGG- 2012 list as four subclades (A0, A0a, A0a1, and A0b). They are not associated with the α-haplogroup. When we compare them at 346 nucleotide sites, they differ from each other at 36 loci. The two A0 lineages differ from each other in three blocks of SNPs (between L92.2 and L348.2, between L979 and L1017, and between L1036 and L1058), and in 153 sites—almost half of the 346 nucleotides—they differ from the BT lineage. A pat- tern of STR-mutations show that A0 lineages are much more ancient than the α-haplogroup (Rozhanskii & Klyosov, 2011), and did not descend from it. The data shown above suggest that the α-haplogroup was the handle of the MSY fork from which the “common African” and non-African lineages diverged and evolved in parallel (see Figure 1); this pattern adequately de- Copyright © 2012 SciRes. 206  A. A. KLYOSOV ET AL. scribes the evolution of the Y-chromosome of H. sapiens for the last 160,000 years. This pattern leaves no room for the Out-of-Africa hypothesis. The deduced ancestral (base) haplotypes of haplogroups A an hat ana- to Conclusion Our consideration o through T, and our an an haplotypes, we di nucleo- tid hy Materials and Methods The SNPs cite1 and 2 and the te of the DNA with certain SNPs or STRs in /gb2/gbrowse/hschrY/ as follows: e.ftdna.com/index.php?name=Draft&parent=root and k d B are shown above; the estimated time to their common ancestors is between 132,000 ± 20,000 and 46,000 ± 5,000 years, respectively (Klyosov & Rozhanskii, 2012a). These two hap- lotypes differ from each other by as many as 18 mutations, which translates to 123,000 years between them. Their common ancestor lived approximately 160,000 ybp (Klyosov & Roz- hanskii, 2012a). One can see that haplogroup B could not possi- bly have descended from haplogroup A. However, the problem would be resolved if the two had a common ancestor who lived 160,000 ybp. We do not know where this common ancestor lived; there certainly is no indication that he lived in Africa. Ne- anderthals did not live in Africa and, as it is known, did not have a melanin-rich skin. Thus, it is not necessary to presume that all lineages of the genus Homo had their origin in Africa. Based on palaeoarchaeological evidence, it appears t mically modern humans are likely to have originated some- where in the vast territory from West Europe across the Russian Plain in the east, and to the Levant in the south. Each of these regions is renowned for the age of the skeletal remains of mod- ern humans dating back to 45,000 - 40,000 ybp (for the latest references see Highham et al., 2011; Benazzi et al., 2011). f haplogroups A alysis of the dynamics of the Y-chromosome nucleotide flow from primates to humans during the evolution of genus Homo has shown that a common ancestor of the majority of present day human males, both Africans and non-Africans, lived 160,000 ± 12,000 years ago. This common ancestor has been identified as belonging to the α-haplogroup, which is equiva- lent or close to haplogroups A1/A1b in the current phylogeny. The archaic lineages (currently designated A0) descend from ancestor who lived at least 180,000 years ago (or much ear- lier). The α-haplogroup and the A0 lineages have nucleotide patterns that are distinct from each other, and both partly retain ancestral MSY chimpanzee nucleotides and partly have them mutated into the respective SNPs. Our research has shown that at least 90% of chimpanzee MSY nucleotides are the same as the nucleotides of living H. sapiens males. This comparison points up the areas of change in H. sapiens, which allows us to use chimpanzee MSY as a proxy for genus Homo’s common α- haplogroup ancestor. It is clear that when they are compared to loci in the DNA of other primates, such as gorillas, orangutans, and macaques, many human Y-chromosome loci have been conserved from our common ancestor. When we studied slow mutating 16-marker scovered that chimpanzees and present day humans had a common ancestor 5.5 ± 0.9 million years before the present. This date is in agreement with dates obtained in genome studies. It is important, since it shows that the DNA genealogy ap- proach can be extended to millions of years in depth. The results of our analysis of haplotypes, conserved es, and SNPs suggest that there is no reason to believe that non-Africans (haplogroup BT and its downstream haplogroups) descended from haplogroups A0, A1a, or other African hap- logroups. These data are adequately described by a model which shows that both the African