 Journal of Biomaterials and Nanobiotechnology, 2012, 3, 519-527 http://dx.doi.org/10.4236/jbnb.2012.324053 Published Online October 2012 (http://www.SciRP.org/journal/jbnb) 519 Overview of Multidrug-Resistant Pseudomonas aerugin osa and Novel Therapeutic Approaches Marilyn Porras-Gómez1, José Vega-Baudrit1, Santiago Núñez-Corrales2 1National Nanotechnology Laboratory LANOTEC, National Center for Advanced Technology CeNAT, San José, Costa Rica; 2E-Science Research Program, Costa Rican Institute of Technology, Cartago, Costa Rica. Email: marilyn.ucr@gmail.com Received July 18th, 2012; revised August 23rd, 2012; accepted September 2nd, 2012 ABSTRACT Gram-negative bacilli Pseudomonas aeruginosa is an important pathogen in hospitalized patients, contributing to their morbidity and mortality due to its multiple resistance mechanisms. Therefore, as therapeutic options become restricted, the search for new agents is a priority. Latterly an accelerated increase in frequency of multidrug-resistant clinical strains has severely limited the availability of therapeutic options. Several in vitro and in vivo studies evaluating the efficacy of different antimicrobials agents and development of vaccines against P. aeruginosa have been reported as novel approaches, such as inhibition of virulence factor expression or inhibition of their metabolic pathways. Keywords: Bacilli; Gram-Negative; Pseudomonas aeruginosa; Multidrug Resistance; Pathogen; Resistance Mechanisms 1. Introduction Pseudomonas aeruginosa is an opportunistic pathogen that may cause severe invasive diseases in critically ill patients. The frequency of infections caused by them is increasing and multidrug-resistant (MDR) strains, resis- tant to almost all available antimicrobials, are emerging in hospitalized patients. Because of its ubiquitous nature, ability to survive in moist environments, and innate resistance to many anti- biotics and antiseptics, P. aeruginosa is a common pathogen in hospitals and particularly in intensive care units. It has become increasingly clear that resistance development in P. aeruginosa is multifactorial, with mu- tations in genes encoding porins, efflux pumps, penicil- lin-binding proteins, and chromosomal β-lactamase, all contributing to resistance to β-lactams, carbapenems, am- inoglycosides, and fluoroquinolones [1]. Strains of P. aeruginosa are the cause of several diseases in nosoco- mial environments, predominantly pneumonia, bactere- mia, meningitis, urinary tract infections, as well as skin and soft-tissue infections [2]. Due to the emergence of MDR pathogens, it is of ultimate importance to develop new antimicrobial drugs. P. aeruginosa has been characterized as one of the most versatile microbial organisms, with a wide span of habitats including soil, disinfectant solution and jet plane fuel [3]. Low permeability of its outer membrane by a complex set of efflux pump systems and secretion of alginate during biofilm formation are major factors that allow the pathogen to become highly virulent and resis- tant to multiple antibiotic agents. Adding to these factors, other baterial exoproducts such as lipopolysaccharides and elastase induce harmful pathogenesis resulting in tissue destruction. Flagellins in P. aeruginosa perform several functions during host infection [4]. Apart from enabling motility, the flagellum of P. aeruginosa plays an indirect role in membrane permeabilization and surfactant protein-me- diated bacterial clearance [5]. Similarly, pili is involved during inflammation due to glycosylation in the interface between pili and host cells. Flagellins are classified into two types: Type-a (polymorphic glycosylated) and type-b (non-glycosylated). 2. Pathogenesis and Colonization Pili, flagella, exoenzyme S, and mucoid exopolysaccha- ride are recognized as major adhesins in P. aeruginosa. Invading pathogens are recognized by Toll-like receptors (TLRs) on epithelial cells and innate immunocytes, both of which are then activated to express inflammatory me- diators. Thereafter, defense systems such as mucociliary clearance, phagocytosis and humoral immunity are pro- moted to neutralize the danger [3]. Invading organisms are first trapped by the mucus layer coating the airway epithelial cells. Airway mucin, the main component of mucus, is a large heterogeneous Copyright © 2012 SciRes. JBNB  Overview of Multidrug-Resistant Pseudomonas aeruginosa and Novel Therapeutic Approaches 520 glycoprotein with carbohydrate side chains consisting of N-acetylglucosamine (GlcNAc), GalNAc, D-mannose, L-fucose, and N-acetylneuraminic acid (NeuAc) which may promote