 Open Journal of Veterinary Medicine, 2012, 2, 104-112 http://dx.doi.org/10.4236/ojvm.2012.23018 Published Online September 2012 (http://www.SciRP.org/journal/ojvm) Production and B i o chemestry -Molecular Analysis of Microbial Community Fermenting Whey as a Potential Probiotic for Use Animals Eduardo N. Esteban1, Mirentxu Indart2, Silvia Cerone2, G. de Yaniz2, Ana G. Inza2, Herminia Landi3, Silvina Mogni2, Marcela Juliarena1, Leticia Igarza2* 1Department of A n i mal Health, Fa c u l ty of Veterinary Science, Universidad Nacional del Centro de la Provincia de Buenos Aires, Tandil, Argentina 2Department of Physiology and Pathology, Faculty of Veterinary Medicine, Universidad Nacional del Centro de la Provincia de Buenos Aires, Tandil, Argentina 3Department of Animal Production, Faculty of Veterinary Science, Universidad Nacional del Centro de la Provincia de Buenos Aires, Tandil, Argentina Email: *letigar@vet.unicen.edu.ar Received May 29, 2012; revised June 28, 2012; accepted July 10, 2012 ABSTRACT The aims of this work were: To achieve a simple and low cost propagation of potential probiotic agents using plain whey as a culture medium, study the diversity of the members of the bacterial community (MC) in plain whey and to evaluate the probiotic capacity of this MC. After a systematic selection of agents according to their growing capacity in whey, the constituted MC was considered as a unit. Biochemical characterization of the lactic acid bacteria were performed using the API system. Molecular characterization of the lactic acid bacteria was realized using AFLP™ DNA- fi ngerprinting, partial 16S rDNA sequence analysis and PCR-denaturing gradient gel electrophoresis (PCR-DGGE). The physiological characterization of yeast was determined with the automated microplate method Allev/Biolomics and using yeast charac- terization system based on standard taxonomic criteria. The identification molecular was realized by PCR-fingerprinting. The resistance of MC to pH and bile salts were evaluated. The MC was composed of agents from different separated Dominium like Bacteria (Lactobacillum) and Eukaria (yeast). They are multispecies and also multistrain assuring high biodiversity. The MC grew at low pH and different co ncentrations bile salts. Keywords: Probiotic; Multispecies; Multistrain; Plain Whey 1. Introduction Whey is a cheese industry effluent and a powerful envi- ronmental contaminant, in most cheese making regions in Argentina. In recent years, the recovery and marketing of whey components like proteins, minerals and lactose has diminished the quantities of th is effluent discarded to the environment [1]. The fermentation of whey by dif- ferent procedures as an alternative to produce single cell protein, beverages, ethanol and methane has been com- municated by various authors [2-4]. In our case the pro- duction of a microbial community fermenting whey could be utilized as potential probio tic in animals. A pro- biotic is a “live microbial feed supplement which benefi- cially affects the host animal by improving its intestinal microbial balance” [5]. Thus, using a probiotic means an intervention on the intestinal ecosystem. Probiotic bacte- ria would have antagonistic impact against intestinal in- fectious bacteria just by competitive exclusion [6], mo du- lating the intestinal medium, favoring growth of friendly bacteria or producing natural antibiotics or bacteriocines [7]. The aim of this work was: To conform a microbial community of simple and low cost propagation with po- tential probiotic agen ts using plain whey as a culture me- dium, to analyze the species that constitute the micro bial community (MC) by means isolating and identifying strains through the application biochemistry and molecu- lar techniques and to evaluate the probiotic capacity of this MC. 2. Materials and methods Development of the microbial community: Fresh pooled whey from cheese producing companies located in Tandil, Argentina, was collected and transported to destination in food grade stainless steel tanks. Average *Corresponding author. C opyright © 2012 SciRes. OJVM  E. N. ESTEBAN ET AL. 105 protein concentration was 3.5 ± 0.5 mg/ml and pH 5.7 ± 0.3. Immediately after arriving, whey was centrifuged up to 2800 xg keeping a continuous flow of 4.167 l/min, at room temperature. Fat free whey was sterilized by micro filtering successively through 1 µ, 0.5 µ and 0.1 µ pore size Koch membranes and was used for MC develop- ment. We examined whey fermentation capacity of some Caucasian Kefir agents and then added local lactic acid bacteria obtained from pooled whey in local cheese pro- ducing plants. Criteria for the addition of new local strains were as follows: The final pH had to be lower and the biomass (g/l) had to be equal or larger than obtained by Kefir agents. It was considered a satisfactory commu- nity fermenting whey which consistently d rops pH down to 3.6 ± 0.1 and biomass increases to 6 ± 0.5 g/l after 24 h culture. Cultures were incubated 24 h in stationary conditions at 37˚C. Biomass was harvested by centrifu- gation at 10˚C and 2800 xg keeping a continuous flow of 4.167 l/min. Freeze-drying: Compact biomass obtained by MC centrifugation was suspended (1:2) in 10% (w/v) recon- stituted skim milk and 5% (w/v) sodium glutamate (pH was adjusted to 5 with 1M NaOH). Bacterial suspension was freeze-drier. After the freeze-drying cycle has been complete, dried MC was stored at 4˚C