lineages and non-African ones diverged from the α-haplogroup approximately 160,000 years before the present and that the Y-chromosomes of the two groups have evolved independently since then. Of course, the Y-chromosome constitutes only a small part of the whole ge- nome, and the term “evolve” here is related to the human MSY only. It can serve, however, a very useful “probe” to trace the human evolution with all reservations concerning the genome. We have no indication of where the common ancestor of the α- haplogroup lived; he could just as easily have been from Europe, Asia, or the Middle East, as from Africa. We believe, however, that only the A0 lineages represent the “truly African” autochthonous inhabitants (along with the respective female lineages), and interbreeding with them of incoming lineages of haplogroups A and B (as well as some lineages of haplogroups E, R1b, etc.), again, with the respective female lineages, created the very diverse current African population. Only further stud- ies will show whether this hypothesis is valid. We believe that all the presuppositions of the Out-of-Africa pothesis fail to hold up under simple scrutiny. This study shows that the Out-of-Africa hypothesis has not been ade- quately substantiated. The common assertion that “anatomically modern humans came out of Africa some 70,000 years ago” has never been convincingly calculated or obtained otherwise. The Out-of-Africa hypothesis has never been proved; our research suggests that it is incorrect with respect to anatomically modern humans. d in this study (see, e.g. Tables xt) were identified by Thomas Krahn of FTDNA Genomics Research Center (indicated by beginning letter L, such as L985), Rozaria Scozzari and Fulvio Cruciani of Universita La Sapi- enza, Rome, Italy (letter V), Peter Underhill of Stanford Uni- versity (letter M) and Michael Hammer of University of Ari- zona (letter P) and publicly available in various databases (see below). Since sequences of extended fragments of Y-chro- mosome are published in databases only for humans, chimpan- zee, and macaque, search for homologous fragments for gorilla and orangutan has been conducted in genomes of the primates, listed in the European Nucleotide Archive (ENA) database, employing Whole Genome Shotgun (WGS). Human and pri- mate gene sequences were aligned using the National Center for Biotechnology Information (NCBI) GenBank and ENA (the links are listed below). Search for fragments human Y-chromosome has been conducted using the FTD- NAY-chromosome Browser http://ymap.ftdna.com/cgi-bin (1) introduce the target SNP or STR into the field Landmark or Region, click on Search, chose a size of the target fragment (100 bp is recommended for an SNP), copy that fragment and find a homologous one in GenBank or ENA, using option BLAST. SNPs in particular haplogroups were those identified at sites http://ytre http://www.isogg.org/tree/. Haplotypes of haplogroup A to compose the tree in Figure 2 are listed in the FTDNA Project. The base haplotypes of haplogroup A and B were determined in (Klyosov & Rozhanskii, 2012a). Repeat motifs for the STRs (http://www.smgf.org/ychromoso me/marker_details.jspx? mar er and http://www.genebase.com/in/dnaMarkerDetail.php) are Copyright © 2012 SciRes. 207  A. A. KLYOSOV ET AL. listed below: DYS426 = 11 TGAAAGCATGACCACTTCATT n repeats (GTT) in the greatest number of haplogroups: am Chimpanzee (Y-Chromosome) ght repeats (GTT)] e (GTT)7 on the 14th chromosome, or DYS388 = 12 TGATTAGCCGTTTAGCGATAT test number of haplogrou- ps Chimpanzee (Y-Chromosome) DYS392 = 11 GTCATCGCAGTGGCCCAAGTGA repeats (TAT) in the greatest number of haplogroups: am CCCAAGTAA GTTGCTCCA [T GAGAATGATACTGCCTAAGCC AAGGCTGCAGTGAGCTGTGAT f hap- lo ups world- w GCCTAAGCC AGTGTGATC D CACATTGTTGTTAAGCCCAGC CAATCTCCCTGTGGTCGGGGC r of hap- lo s in haplogroups world- w osome has no DYS454 GTGGGGAATAGTTGAACGGTA TTTTCTTTTCTTTTCTTTTCT CACCACAACCTCCACTTCCCAGGTTCAAGC CTCAAAGTA TATTGTGTTGTTGTTGTTGTTGTTGTTGTT GTTGTTGTTGACACAAAGTCTCGTCTTGTC ACC [Eleve ong 41 base haplotypes in haplogroups world-wide there are 33 with the allele 11 (including the beta-haplogroup), and eight with 12 (mainly haplogroup P and its downstream haplogroups) (Klyosov & Rozhanskii, 2012)] CTCAAAGTATGAAAGCATGACCACTTCATT TAGTTGTTTTTTTGTTGTTGTTGTTGTTGT TGTTGTTGACACAAAGTCTCGTCTTGTCAC C [Ei NOTE: Macaques hav angutans have (GTT)7 on the 17th chromosome GAATTCATG ATACATATTATGAAACATTATTATTATTAT TATTATTATTATTATTATTATTTGAGACGG ACTCTCGCTCTGTCGCCCAG [Twelve repeats (ATT) in the grea : among 41 base haplotypes in haplogroups world-wide there are 28 with the allele 12 (including the beta-haplogroup), seven with 13, two with 14 (I and I1), one 16, one 15 (J1 and J2, re- spectively), one 11 (A), and one with 10 (B2a2) (Klyosov & Rozhanskii, 2012)]. GAATTCATGTGAGTTAGCCGTTTAGCGATA TATACATATTATGAAACATTATTATTATAT ATTATTATTATTATTATTATTATTATTATT GAGACGGACTCTCGCTCTGTCGCCCAG [15 repeats (ATT)] TAGAGGCA TCTTGCAACATCTCCATCCATGTTGCTCCA AAGGACCCAATTTTACTGTAAATGGTTGTA TAGTATTTTATGGTCTACATAGACCATATT TACCATATGTTCATCCATATTTTCTTCATT AATCTAGCTTTTAAAAACAACTAATTTGAT TTCAAGTGTTTGTTATTTAAAAGCCAAGAA GGAAAACAAATTTTTTTCTTGTATCACCAT TTATTTATTATTATTATTATTATTATTATT ATTATTATTTACTAAGGAATGGGATTGGTA GGTC [Eleven ong 41 base haplotypes in haplogroups world-wide there are 27 with the allele 11 (including the beta-haplogroup), six with 13, three with 12, three with 14, one with 10 (R2) and one with 7 (D3a) (Klyosov & Rozhanskii, 2012)]. Chimpanzee (Y-Chromosome) TAGAGGCAGTCATTGCAGTGG TCTTGCAACATCTCCATCCAT AAGGACCCAATTTTACTGTAAATGGTTGTA TAGTATTTTATGGTCTACATAGACCATATT TACCATATGTTCGTCCATATTTTCTTCATT AATCTAGCTTTTAAAAACAACTAATTTGAT TTCAAGTGTTTGTTATTTAAAAGCCAAGAA GGAAAACAATTTTTTTCTTGTATCACCACT TATTTATTATTATTATTATTATTATTATTA TTATTTACTAAGGAATGGGATTGGTAGGT en repeats (TAT)] DYS455 = 11 CTGAGCCGA CACAAGGTC CACCCGAGGGCACTCCAGCCTGGGCAACAC TGTGAGACCATATATCTAAAATAAATAAAT AAATAAATAAATAAATAAATAAAT AAATAA ATAACGGAAGAACACTCGTTTCCACCCC [Eleven repeats (AAAT) in the greatest number o groups: among 41 base haplotypes in haplogro ide there are 33 with the allele 11, four with 10, two with 9, and one with 8 (I1) (Klyosov & Rozhanskii, 2012)]. Chimpanzee (Y-Chromosome) CTGAGCCGAGAGAATGATACT CACAAGGTCAAGGCTGCAGTG ACCCGAGGGCACTCCAGCCTGGGCAACACT GTGAGAGCATATATCTAAAATAAATAAATA AATAACGGAAGAA [4 repeats (AAAT] YS454 = 11 GACTGACCT AACATATCA ACAGGCAAAAGCAAAATA AATAAATAAATA AATAAATAAATAAATAAATAAATA AATAAC CTAGGTGCTAATCCAAGTGATATGTTACAA TGTTTCCTGTTGACACAACCCAACCTGGGT GAAGTGAAGAGCTACATGTC [Eleven repeats (AAAT) in the greatest numbe groups: among 41 base haplotype ide there are 32 with the allele 11, eight with 12, and one with 13 (T1a) (Klyosov & Rozhanskii, 2012)]. Chimpanzee (Y-Chromosome) NOTE: The chimpanzee Y-Chrom homologous fragments. DYS438 = 10 CCAAAATTA AACAGTATA TTTCTTTTCTTTTCTTTTCTTTTCTTTTCT ATTTGAAATGGAGTTTCACTCTTGTTGCCC AGGCTGAAATGCAATGGTGTGATCTCGACT Copyright © 2012 SciRes. 208  A. A. KLYOSOV ET AL. GATTCTCCTGCATCAGCCTCCCAGGTAGCT GGGATTATAGGCGTCTGCCACCACGCCCAG CTAATTTTTTGTGTTTTTAGTAGAGACAGG GTTTCACCATGTTGGTGAGGCTGGTCTCGA ACTCCAGACCCTGGGTGATC [Ten repeats (TTTTC) in the greatest number of haplogroups: am there are 27 ta-haplogroup), eight with 11 A TTCTTTTGT CACTCTTGT - m D CATTCAAATCCTCTCCCACT TAAATAAATAAATAAATAAATA AATAAATAAATAAAAAGCCTTT f hap- lo world- wncluding the beta-hap- lo osome has no DYS531 ho D ACTCTAGCCTGGATGACACAAC ATCTCAAATAAATAAATAAATA pes out of; 10 in F; anskii, 2012)]. TGACAGAAC AAATAAATA GACAACAATTGTGTGTAAGTGACAAAATGT D AACTGAAAATCAT TTTTGTTTTGTTTTG TGTTTTGTTTTGTTTTGTTTTT TTCTTGCTCTGTCACCC plotypes ou & Roz- ha C GAGACTCTTCCATCTAAATAA ATAAATAAATAAATAAATAAAT types out of Rozhan- sk C GCCCGCTGCTCTCCAGCCTGG TCTAAATAA ATAAATAAAT D CACTCCAGCCTGGCG AAT TAATAATAATAAT GCCTCTTTG AGTGCCTC groups] C TAATAGTAAT TTGCTGAAC llow the hu -chromo- some, however, shows (TAA)4 on the 3rd chromosome. ong 41 base haplotypes in haplogroups world-wide with the allele 10 (including the be , and six with 9 (Klyosov & Rozhanskii, 2012)]. Chimpanzee (Y-Chromosome) CCAAAATTAGTGGGGAATAGTTGAATGGT AACAGTATATTTTCTTTTCTT TTTCTATTTGAAATGGAGTTT TGCCCAGGCTGAAATGCAATGGTGTGATCT CGACTCACCACAACCTCCACTTCCCAGGTT CAAGCGATTCTCCTGCATCAGCCTCCCAGG TAGCTGGGATTATAGGCGTCTGCCACCATG CCCAGCTAATTTTTTGTGTTTTTAGTAGAG ACAGGTTTTCACCATGTTGGTGAGGCTGGT CTTGAACTCCAGACCCTGGGTGATC [Repeats (TTTTC)3(TTTTG)1(TTTTC)1] [described in (Gus ao et al., 2002)] YS531 = 11 GACCCACTGG GCAAAAAA AATAAATA CGTCTACAAAGAAAGGGAGCA [Eleven repeats (AAAT) in the greatest number o groups: among 41 base haplotypes in haplogroups ide there are 30 with the allele 11 (i group), 10 with 10, and one with 12 (I2*) (Klyosov & Roz- hanskii, 2012)]. Chimpanzee (Y-Chromosome) GACCCACTGGCATCCAAATAACCTCCCTTG GCAAAAAAAAAA… NOTE: The chimpanzee Y-Chrom mologous fragments. YS578 = 8 GAGGCGGAACTTTCAGTGAGCCGAGATCAC GCCACTCC AAAACTCC AATAAATAAATAAATAAA GTAAGTAAGACA GACAACAACTGTGTGTAAGTGACAAAATGT CCAGGGTTGTTGAAGC [Eight repeats (AAAT) in 36 haplogroup base haploty 41 world-wide, except 9 in C3, F3 and O, NO and O d 7 in J2 (Klyosov & Rozhan Chimpanzee (Y-Chromosome) GAGGCGGAACTTTCAGTGAGCCGAGATCGC GCCACTCCGCTCTAGCCTGGG AAGACTCCATCTTAAATAAAT AATAAATAAATAAATAAAT AAATAAAGACA CCAGGGTTGTTGAAGC [Nine repeats (AAAT)] YS590 = 8 GAACATAGTCGGGCT AGTTGGGCAAGTTTT GT TTTTGTTT GAGACGGA [Eight repeats (TTTTG) in 39 haplogroup base ha t of 41 world-wide, except 7 in A and L (Klyosov nskii, 2012)]. himpanzee (Y-Chromosome) NOTE: The chimpanzee Y-Chromosome has no DYS590 homologous fragments. DYS641 = 10 CTTGAGCCCAGGAAGCATAGGTTGCAGTGA GCTGAGATCGCCTGCTGCTCTCCAGCCTGG TGATAGAGA ATAAATAA AAATGCTCCACAGCTAGGTGATATTGTTAA TTGTTAGGTAACAGTTATTACAGACAAGAA AGCCTAATTTACAAATAAAGGACAAAATTC ATCGTGTGG [Ten repeats (TAAA) in 39 haplogroup base haplo 41 world-wide, except 7 in NO and O (Klyosov & ii, 2012)]. himpanzee (Y-Chromosome) CTTGAGCCCAGGAAGCAGAGGTTGCAGTGA GCTGAGATT TGATAGAGAGAGACTCTTCCG ATAAATAAATAAATAAAT AA AAAAAGTTGCTCCACAGCTAGGTGATATTG TTAATTGTTAGGTAACAGTTATTACAGACA AGAAAGCCTAATTTACAAATCCAGGACAAA ATTGCATCGTGTGG [Ten repeats (TAAA)] YS472 = 8 AGATTGTCCCACCT ACACAGGAAGGTTCCATCTCAAATAGT G AATAATAA CTGAACAC [Eight repeats (AAT) in base haplotypes of all haplo himpanzee (Y-Chromosome) AGATTGTCCCACCTGCACTCCAGCCTGGCG ACACAGGAAGGTTCCATCTCAAATAGTAAT AATAATAATAATGATAATAA AATAATAATAATAATGCCTCT ACAGTGCCTC [Five repeats (AAT) in this particular case if we fo man homology]. NOTE: Macaques does not show homology on Y Copyright © 2012 SciRes. 209  A. A. KLYOSOV ET AL. O he 11th chromosome. AA CTATTGGAGGAGGAGAACCCT AGAAGAAGAGAGAAACAGGCT pes out of 20 C TATAGAAGGCAAGCAAGCTAA GAGAACCCT GAAACAGGCT D AATAAATCTAAAGCACATAAA AATAAAATAAAATAAAATAAAA of hap- lo world- weta-hap- loh 13 (Klyosov & R GCACATAAA TAAAAAAAAC D AGCAG TGTTGTTGTTGT TATCACCTCTAACGCAGCTCG CTGTGTCCTGCTTCCATTGGC f hap- lo s world- we beta-hap- lolyosov & Rozhanskii, 2012)]. TGTCCTGCT GC D ATTATTATTATT AGTCTTGCTCTGTCACCCAGGC GTGGCGTGATCTCAGCTCAGT of hap- lo s world- wbeta-hap- lo3, three with 15 and one with 16 (Klyosov & AGTCTTGCTCTGTCAC ATCTCGG D TTTCTATTGCAGCA ACATAGGTGTGTC TGTGTGTGTGTGTAAGCATGTA CGGTTGAACAAACTAGGATCAC (Redd et al er, among 41 are 33 w beta-haplogroup) and eight with 7 (K ATAGGTGTGTG TGTGTGTG TGTTGAACA rangutan shows (TAA)4 on the 18th chromosome and (TAA)2 (TGG)1 (TAA)4 on t DYS425 = 12 GTGGGCTGAGAAATTTCTGGAGACACTTCT CTTTCTGTCTATAGAAGGCAAGCTAGCT CTCTCTTCA GGATTGGAG CTAGAATTTAGAAAAATGTTGTTGTTGTTG TTGTTGTTGTTGTTGTTGTTGTTTAATTTC CATTTTACCTCCAGAATTACT [Twelve repeats (TGT) in 36 haplogroup base haploty 41, except G2a, G2c, H, H1, L (Klyosov & Rozhanskii, 12)]. himpanzee (Y-Chromosome) GTGGGCTGAGAAATTTCTGGAAACATTTCT CTTTCTGTC CTCTCTTCACTATTGGAGGAG GGATTGGAGAGAAGAAGAGA CTAGAATTTAGAAAAATGTTGTTGTTGTTG TTGTTGTTGTTGTTGTTGTTGTTTAATTTC CATTTTACCTCCAGAATTACT [Ten repeats (TGT)] YS594 = 10 GATGTGCCTAATG AAAATGGTAAATATTATATTATATTGTATA CCACAGAATGTATACTT TATAACAAT AGAAATAA TAAAATAAAATAAAATAAAATAA AAAA AAC AGAAAATACTCGACTATTGGTAAGAACGTT GCAGCCCCAATGGAAAAGAGCCAGAAAGTT ACTCAAAGAAATAAAGAAATAGGAAGTTGG ACTGAGGACACGATTAACACCAGGG [Ten repeats (TAAAA) in the greatest number groups: among 41 base haplotypes in haplogroups ide there are 22 with the allele 10 (including the b group), 14 with 11, four with 12 and one wit ozhanskii, 2012)]. Chimpanzee (Y-Chromosome) GATGTGCCTAATGCCACAGAATGTATACTT AAAATGGTAAATATTATATTATATTGTATA TATAACAATAATAAATCTAAA AGAAATAAAATAAAATAA AA AGAAAATACTCGACTATTGGTAAGAACGTT GCAGCCCCAATGGAAAAGAGCCAG AAAGTT ACTCAAAGGAATAAAGAAATAGGAAGTTGG ACTGAGGACACGATTAGCACCAGGG [4 repeats (TAAAA)] YS436 = 12 CCAGGAGAGCACA GGTTGTTGTTGTTGTTGT CACAAAAAGGAA TGTTGTTTT TCCCTTTTA TGAAGTTGGATTGC [Twelve repeats (GTT) in the greatest number o groups: among 41 base haplotypes in haplogroup ide there are 36 with the allele 12 (including th group) and five with 11 (K Chimpanzee (Y-Chromosome) CCACGAGAGCACACACAAAAAGGAAAGCAG GGTTGTTGTTGTTATGTTTATCACCTCTAA CGCAGCTCATCCCTTTTACTG TCCATTGGCTGAAGTTGGATT [4 repeats (GTT)] YS490 = 12 AGTATGTCCTTG ATTATTATTATTATTATT ACAACATTCTTTATTATT TGAGACGG TGGAGTGCA GCAAGCTCTG [Twelve repeats (TTA) in the greatest number groups: among 41 base haplotypes in haplogroup ide there are 33 with the allele 12 (including the group), four with 1 Rozhanskii, 2012)]. Chimpanzee (19th Chromosome) AGTATGTCCTTACCTTATTATTATTATTAT TATTATTGAGACAG CCAGGCTGGAGTTCAGTGGCATG CTCACTGCAGCCTCTG [Seven repeats (TTA)] YS450 = 8 AGAGACCAGCTTGG TTTTGAGAGAACGTGAA TA CTGTTGTG TACATAAC TCACACAATGGAATATGATGCAGCTGTTTG TAGATCTGGTAGAAATTATTCTGAATCTAT CGGTTCCTGGGCCGTTTTTTATTTTATTTT ATTTTATTTTATTTTATTTTATTTTATTTT ATTTCATTTTATATTTTATTTTATTTTATT TTTATTTTATTTTTGGCTGGTGGGTTATTT ATTACTGATTCCATT [Fourteen repeats (TTTTA) are shown according to ., 2002) as (TTTTA)9(TTTTA)3 (TTTTA)2; howev base haplotypes in haplogroups world-wide there ith the allele 8 (including lyosov & Rozhanskii, 2012)]. Chimpanzee (Y-Chromosome) AGAGGCCAGCTTGGCATTTCCACTGCAGCT TTTTGAGAGAACATGAAAC TGTTTGTATGTGTGTTTGTGTG TGTAAGCATACACACAGAACC AACTAGGATCACCCACACAATGGAATATGA TGTAGCTTTTTGTAGATCTGGTAGAATTTT CCAAATCTATCAGTTTCTGGGCTTTTTTTT … Copyright © 2012 SciRes. 