colonization, whereas binding to the gly- colipids may cause an inflammatory response [6 ]. These exposed oligosaccharide residues become adhesive re- ceptors for P. aeruginosa and others microbes. The diversity of oligosaccharide side chains on glycol- proteins or glycolipids in mucin may determine which organisms will effectively bind to it [7]. P. aeruginosa has a variety of lectin-like adhesions (Figure 1), include- ing pili, mucoid exopolysaccharide, and non-pilus ad- hesins, represented by exoenzyme S, that have binding domains similar to that of the pilus. Flagella motility and pili, which mediate twitching motility in P. aeruginosa, are thought to be the prevailing adhesins for the initial attachment required for colonization of the airway tract [3]. Alginate expression is observed afterward the initial attachment of P. aeruginosa to a solid surface. Alginate may be involved in cementing the primary adherence of the non-pilus adhesion found on the surface of P. aeruginosa so it can bind to respiratory cells or mucin in the absence of other adhesions [3]. When the organisms trapped in mucus multiply faster than the removal rate the production of exoproducts increases, most of which are virulent and result in decreased mucociliary transport and airway epithelial cell function. The latter results in enhanced mucus inactivity and cell surface colonization. Following, quorum sensing and biofilm formation start. Intracellular communication is involved in P. aeruginosa biofilm development [8]. The phenomenon of quorum sensing or cell-to-cell communication requires self-gen- erated signal molecules, named autoinducers. As cell density increases, there is a proportional increase in autoinducer production. P. aeruginosa has at least two quorum-sensing systems. Each system includes a gene encoding a transcriptional activator, LasR or RhlR and a gene encoding an autoinducer lasI or rhlI. These systems contribute to the development of the biofilm. In addition, Figure 1. Schematic interactions of the possible roles of P. aeruginosa lectins during host recognition and biofilm for- mation [10]. the LasR-las I and RhlR-rhlI quorum-sensing systems regulate the expression of various virulence genes in a density-dependent fashion [3]. In animal models of acute and chronic infections with P. aeruginosa containing a mutation in quorum-sensing genes, less tissue destruction was induced and less mor- tality was observed compared with findings in wild-type strains [9]. Bacteria inside a mature biofilm exhibit in- creased resistance to antibacterials and phagocytic cells, and are less stimulatory to the mucosa; these facts are found in bacterial colonization grown particularly in in- appropriate environments. It can be said that one of the ways the pathogen and host coexist is established at the site of colonization. In a chronic colonization with P. aeruginosa biofilms, the lungs show chronic inflammation that is associated with the development of lymphocyte follicles around respiratory bronchioles and with the influx of PMNs into airway lumens [3]. 3. General Mechanisms against Multiple Drugs 3.1. Multidrug Resistance P. aeruginosa is naturally resistant to a significant num- ber of antimicrobials (Table 1). Furthermore, they easily acquire resistance to new antibacterial agents by muta- tional changes or acquisition of genetic material. In a study, P. aeruginosa strains isolated presented resistance to carbenicillin and gentamicin. P. aeruginosa is intrin- sically less susceptible to the fluoroquinolones and usu- ally it is moderately susceptible or resistant [11]. Resistance of P. aeruginosa to commonly used thera- peutic agents has increased in recent years. MDR can be defined as resistance to at least four classes of antibiotics used during treatment of these infections: third-genera- tion cephalosporins, fluoroquinolones, aminoglycosides, and carbapenems [12]. Emergence of MDR strains is often due to selective pressure of antimicrobial therapy. Genetic studies con- firm the selection of resistant mutants and their subse- quent spread. Outbreaks caused by MDR P. aeruginosa Table 1. Natural resistance of P. aeruginosa to antibiotics [2]. Bacteria Natural resistance P. aeruginosa ampicillin amoxicillin amoxicillin/clavulanate first-generation cephalosporins second-generation cefotaxime ceftriaxone nalidixic acid trimethoprim Copyright © 2012 SciRes. JBNB  Overview of Multidrug-Resistant Pseudomonas aeruginosa and Novel Therapeutic Approaches Copyright © 2012 SciRes. JBNB 521 may follow an increased use of third-generation cepha- losporins or carbapenems