210  A. A. KLYOSOV ET AL. NOTE: The chimpanzee Y-Chromosome has no DYS454 ho- m D17 = 12 TCTTTTTATTATTAT TATTATTATTATTATTATACT AGGGTACATGTGCACAATGTG of hap- lo s world- wta-hap- loith 11 and only one with 14 (K TTCTTCTTCT TCAAGTTTT CAGGTTAGT ] D TGCAGTGAGCTGAGATTGGGTG CAGCCTGGGCAACAAAAGCAA lo ps world- w beta-hap- logroup), 10 with 12, three with 10 and one with 9 (Klyosov & R TGAGGCAGGAGAGTCACTTGAAACTGGAAG GATTGTATC GAGCAA ology] D GATACCACCTTAAGTATATAC TTCTGTACTTCTACCTGCTTC lo ps world- w beta-hap- lo)]. C AATATATAC CATGCTTC D TCTTTCCATGTTGCTCAGGCTA TCCTGAGCTGACATGATCCTT , C, D, E, G C ATTGATAGAG CTAGTCTCA CTTCCTCCT D ht roup_A/default.as px 12 sogg.org/tree/. ologous fragments. YS6 AGCATGATGCCTTCAGCTTTGTTCTTTCTG CTTAGTACTGTGTTT TATTATTAT TTAAGTTTT CAGGTTAGTTAGTTACATACGTATATATGT GCCATGCTGGTGTGCTGCACCCATTAACTT GTCATTTAGCATAAGGTATGTCTCCTAATG CTATCACTCCCCAATCC [Twelve repeats (TTA) in the greatest number groups: among 41 base haplotypes in haplogroup ide there are 25 with the allele 12 (including the be group), 12 with 13, three w lyosov & Rozhanskii, 2012)]. Chimpanzee (Y-Chromosome) AGCATGATGCCTTCAGGTTTGTTCTTTATG CTTAGTATTGTGTTTTCTTC TCTTCTTATTATTATTATACT AGGGTACATGTGCACAATGTG TAGTTACATATGTATATATGTGCCATGCTG GTGTGCTGCACCCATTAACTTGTCATTTAG CATAAGGTATGTCTCCTAATGCTATCACTC CCCAATCC [Repeats (TTA)4 if we follow the human homology YS568 = 11 GTGGCAGACAAAACCCAGTTACTCGGCAGG CTGAGGCAGGAGTGTCACTTGAAACCAGAA GGTGGAGG ACAACACTC AACTCCTTCTCAAAAATAAATAAATAAATA AATAAATAAATAAATAAATAAATA AATAAA TAAGTCCTCTTTTCCATCTTCCTCTTCAAT ACCATTGTTTTCCCACGCTTACTGCAAATT CGCCTAGATGAGTCCCATCCCTTTTCAA [Eleven repeats (TAAA) in the greatest number of hap- groups: among 41 base haplotypes in haplogrou ide there are 27 with the allele 11 (including the ozhanskii, 2012)]. Chimpanzee (Y-Chromosome) GTGGCAGGCATTACCAGCTACTCAGGAGGC GCGGAGGTTGCAGTGAGCCGA AGTACACTCCAGCCTGGGCAACAA AACTCCTTCTCCAAAAATTAAATAAATAAA TAAATAATAACATAAAGAAAGAAAG AAAGA AAGAAAGAAAGAGTCCTCTTTTCCATCTTC CTCTTCCATACCATTGTTCTCCTGCACTTA CTATAAATTTGCCTAGATGAGTCCCACCCT TTTCAA [Four repeats (TAAA)4 if we follow the human hom YS640 = 11 TGGGAAAAACCATGAGATCCTGTCTCAAAA AATAAATAAATAAATAAATAAATAAATAAA TAAATAAATAAATAAATCCATTATTGCCCA ATAGTTTGA TAAACTTCC CAGATGTTCTTTTATATTCCTCTAGTCTTT TTGTGTGTATGAGTGTTTTCATGCATCTGT AACATATTATTTTCATTATAGATGGTTAAT AATGTCTTTAAAATATGAACGGGCTTGACC CTA [Eleven repeats (TAAA) in the greatest number of hap- groups: among 41 base haplotypes in haplogrou ide there are 28 with the allele 11 (including the group) and 13 with 12 (Klyosov & Rozhanskii, 2012 himpanzee (Y-Chromosome) TGGGAAAAACCGTGAGATCCTGTCTCAAAA AATAAATAAACAAATAAATAAATAAATAAA TAAATAAACAAACCAACCCATTATTGCCCA ATAGTTTGAGATACCACCTTA TAAACTTCCTTCTGTACTTCTA TAGATGTTCTTTTATATTCCTCTAGTCTTT TTGTGGGTGTGTGTGTTTTCATGCATCTGT AACATATTATTTTCATTATAGATGGTTAAT AATGTCTTTAAAATATGAACGGGCTTGACC CTA [Repeats (TAAA)2(CAAA)1(TAAA)6 (CAAA)1] YS492 = 11 AGATGAGCCAGGCTTCAGACCCATGCAGCT CCATTTCAAGCCCATATCCTATCATTATTA TTATTATTATTATTATTATTATTATTATTG ATAGAGAG GTCTCAAAC CCTCCTCAGCCTCCAAAAAACCTGGGGTAA CAAGTGCGAGCCATTGTGCCTGACCCCTAC T [Eleven repeats (TTA) in the oldest haplogroups: B (Klyosov & Rozhanskii, 2012)]. himpanzee (Y-Chromosome) AGATGAGCCAGGCTTCAGACCCATGCAGCT CCATTTCAAGCCCATATCCTATCATTATTA TTATTATTATTATTATTATT AGTCTTTCCATGTTGCTCAGG AACTCCTGAGCTGACATGATC CAGCCTCCAAAAAACATGGGGTAACAAGTG CAAGCCATTGTGCCTGACCCCTACT [Nine repeats (TTA)] atabases The FTDNA Haplogroup A Project: a.com/public/Haplogtp://www.familytreedn ?section=yresults. ISOGG-20 http://www. i European Nucleotide Archive (ENA) Copyright © 2012 SciRes. 