for therapy of infections caused by other resistant bacteria [2]. All bacteria rely on a heavily cross-linked peptidogly- can layer for cell shape and morphological stability [13]. The formation of this layer depends on the catalytic ac- tivity of transpeptidase enzymes, which utilize an active site serine and perform their catalytic cycle by way of an acylation/deacylation pathway. Beta-lactam antibiotics inhibit the action of transpeptidases, effectively blocking the transpeptidation reaction and therefore leaving bacte- ria susceptible to cell lysis. Beta-lactamases confer sig- nificant antibiotic resistance to their bacterial hosts by hydrolysis of the amide bond of the four-membered beta- lactam ring. Their mechanism depends heavily on the concentration of zinc ions, required by the four different classes of beta-lactamases during hydrolisis [14]. Bacteria exhibit two control mechanisms when in pre- sence of metallic compounds: One based on sensing of the environment and, in proportion to what is detected, one of regulatory response [15]. The process occurs in the context of chemical gradi- ents, which activate several metabolic pathways acquired by bacteria along their evolutionary history. Enthalpy plays a determinant role in this case: All non-deleterious mutations attempt to minimize metabolic costs. Toxicity of metals in the cellular medium can be either independ- ent of concentration or regulated by it, in which case metals at appropriate concentration levels become cata- lytics. In the case of P. aeruginosa zinc is not only re- quired for metabolic functions but fulfills a definitive role in resistance to beta-lactams, also dependent on chemical gradients. A similar case occurs with iron ac- quisition, allowing the pathogen to degrade host iron binding proteins and act as an extracellular protein, bringing the host cell into metabolic stress [16]. Apart from the two general mechanisms described above, the complexity in the genome of P. aeruginosa provides precise binding mechanisms in different ways. As in the case of beta-lactamases which hydrolize beta- lactams by attacking their moieties, microcalorimetry studies have shown that paralogous genes in P. putida have the capacity to bind in different ways depending on subtle changes in enthalpy [17]. Evidence suggests that selective pressure induced by both natural and clinical factors has forced adaptations based on both general mechanisms (extracellular and within the cell) for mo- lecular recognition as well as specific binding strategies for novel antimicrobials based on physicochemical pa- rameters. 3.2. Efflux Pumps The resistance of MDR strains may be mediated by the active export of the antibiotics out of the bacterial cell by efflux pumps [18]. Evidence from diverse bacterial genomes indicates that approximately 5% - 10% of genes are involved in transport, with a large proportion of them encoding efflux pumps [19]. These can be or- ganized into five superfamilies: small multidrug resis- tance (SMR), multidrug endosomal transporter (MET), major facilitator superfamily (MFS), resistance nodula- tion division (RND) and multi antimicrobial resistance (MAR) [20]. The latter indicates the mechanism has evolved long ago and is shared amongst all Gram-posi- tive and Gramnegative bacteria. P. aeruginosa exhibits several efflux pump systems that allow it to be resistant to several antimicrobial agents [21]. Tab le 2 summarizes some of these systems and their effect on resistance to different agents. 3.3. Other Resistance Mechanisms Another mechanism present in P. aeruginosa is the for- mation of permeability barriers (OM) [18]. Impaired penetration of different substances through the mem- brane (e.g. imipenem) is due to diminished expression of specific OM protein. It has been shown that OM perme- abilizers such as EDTA increase susceptibility to antibi- otics, indicating that the lack of OprD protein leads to a reduction of active antibiotic molecules capable of reach- ing the target penicillin-binding-proteins. Two-component systems (2CS) are common molecu- lar mechanisms that allow diverse bacteria to have adap- tive regulation in response to complex environments, often composed by a sensor histidine kinase and a re- sponse regulator [22]. The sensor kinase is composed of Table 2. Efflux pump systems associated to antibiotics resistance in P. aeruginosa [21]. Efflux pump system Antimicrobial agents MexAB-OprM fluoroquinolones, beta-lactams, tetracyclines, macrolides, chloramphenicol, novobiocin, trimethoprim, sulphonamides MexEF-OprN fluoroquinolones, chloramphenicol, trimethoprim, imipenem