211  A. A. KLYOSOV ET AL. http://www.ebi.ac.uk/ena/ The National Center for Biotechnology Information (NCBI) GenBank (http://www.ncbi.nlm.nih.gov/) ts ge expansion from Africa. Science, 332, 346-349. doi:10.1126/science.1199295 Acknowledgemen The authors are indebted to Dr. Judith Remy Leder and Dr. Alexander S. Zolotarev for their valuable help with the prepara- tion of the manuscript. REFERENCES Atkinson, Q. D. (2011). Phonemic diversity supports a serial founder effect model of langua Behar, D. M., Villems, R.Smith, J., Pereira, L., Metspalu, E., Scozzadawn of human matri- , Soodyall, H., Blue- ri, R. et al. (2008). The lineal diversity. The American Journal of Human Genetics, 82, 1130-1140. doi:10.1016/j.ajhg.2008.04.002 enazzi, S., Douka, K., Fornai, C., Bauer, C. C., Kullmer, O., Svoboda, J. et al. (2011). Early dispersal of moder B n humans in Europe and im- plications for Neanderthal behaviour. Nature, 479, 525-528. doi:10.1038/nature10617 ampbell, M. C., & Tishkoff, S. A. (2010). The evolution of human genetic and phenotypic variation in Africa C . Current Biology, 20, R166-R173. doi:10.1016/j.cub.2009.11.050 arrigan, D., & Hammer, M. F. (2006). Reconstructing human origins in the genomic era. Nature Reviews, 7, 669-680. C doi:10.1038/nrg1941 Cavalli-Sforza, L. L., Piazza, A., Menozzi, & P., Mountain, J. (1988). Reconstruction of human evolution: Bringing together genetic, archaeological, and linguistic data. Proceedings of the National Academy of Sciences, 85, 6002-6006. doi:10.1073/pnas.85.16.6002 hiaroni, J., Underhill, P. A., & Cavalli-Sforza Y-chromosome divers C, L. L. (2009). ity, human expansion, drift, and cultural evolu- tion. Proceedings of the National Academy of Sciences, 106, 20174- 20179. doi:10.1073/pnas.0910803106 hromosomal Laboratories (2005). Y chromosomal haplogroup and ancient origins. C C http://www.chromosomal-labs.com/ancestry/yhaplogroup.pdf ruciani, F., Santolamazza, P., Shen, P., Macaulay, V., Moral, P., Olckers, A., Modiano, D. et al. (2002). A back migrartion from Asia to Sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. The American Journal of Human Genetics, 70, 1197-1214. doi:10.1086/340257 ruciani, F., Trombetta, B., Sellitto, D., Mas-Saia, A., Destro-BisCol, G., Watson, E. et al. (2010). Human Y-chromosome haplogroup R-V88: A paternal genetic record of early mid Holocene trans-Saharan con- nections and the spread of Chadic languages. European Journal of Human Genetics, 231, 1-8. ruciani, F., Trombetta, B., Massaia, A., Destro-CBisol, G., Sellitto, D., & Scozzari, R. (2011). A revised root for the human Y chromosomal phylogenetic tree: The origin of patrilineal diversity in Africa. The American Journal of Human Ge n e t i c s , 88, 1-5. doi:10.1016/j.ajhg.2011.05.002 urnoe, D., Xueping, J., HerriesC, A. I. R., Kanning, B., Tacon, P. S. C., Zhende, B., Fink, D. et al. (2012). Human remains from the Pleisto- cene-holocene transition of Southwest China suggest a complex evolutionary history for East Asians. PLOS One, 7, e31918. doi:10.1371/journal.pone.0031918 monds, C. A., Lillie, A. S., & CaEdvalli-Sforza, L. L. (2004) Mutations arising in the wave front of an expanding population. Proceedings of the National Academy of Sciences, 101, 975-979. doi:10.1073/pnas.0308064100 ernandes, V., Alshamali, F., Alves, M., Costa, M. D., Pereir Silva, N. M. et al. (2012). The Arab Fa, J. B., G T., Stenzel, U., al. (2010). A draft sequence of the ian cradle: Mitochondrial relicts of the first steps along the Southern route out of Africa. The Ameri- can Journal of Human Genetics, 90, 347-355. reen, R. E., Krause, J., Briggs, A. W., Maricic, Kircher, M., Patterson, N. et Neandertal genome. Science, 328, 710-722. doi:10.1126/science.1188021 rine, F. E., Bailey, R. M., Harvati, K., Nathan, R. P., Morris, A. G., Henderson, G. M., Robot, I. et al. (2007). La G te Pleistocene human skull from Hofmeyr, South Africa, and modern human origins. Science, 315, 226-229. doi:10.1126/science.1136294 rine, F. E., Gunz, P., Betti-Nash, L., Neubaue (2010). Reconstruction of the Gr, S., & Morris, A. G. late Pleistocene human skull from Hofmeyr, South Africa. Journal of Huma n Evolution, 59, 1-15. doi:10.1016/j.jhevol.2010.02.007 usmao, L., Gonzales-Neira, A., Alves, C., Lareu M, Costa S, Amorim A, & Carracedo A. (2002). Chimpanzee homologou G s of human Y H aran African hominids. South Af- specific STRs. A comparative study and a proposal for nomenclature. Forensic Science Internships, 126, 129-136. abgood, P. J. (1989). An examination of regional features on middle and early late Pleistocene Sub-Sah rican Archaeological Bulletin, 44, 17-22. doi:10.2307/3888315 anihara, T., Ishida, H., & Dodo, Y. (2003). Characterization of bio- logical diversity through analysis of discrete cranial traits. American Journal of Physical Anthropology, 121, 241-251. H doi:10.1002/ajpa.10233 ellenthal, G., Auton, A., & Falush, D. (2008). Inferring human colo- nization history using a copying model. PLOS Genetics, 4, e100 H 