MexXY-OprM fluoroquinolones, aminoglycosides, tetracyclines, erythromycin MFP-RND-OMF multidrug efflux pump systems fluoroquinolones, tetracyclines  Overview of Multidrug-Resistant Pseudomonas aeruginosa and Novel Therapeutic Approaches 522 at least one signal recognition domain coupled to an autokinase domain in an input-transmitter arrangement. Two hypothesis exist regarding the evolution of 2CS. The co-evolution model proposes that 2CS genes have appeared as a result of duplication and further different- tiation of these in bacterial genomes. The recruitment model on the other hand proposes that some of the 2CS operons have appeared as a result of an assembly of a sensor gene and a regulator gene from heterologous 2CS genes. Both are supported by evidence from phylogenetic analysis and gene regulatory network modeling in the P. aeruginosa PA01 strain. Finally, it has been shown that cytotoxicty is an im- portant mechanism that contributes to high morbidity and mortality in P. aeruginosa infections, particularly in cys- tic fibrosis [23]. Along with mucoidy resultant from the release of alginate, P. aeruginosa synthesizes a secretory apparatus (Type III) that allows it to inject toxins from their cytoplasm into the target cell. The latter mechanism allows mucoid bacteria to lyse the host’s macrophages and overcome various defense such as in the case of cys- tic fibrosis lung infection. 3.4. Genomic Profile of P. aeruginosa Complete sequencing of the P. aeruginosa PA01 strain (Figure 2) revealed a complex organism, comparable in terms of genome length (6.3 Mb) and amount of open reading frames (5570 ORFs) to Saccharomyces cere- visiae [24]. Comparison with E. coli revealed a strong evolutionary link between both. This fact is based on evidence of almost one half of the ORFs in P. aeruginosa having an E value of 10e−5 in BLASTP alignments with E. coli segments despite only 40% of amino-acid identity in the common coding regions. Also, low gene replica- tion across the genome indicates that such a compara- tively large genome has vast potential of functional di- versity, which may support rich mechanisms responsible for multidrug resistance thanks to environmental versatil- ity. Available genome data supports the hypothesis of the existence of evolutionary paths where resistance to natu- rally available antibiotics was acquired. The wide collection of genes in P. aeruginosa can be mapped to diverse functions. Table 3 shows some rele- vant data regarding the relation between the gene pool and mechanisms ranging from cell regulation to chemo- sensing and chemotaxing. It is interesting to note that the limited ability to grow on sugars has forced P. aerugi- nosa down to an evolutionary path where it adapted to Figure 2. Circular representation of the P. aeruginosa PA01 genome [24]. Table 3. Number of genes associated to functions related to multidrug resistance in P. aeruginosa [24]. Function No. of genes Cellular act i v it ies and pro d u c ts Cell regulation 521 Transcriptional regulators, two-component regulatory system proteins Cell-surface exposure 71 OprD family of specific porins, TonB family of gated porins (metal-related uptake), OprM family of outer membrane proteins (efflux and secretion) Import of nutrients 555 300 cytoplasmic membrane transport systems Metabolism 626 Amino acid biosynthesis and metabolism, carbon compound catabolism, central intermediary metabolism, energy metabolism, fatty acid and phospholipid metabolism, nucleotide biosynthesis and metabolism Intrinsic drug resistance and efflux systems 34 Resistance/nodulation/cell division, major facilitator superfamily, small multidrug resistance family, multidrug and toxic compound extrusion family, ATP-binding cassette family Protein secretion 83 Secretion of alkaline protease, general secretion pathway, contact-dependent delivery of proteins into the cytoplasm of host cells, exoenzymes S, T and Y Chemotaxis 43 Flagella-mediated swimming towards chemoattractors, attraction to sugars, amino acids and inorganic phosphate, repulsion to thiocyanic and isothiacyanic esters Copyright © 2012 SciRes. JBNB  Overview of Multidrug-Resistant Pseudomonas aeruginosa and Novel Therapeutic Approaches 523 use an ample range of carbon compounds. Recent advances in in silico genomic approaches have provided an opportunity to specifically highlight poten- tial drug targets and has facilitated a paradigm shift from direct antimicrobial screening programs toward rational target-based strategies, where drug discovery starts at the level of the gene [25]. According to the generally used definition, essential genes cannot be deleted from an organism without a le- thal effect, being more evolutionarily conserved than nonessential genes. If genes are grouped by their func- tional classification, variation is lowest for genes in- volved in transcription, RNA processing, degradation, translation, post-translational modification, degradation and cell division, which are enriched for essential genes (Figure 3). In addition to gene essentiality, a high gene expression rate has previously been shown to correlate with low sequence variation, and it was proposed that the underlying driving force for the slower evolution of es- sential genes is that most of the highly expressed genes are also indispensable in general [25]. 4. Current Approaches against Multidrug Resistance 4.1. New Strategies with Known Antimicrobials Polymyxin B agents were used in the therapy of infec- tions in the 1970s, but due to reported toxicity and the subsequent development of less toxic drugs such as nephrotoxicity, ototoxicity and neuromuscular blockade, their use has been discontinued. Polymyxin B is a poly- peptide antibiotic produced by a strain of Bacillus po- lymyxa and is primarily used for resistant Gram-negative infections. Now, with the emergence of MDR strains, their clinical use is being reconsidered [2]. Several re- ports in the past five years showed that colistin toxicity is not as frequent as previously reported [26]. Renal failure was rare and usually reversible, while neurotoxicity was not reported. Furthermore, colistin (polymyxin E) has been used in several cases as a salvage agent during therapy of infec- tions caused by strains resistant to all available antim- icrobials [26]. However, clinical strains with reduced susceptibility to polymyxin B have been reported [27]. Colistin, in combination with antibiotics from other classes, may be a useful agent for the treatment of infec- tions caused by pandrug-resistant P. aeruginosa [28]. Aztreonam may be used in the therapy of infections caused by P. aeruginosa. Combination therapy of az- treonam with other antimicrobials may be effective. A two-drug (aztreonam and amikacin) and a three-drug com- bination (aztreonam, ceftazidime, and amikacin) were very active against MDR strains of P. aeruginosa in an in vitro study [29]. Imipenem and meropenem are carbapenems com- monly used in hospital practice. Many reports confirm their usefulness in the therapy of nosocomial infections caused by MDR Gram-negative bacilli. Apart from imipenem and meropenem, new carbapenems are being evaluated for their efficacy against MDR pathogens [2]. Carbapenems may be administered as monotherapy, but with the emergence of MDR P. aeruginosa, combination Figure 3. Protein evolution rates in genes of different functional categories [25]. Copyright © 2012 SciRes. JBNB 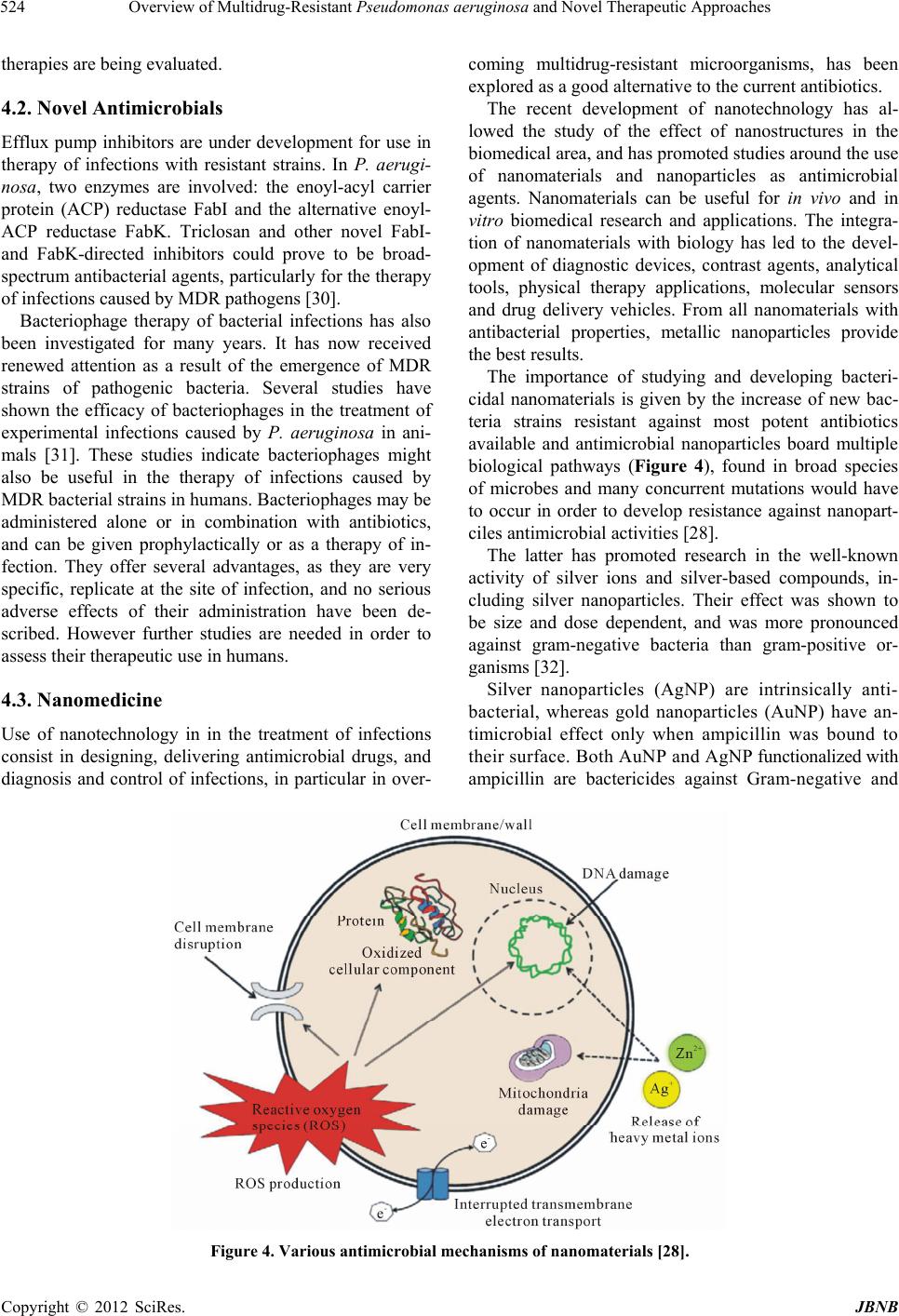 Overview of Multidrug-Resistant Pseudomonas aeruginosa and Novel Therapeutic Approaches 524 therapies are being evaluated. 4.2. Novel Antimicrobials Efflux pump inhibitors are under development for use in therapy of infections with resistant strains. In P. aerugi- nosa, two enzymes are involved: the enoyl-acyl carrier protein (ACP) reductase FabI and the alternative enoyl- ACP reductase FabK. Triclosan and other novel FabI- and FabK-directed inhibitors could prove to be broad- spectrum antibacterial agents, particularly for the therapy of infections caused by MDR pathogens [30]. Bacteriophage therapy of bacterial infections has also been investigated for many years. It has now received renewed attention as a result of the emergence of MDR strains of pathogenic bacteria. Several studies have shown the efficacy of bacteriophages in the treatment of experimental infections caused by P. aeruginosa in ani- mals [31]. These studies indicate bacteriophages might also be useful in the therapy of infections caused by MDR bacterial strains in humans. Bacteriophages may be administered alone or in combination with antibiotics, and can be given prophylactically or as a therapy of in- fection. They offer several advantages, as they are very specific, replicate at the site of infection, and no serious adverse effects of their administration have been de- scribed. However further studies are needed in order to assess their therapeutic use in humans. 