0078. doi:10.1371/jpurnal.pgen.1000078 enn, B. M., Gignoux, C. R., Jobin, M., Granka, J. M., Macpherson, J. M., Kidd, J. M., Rodríguez-Botigué, L. et al. gatherer genomic divers H (2011). Hunter- ity suggests a southern African origin for modern humans. Proceedings of the National Academy of Sciences, 108, 5154-5162. doi/10.1073/pnas.1017511108 igham, T., Compton, T., Stringer, C.H, Jacobi, R., Shapiro, B., Trinkaus, E., Chandler, B. et al. (2011). The earliest evidence for anatomically modern humans in North-Western Europe. Nature, 479, 521-524. doi:10.1038/nature10484 offecker, J. F. (2011). The early upper Paleolithic of Eastern Europe reconsidered. Evolut iona ry Anthropology, 20, 24 H -39. doi:10.1002/evan.20284 ublin, J.-J. (2011). African origin. Nature, 476, 395. H doi:10.1038/476395a udhes, J. F., Skaletsky, H.,H Pyntikova, T., Graves, T. A., van Daalen, structure and , 536-539. doi:10.1038/nature08700 S. K. M., Minx, P. J., Fulton, R. S. et al. (2010). Chimpanzee and human Y-chromosomes are remarkably divergent in gene content. Nature, 463 ., Graves, T. A., , S. et al. (2012). Strict evolutionary con- Hudhes, J. F., Skaletsky, H., Brown, L. G., Pyntikova, T Fulton, R. S., Dugan servation followed rapid gene loss on human and rhesus Y-Chromosomes. Nature, 483, 82-86. doi:10.1038/nature10843 udjashov, G., Kivisild, T., Underhill, P. A., Endicott, P., Sanchez, J. J., Lin, A. A., Shen, P. et al. (2007). Revealing the prehistoric H settle- ment of Australia by Y-chromosome and mtDNA analysis. Proceed- ings of the National Academy of Sciences, 104, 8726-8730. doi:10.1073/pnas.0702928104 bling, M. A., & Tyler-Smith, C. (2003) The human Y-chromosoJome: An evolutionary marker comes of age. Nature Reviews, 4, 598-612. doi:10.1038/nrg1124 lyosov, A. A. (2009). DNA genealogy, mutation rates, and some historical evidences written in Y-chromosome. I. Basic pr and the method. Journal of Gen K inciples etic Genealogy, 5, 186-216. Klyosov, A. A., & Rozhanskii, I. L. (2011). An archaic lineage of hap- logroup A. Proceedings of the Russian Academy of DNA Genealogy, 4, 1495-1502. Klyosov, A. A. (2012). Ancient history of the Arbins, bearers of hap- logroup R1b, from central Asia to Europe, 16,000 to 1500 years be- fore present. Advances in Anthropology, 2, 87-105. doi:10.4236/aa.2012.22010 lyosov, A. A., & Rozhanskii, I. L. (2012a). Re-examining the out of Africa theory an K d the origin of Europeoids (Caucasoids) in light of DNA genealogy. Advances in Anthropology, 2, 80-86. doi:10.4236/aa.2012.22009 lyosov, A. A., & Rozhanskii, I. L. (2012b). Haplogr proto indo-Europeans and th Koup R1a as the e legendary Aryans as witnessed by the Copyright © 2012 SciRes. 212  A. A. KLYOSOV ET AL. Copyright © 2012 SciRes. 213 DNA of their current descendants. Advances in Anthropology, 2, 1-13. doi:10.4236/aa.2012.21001 i, H., & Durbin, R. (2011). Inference of human popula from individual whole-genom Ltion history e sequences. Nature, 475, 493-496. doi:10.1038/nature10231 ajumder, P. P. (2010). The human genetic history of South Asia. Current Biology, 20, R184-R187. M doi:10.1016/j.cub.2009.11.053 ellars, P., & French, J. C. (2011M). Tenfold population increase in Western Europe at the Neandertal-to-modern human transition. Sci- ence, 333, 623-627. doi:10.1126/science.1206930 ellars, P. (2011). The earlieMst modern humans in Europe. Nature, 479, 483-485. doi:10.1038/479483a oorjani, P., Patterson, N., Hirschhorn, J. N., Keinan, A., Hao, L.M, Atzmon, G., Burns, E. et al. (2011). The history of African gene flow into Southern Europeans, Levantines, and Jews. PLOS Genetics, 7, e1001373. doi:10.1371/journal.pgen.1001373 Nei, M., & Takezaki, N. (1996). The root of the phylogenetic tree of human populations. Molecular Biology and Evolution, 13, 170-177. doi:10.1093/oxfordjournals.molbev.a025553 ttoni, C., Primativo, G., Kashani, B. H., Achilli, A., Martinez-Labarga, C., Biondi, G., Torroni, A., & Rickards, O. (2010). Mitochondrial haplogroup H1 in North Africa: An early h O olocene arrival from Iberia. Plos One, 5, e13378. doi:10.1371/journal.pone.0013378 atin, E., Laval, G., Barreiro, L. B., Salas, A., Semino, O., Santachiara-Benerecetti, S., Kidd, K. K. et a P l. (2009). Inferring the P y of demographic history of African farmers and Pygmy hunter-gatherers using a multilocus recequencing data set. PLOS Genetics, 5, 1-13. erelman, P., Johnson, W. E., Roos, C., Seuanez, H. N., Horvath, J. E., Moreira, M. A. M., & Kessing, B. (2011). A molecular phylogen living primates. PLOS Genetics, 7, e1001342. doi:10.1371/journal.pgen.1001342 inhasi, R., Higham, T. F. G., Golovanova, L. V., & Doronichev, V. B. (2011). Revised age of late Neanderthal occupation and the end o P f Puaud, S. J., Valladas, (2011). The oldest anatomically the middle Paleolithic in the northern Caucasus. Proceedings of the National Academy of Sciences, 108, 8611-8616. rat, S., Pean, S. C., Crepin, L., Drucker, D. G., P H., Lasnickova-Galetova, M. et al. modern humans from far South-East Europe: Direct dating, culture and behavior. PLOS One, 6, e20843. doi:10.1371/journal.pone.0020834 rüfer, K., Munch, K., Hellmann, I., Akagi, K., MPiller, J. R., Walenz, R . Forensic value of 14 novel e. Forensic Science Internships, B., Koren, S. et al. (2012). The bonobo genome compared with the chimpanzee and human genomes. Nature, 486, 527-531. edd, A. J., Agellon, A. B., Kearney, V. A., Contreras, V. A., Karafet, T., Park, H., de Knijff, P. et al. (2002) STRs on the human Y-chromosom 130, 97-111. doi:10.1016/S0379-0738(02)00347-X ightmire, G. P. (2009) Middle and later Pleistocene hominins in Af- rica and Southwest Asia. Proceedings of the National RAcademy of Sciences, 106, 16046-16050. doi:10.1073/pnas.0903930106 ogers, A. R., & Jorde, L. B. (1995). Genetic evidence of modern human origins. Human B i ol ogy, 67, 1-36. ozhanskii, I. L., & Klyosov, A. A. (2011). Mutation R R rate constants in DNA genealogy (Y-Chromosome). Advances in Anthropology, 1, 26-34. doi:10.4236/aa.2011.12005 ahoo, S., Singh, A., Himabindu, G., Banerjee, J., Sitalaximi, T., Gaikwad, S., Trivedi, R. et al. S (2006). A prehistory of Indian Y-chromosomes: Evaluating demic diffusion scenarios. Proceedings of the National Academy of S ciences, 103, 843-848. doi:10.1073/pnas.0507714103 cally, A., Dutheil, J. Y., Hillier, L. W., Jordan, G. E. Herrero, J., Hobolth, A., et S, Goodhead, I., al. (2012). Insights into hominid evolution from the gorilla genome sequence. Nature, 483, 169-175. doi:10.1038/nature10842 hi, W., Ayub, Q., Vermeulen, M., Shao, R.-G., Zuniga, S., van der Gaag, K., de Knijff, P. et S al. (2010). A worldwide survey of human male demographic history based on Y-SNP and Y-STR data from the HGDP-CEPH populations. Molecular Biology and Evolution, 27, 385-393. doi:10.1093/molbev/msp243 mms, T. M., Martinez, E., Herrera, K. J., Wright, M. R., Perez, O. A., Hernandez, M., Ramirez, E. C., McC Si artney, Q., & Herrera, R. J. (2011). Paternal lineages signal distinct genetic contributions from British loyalists and continental Africans among different Bahamian islands. American Journal of Physical Anthropology, 146, 4594-608. doi:10.1002/ajpa.21616 ares, P., Ermini, L., Thompson, N., Normina, M., Rito, T., Rohl, A., Salas, A., Oppenheimer, So S., Macaulay, V., & Richards, M. B. (2009). Correcting for purifying selection: An improved human mitochon- drial molecular clock. Journal of Human Genetics, 84, 740-759. doi:10.1016/j.ajhg.2009.05.001 tewart, J. R., & Stringer, C. B. (2012). Human evolution out of Afri The role of refugia and climate c Sca: hange. Science, 335, 1317-1321. doi:10.1126/science.1215627 toneking, M, & Delfin, F. (2010). The human genetic history of E Asia: Weaving a complex tape Sast stry. Current Biology, 20, R188-R193. doi:10.1016/j.cub.2009.11.052 tringer, C. B., & Andrews, P. (1988). Genetic and fossil evidence for the origin of modern humans. Sc Sience, 239, 1263-1268. doi:10.1126/science.3125610 un, J. X., Helgason, A., Masson, G., Ebenesersdóttir, S. Mallick, S., Patterson, N. et a S S., Li, H., l. (2011). A direct characterization of T human mutation. 61th Annual Meeting, American Society of Human Genetics/ICHG, 11-15 October 2011, Montreal. attersall, I. (2009). Human origins: Out of Africa. Proceedings of the National Academy of Sciences, 106, 16018-16031. doi:10.1073/pnas.0903207106 nderhill, P. A., Shen, P., Lin, A. A., Jin, L., Passarin H., Kauffman, E. et al. (2000) Uo, G., Yang, W. . Y-chromosome sequence variation and the history of human populations. N ature Genetics, 26, 358-361. doi:10.1038/81685 ue, Y., Zerjal, T., Bao, W., Zhu, S., Lim,S.-K., Shu, Q. et al. (2005). Recent spread of a X Y-chromosomal lineage in Northern China and Mongolia. The American Journal of Human Genetics, 77, 1112-1116. doi:10.1086/498583 otova, V., Lefebvre, J.-F., Moreau, C., Gbeha, E., Hovhannesyan, K., Bourgeois, S., & Be Y darida, S. (2011). An X-linked haplotype of Neandertal origin is present among all non-African populations. Molecular Biology and Evolution, 28, 1957-1962
|