4.3. Nanomedicine Use of nanotechnology in in the treatment of infections consist in designing, delivering antimicrobial drugs, and diagnosis and control of infections, in particular in over- coming multidrug-resistant microorganisms, has been explored as a good alternative to the current antibiotics. The recent development of nanotechnology has al- lowed the study of the effect of nanostructures in the biomedical area, and has promoted studies around the use of nanomaterials and nanoparticles as antimicrobial agents. Nanomaterials can be useful for in vivo and in vitro biomedical research and applications. The integra- tion of nanomaterials with biology has led to the devel- opment of diagnostic devices, contrast agents, analytical tools, physical therapy applications, molecular sensors and drug delivery vehicles. From all nanomaterials with antibacterial properties, metallic nanoparticles provide the best results. The importance of studying and developing bacteri- cidal nanomaterials is given by the increase of new bac- teria strains resistant against most potent antibiotics available and antimicrobial nanoparticles board multiple biological pathways (Figure 4), found in broad species of microbes and many concurrent mutations would have to occur in order to develop resistance against nanopart- ciles antimicrobial activities [28]. The latter has promoted research in the well-known activity of silver ions and silver-based compounds, in- cluding silver nanoparticles. Their effect was shown to be size and dose dependent, and was more pronounced against gram-negative bacteria than gram-positive or- ganisms [32]. Silver nanoparticles (AgNP) are intrinsically anti- bacterial, whereas gold nanoparticles (AuNP) have an- timicrobial effect only when ampicillin was bound to their surface. Both AuNP and AgNP functionalized with ampicillin are bactericides against Gram-negative and Figure 4. Various antimicrobial mechanisms of nanomaterials [28]. Copyright © 2012 SciRes. JBNB  Overview of Multidrug-Resistant Pseudomonas aeruginosa and Novel Therapeutic Approaches 525 Gram-positive bacteria. Most importantly, when AuNP and AgNP are functionalized with ampicillin they be- came potent bactericidal agents with unique properties that subverted antibiotic resistance mechanisms of multi- ple-drug-resistant bacteria as P. aeruginosa [33]. Currently nanoparticles such as chitosan nanoparticles, quantum dots, dendrimers and liposomes are under study as antimicrobial agents. Polymyxin B-loaded liposomes represent a successful example of liposomal antimicro- bial drug delivery [34]. As mentioned before, polymyxin B has been recognized as a viable treatment for P. aeruginosa related infections. However, its systemic use has been limited due to toxic side effects. It has been re- ported that liposomal encapsulation of polymyxin B dra- matically diminishes side effects and improves its antim- icrobial activity against resistant strains of P. aeruginosa [35]. The action mechanism of liposomal polymyxin B against bacteria has been identified as membrane fusion. Membrane fusion between liposomes and bacteria is a rapid and spontaneous process driven by non-covalent forces such as van der Waals force and hydrophobic in- teractions that minimize the free energy within the sys- tem. Antibiotic efflux is a widely accepted mechanism of microbial drug resistance, in which proteinaceous trans- ports located in bacterial membranes preferentially pump antimicrobial drugs out of the cells. When liposomes fuse with cell membranes, a high dosage of drug contents is immediately delivered to the bacteria, potentially sup- pressing the antimicrobial resistance of the bacteria by overwhelming the efflux pumps, thereby improving drug’s antimicrobial activity [34]. 5. Concluding Remarks The pathogenesis of chronic airway infection of P. aeruginosa has been discussed, and the factors affecting the progress of the pathogenic manifestations described in detail. This review focuses at first on bacterial adher- ence, biofilm formation, immunological disorders and current and novel therapeutic treatment. Resistance to antibiotics exhibited by some strains of pathogenic bacteria pose a serious challenge in combat- ing infectious diseases. As it is known, application of more powerful antibiotics can lead to limited and tempo- rary advances and eventually contribute to developing greater resistance. Resources against multidrug-resistant pathogenic infections are now limited. Due to their promising antimicrobial properties, nano- materials and nanoparticles are currently being studied as potential, highly potent antimicrobial agents for a variety of medical applications. Different kinds of nanoparticles have been investigated for carrying and delivering anti-biotics. Also, nanoparticles enable combining multiple approaches in order to enhance antimicrobial activity and overcome the various resistance mechanisms in P. aeruginosa. Further investigation is required indeed. In particular, the performance of nanomaterials and nanoparticles is an interdisciplinary endeavor. Tools from pathology, im- munology, biocompatible materials, polymers, nanotoxi- cology, pharmacology and nanotechnology are essential for such task. REFERENCES [1] B. Ozer, M. Tatman-Otkun, D. Memis and M. Otkun, “Cha- racteristics of Pseudomonas aeruginosa Isolates from In- tensive Care Unit,” Central European Journal of Medi- cine, Vol. 4, No. 2, 2009, pp. 156-163. doi:10.2478/s11536-008-0090-2 [2] M. Wróblewska, “Novel Therapies of Multidrug-Resistant Pseudomonas aeruginosa and Acinetobacter spp. Infec- tions: The State of the Art,” Archivum Immunologiae et Therapia Experimentalis, Vol. 54, No. 2, 2006, pp. 113- 120. doi:10.1007/s00005-006-0012-4 [3] H. Kobayashi, O. Kobayashi and S. Kawai, “Pathogenesis and Clinical Manifestations of Chronic Colonization by Pseudomonas aeruginosa and Its Biofilms in the Airway Tract,” Journal of Infection and Chemotherapy, Vol. 15, No. 3, 2009, pp. 125-142. doi:10.1007/s10156-008-0691-3 [4] A. Verma and R. Rampal, “Glycosylation Islands of Pseu- domonas Species,” In: J. L. Ramos and A. Filloux, Eds., Pseudomonas, Springer Netherland, Dordrecht, 2007, pp. 31-56. [5] S. Zhang, F. McCormack, R. Levesque, G. O’Toole and G. Lau, “The Flagellum of Pseudomonas aeruginosa Is Re- quired for Resistance to Clearance by Surfactant Protein A,” Public Library of Science, Vol. 2, No. 6, 2007, p. e564. [6] H. C. Krivan, D. D. Roberts and V. Ginsburg, “Many Pulmonary Pathogenic Bacteria Bind Specifically to the Carbohydrate Sequence GalNAc Beta 1-4Gal Found in Some Glycolipids,” Proceedings of the National Academy of Sciences, Vol. 85, No. 16, 1988, pp. 6157-6161. doi:10.1073/pnas.85.16.6157 [7] G. Lamblin, M. Lhermitte, A. Klein, N. Houdret, A. Scharfman, R. Ramphal and P. Roussel, “The Carbohy- drate Diversity of Human Respiratory Mucins: A Protec- tion of the Underlying Mucosa?” American Review of Respiratory Diseases, Vol. 144, No. 3, 1991, pp. s19-s24. [8] D. G. Davies, M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton and E. P. Greenberg, “The Involvement of Cell-To-Cell Signals in the Development of a Bacterial Biofilm,” Science, Vol. 280, No. 5361, 1998, No. 295- 298. [9] R. Smith and B. Iglewski, “Pseudomonas aeruginosa Quorum-Sensing Systems and Virulence,” Current Opi- nion in Microbiology, Vol. 6, No. 1, 2003, pp. 56-60. doi:10.1016/S1369-5274(03)00008-0 [10] A. Imberty, M. Wimmerová, E. P. Mitchell and N. Gil- boa-Garber, “Structures of the Lectins from Pseudomonas aeruginosa: Insights into the Molecular Basis for Host Copyright © 2012 SciRes. JBNB  Overview of Multidrug-Resistant Pseudomonas aeruginosa and Novel Therapeutic Approaches 526 Glycan Recognition,” Microbes and Infection, Vol. 6, No. 2, 2004, pp. 221-228. doi:10.1016/j.micinf.2003.10.016 [11] M. Araque and E. Velazco, “In Vitro Activity of Flerox- acin against Multiresistant Gram-Negative Bacilli Isolated from Patients with Nosocomial Infections,” Intensive Care Medicine, Vol. 24, No. 8, 1998, pp. 839-844. doi:10.1007/s001340050675 [12] J. A. Karlowsky, D. C. Draghi, M. E. Jones, C. Thorns- berry, I. R. Friedland and D. F. Sahm, “Surveillance for Antimicrobial Susceptibility among Clinical Isolates of Pseudomonas aeruginosa and Acinetobacter baumannii from Hospitalized Patients in the United States, 1998 to 2001,” Antimicrobial Agents and Chemotherapy, Vol. 47, No. 5, 2003, pp. 1681-1688. doi:10.1128/AAC.47.5.1681-1688.2003 [13] M. S. Wilke, A. L. Lovering and N. C. J. Strynadka, “β- Lactam Antibiotic Resistance: A Current Structural Per- spective,” Current Opinion in Microbiology, Vol. 8, No. 5, 2005, pp. 525-533. doi:10.1016/j.mib.2005.08.016 [14] G. Estiu, D. Suárez and K. M. Merz Jr., “Quantum Me- chanical and Molecular Dynamics Simulations of Ureases and Zn Beta-Lactamases,” Journal of Computational Che- mistry, Vol. 27, No. 12, 2006, pp. 1240-1262. doi:10.1002/jcc.20411 [15] R. Choudhury and S. Srivastava, “Zinc Resistance Me- chanisms in Bacteria,” Current Science, Vol. 81, No. 7, 2001, pp. 768-775. [16] M. L. Vasil, “How We Learnt about Iron Acquisition in Pseudomonas aeruginosa: A Series of Very Fortunate Events,” Biometals, Vol. 20, No. 3-4, 2007, pp. 587-601. doi:10.1007/s10534-006-9067-2 [17] T. Krell, A. Busch, M.-E. Guazzaroni, J. Lacal, M.-T. Gallegos and W. Téran, “The Use of Microcalorimetry to Study Regulatory Mechanisms in Pseudomonas,” In: J. L. Ramos and A. Filloux, Eds., Pseudomonas, Springer Ne- therland, Dordrecht, 2007, pp. 255-277. [18] A. Kadry, “Lack of Efflux Mechanism in a Clinical Iso- late of Pseudomonas aeruginosa Highly Resistant to Beta-Lactams and Imipenem,” Folia Microbiologica, Vol. 48, No. 4, 2003, pp. 529-533. doi:10.1007/BF02931336 [19] M. A. Webber and L. J. V. Piddock, “The Importance of Efflux Pumps in Bacterial Antibiotics Resistance,” Jour- nal of Antimicrobial Chemotherapy, Vol. 51, No. 1, 2003, pp. 9-11. doi:10.1093/jac/dkg050 [20] F. Van Bambeke, E. Balzi and P. M. Tulkens, “Antibiotic Efflux Pumps,” Biochemical Pharmacology, Vol. 60, No. 4, 2000, pp. 457-470. doi:10.1016/S0006-2952(00)00291-4 [21] K. Poole, “Multidrug Efflux Pumps and Antimicrobial Resistance in Pseudomonas aeruginosa and Related Or- ganisms,” Journal of Molecular Microbiology and Bio- technology, Vol. 3, No. 2, 2001, pp. 255-264. [22] Y.-T. Chen, H. Y. Chang, C. L. Lu and H.-L. Peng, “Evolutionary Analysis of the Two-Component System in Pseudomonas aeruginosa PAO1,” Journal of Molecular Evolution, Vol. 59, No. 6, 2004, pp. 725-737. doi:10.1007/s00239-004-2663-2 [23] J. F. Guespin-Michel, G. Bernot, J. P. Comet, A. Méireau, A. Richard, C. Hulen and B. Polack, “Epigenesis and Dynamic Similarity in Two Regulatory Networks in Pseu- domonas aeruginosa,” Acta Biotheoretica, Vol. 52, No. 4, 2004, pp. 379-390. doi:10.1023/B:ACBI.0000046604.18092.a7 [24] C. K. Stover, X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory and M. V. Olson, “Complete Genome Sequence of Pseudomonas aeruginosa PAO1, an Opportunistic Pa- thogen,” Nature, Vol. 406, No. 6799, 2000, pp. 959-964. doi:10.1038/35023079 [25] A. Dötsch, F. Klawonn, M. Jarek, M. Scharfe, H. Blöcker and S. Häussler, “Evolutionary Conservation of Essential and Highly Expressed Genes in Pseudomonas aerugi- nosa,” BMC Genomics, Vol. 11, No. 1, 2010, p. 234. doi:10.1186/1471-2164-11-234 [26] M. E. Falagas, I. A. Bliziotis, S. K. Kasiakou, G. Samonis, P. Athanassopoulou and A. Michalopoulos, “Outcome of Infections Due to Pandrug-Resistant (PDR) Gram-Bacte- ria,” BMC Infectious Diseases, Vol. 5, No. 1, 2005, p. 24. doi:10.1186/1471-2334-5-24 [27] D. Landman, S. Bratu, M. Alam and J. Quale, “Citywide Emergence of Pseudomonas aeruginosa Strains with Re- duced Susceptibility to Polymyxin B,” Journal of Anti- microbial Chemotherapy, Vol. 55, No. 6, 2005, pp. 954- 957. doi:10.1093/jac/dki153 [28] A. Huh and Y. Kwon, “Nanoantibiotics: A New Paradigm for Treating Infectious Diseases Using Nanomaterials in the Antibiotics Resistant Era,” Journal of Controlled Re- lease, Vol. 156, No. 2, 2011, pp. 128-145. doi:10.1016/j.jconrel.2011.07.002 [29] S. Oie, T. Uematsu, A. Sawa, H. Mizuno, M. Tomita, S. Ishida, Y. Okano and A. Kamiya, “In Vitro Effects of Combinations of Antipseudomonal Agents against Seven Strains of Multidrug-Resistant Pseudomonas aeruginosa,” Journal of Antimicrobial Chemotherapy, Vol. 52, No. 6, 2003, pp. 911-914. doi:10.1093/jac/dkg478 [30] T. Hoang, H. Schweizer, “Characterization of Pseudo- monas aeruginosa Enoyl-Acyl Carrier Protein Reductase (Fabl): A Target for the Antimicrobial Triclosan and Its Role in Acylated Homoserine Lactone Synthesis,” Jour- nal of Bacteriology, Vol. 181, No. 17, 1999, pp. 5489- 5497. [31] J. S. Soothill, “Bacteriophage Prevents Destruction of Skin Grafts by Pseudomonas aeruginosa,” Burns, Vol. 20, No. 3, 1994, pp. 209-211. doi:10.1016/0305-4179(94)90184-8 [32] M. Singh, S. Singh, S. Prasad I. Gambhir, “Nanotechnol- ogy in Medicine and Antibacterial Effect of Silver Na- noparticles,” Digest Journal of Nanomaterials and Bio- structures, Vol. 3, No. 3, 2008, pp. 115-122. [33] A. N. Brown, K. Smith, T. A. Samuels, J. Lu, S. O. Obare and M. E. Scott, “Nanoparticles Functionalized with Am- picillin Destroy Multiple-Antibiotic-Resistant Isolates of Copyright © 2012 SciRes. JBNB  Overview of Multidrug-Resistant Pseudomonas aeruginosa and Novel Therapeutic Approaches Copyright © 2012 SciRes. JBNB 527 Pseudomonas aeruginosa and Enterobacter aerogenes and Methicillin-Resistant Staphylococcus aureus,” Ap- plied and Enviromental Microbiology, Vol. 78, No. 8, 2012, pp. 2768-2774. doi:10.1128/AEM.06513-11 [34] L. Zhang, D. Pornpattananangkul, C.-M. Hu and C.-M. Huang, “Development of Nanoparticles for Antimicrobial Drug Delivery,” Current Medicinal Chemistry, Vol. 17, No. 6, 2010, pp. 585-594. doi:10.2174/092986710790416290 [35] M. Alipour, M. Halwani, A. Omri and Z. Suntres, “An- timicrobial Effectiveness of Liposomal Polymyxin B against Resistant Gramnegative Bacterial Strains,” Inter- national Journal of Pharmaceutics, Vol. 355, No. 1-2, 2008, pp. 293-298. doi:10.1016/j.ijpharm.2007.11.035
|