unde r va cuum. Isolation and biochemical characterization of the lac- tic acid bacteria were carried out by the Centro de refer- encia de Lactobacillus, Tucumám, Argentina (CERELA). Lactic acid bacteria isolation: A twenty grams sam- ple of the compact biomass obtained by MC centrifuga- tion were added to 180 ml of saline-2 peptone water (2% of NaCl, 0.1% of bacteriological peptone and 1% of Tween 80) and mixed for 3 min into a sterile stomacher bag (Stomacher Lab-Blender 400, A. J. Seward Lab., London, England) Serial dilutions were made in 1‰ peptone water and plated on MRS [8] agar, MRS pH 5.4, LAPTg agar [9], KF agar and ST medium, all supplemented with cyclo- heximide (25 µg/ml) to suppress yeast growth. Plates were incubated at 30˚C and 37˚C anaerobically for 6 days. Isolated colonies that differed microscopically were randomly selected. The isolates were propagated in MRS broth and purified. Gram positive rods and negative to catalase reaction were selected for further studies as pre- sumptive Lactobacillus species. Biochemical characterization: Bacteria were first clustered on the basis of cellular morphology, growth at 15˚C, 30˚C, 37˚C and 45˚C, 0.1% and 0.3% methylene blue, nitrate reduction, indol production, ammonia pro- duction from arginina, esculin hydrolysis, CO2 production from glucose and gluconate, ability to form diacetyl from citrate. Carbohydrate fermentation tests of selected lacto- bacilli were performed using the API 50CH system (bio- Merieux, Marcyl’ Etoile France) according to the manu- facturer instructions. Tests preparations were incubated at 37˚C and final readings were made after 48 h. Fermenta- tion profiles were analyzed by APILAB Plus version 4.0 program (bioMerieux, Ma rcy l’ Etoile France). Molecular characterization of the lactic acid bacte- ria: AFLP™ DNA-fingerprinting, partial 16S rDNA se- quence analysis and PCR-denaturing gradient gel elec- trophoresis (PCR-DGGE) were carried out by the BCCM/ LMG Bacteria Collection (Ghent University. Belgium). AFLP™ (The AFLP™ technology is subject to patents and patent applications and AFLP is a registered trademark, all owned by Keygene N. V., the Netherlands) is a PCR based technique for whole genome DNA fingerprinting via the selective amplification of restriction fragments [10]. Five lyophilized individual specie biochemically char- acterized and 2 lyophilized samples of the whole MC, obtained 4 months apart, were sent to the BCCM/LMG. The twelve lyophilized cultures were recovered and checked for purity on MRS (Oxoid CM361) after an- aerobic incubation at 37˚C for 72 h. One well isolated colony was picked for further cultiv ation and subsequent analyses. Elementary bacteriological tests (cell mor- phology, gram stain, oxidase and catalase reactions) were performed for purity check. DNA was prepared using the method of Gevers et al. 2001 [11], slightly modified. Purified total DNA was digested by two restriction enzymes (4 and 6 base cutter). In this way, only a limited number of fragments with two different ends and of suitable size for efficient PCR were obtained. Small ds DNA molecules (15 - 20 bp) contain- ing one compatible end were ligature to the appropriate “sticky end” of the restriction fragments. Both adaptors are restriction half site-specific and have different se- quences. These adaptors serve as binding sites for PCR primers. In the current analyses, the following restriction enzymes and adaptors were used: Restriction enzyme: EcoR I [hexacutter] Adaptor: 5 ’-CTCGTAGACTGCGTAC C -3’ 3’-CTGA CGCATGGTTAA-5’ Restriction enzyme: Taq I [tetracutter] Adaptor: 5 ’-GACGAT G AGTCCTGAC - 3’ 3’-TACT CAGGACTGGC-5’ Selective amplification of some of the restriction fragments: PCR primers were specifically hybridized with the adaptor ends of the restriction fragments. Since the primers contain at their 3’-end one or more so-called “selective bases” that extend beyond the restriction site into the fragment, only those restriction fragments that have the appropriate complementary sequence adjacent to the restriction site will be amplified. Following primer combination was used: E01:5’-GACTGCGTACCAATTCA-3’, T11:5’-GTTT Copyright © 2012 SciRes. OJVM  E. N. ESTEBAN ET AL. 106 CTTATGAGTCCTGACCGAA-3’. PCR products were separated according to their length using an ABI Prism® 3130XL Genetic Analyzer. Fragments that contain an adaptor specific for the restriction half site created by the 6-bp cutter are visualized due to the 5’-end labeling of the corresponding primer with the fluorescent dye FAM. The resulting electrophoreses patterns were normal- ized and subjected to a band pattern recognition proce- dure using the GeneMapper 4.0 software (Applera, USA). A normalized table of peaks, containing fragments of 20 to 600 base pairs, was transferred into the BioNumer- ics™ 4.61 software. For numerical analysis, a data inter- val was delineated between the 40 and 580 bp bands of the internal size standard. The profiles were compared with the reference profiles of the lactic acid bacteria (in- cluding bifidobacteria) as currently available in BCCM database. Clustering of the patterns was done using the Dice coefficient and the Upgma algorithm. Partial 16S rDNA sequence analysis and phyloge- netic study: DNA prepared for the PCR-DGGE analyses was used. 16S rRNA genes were amplified by PCR using the forward primer 16F27 (pA) 5’AGA GTT TGA TCC TGG CTC AG 3’ and 6R1522 (pH) 5’AAG GAG GTG ATC CAG CCG CA 3’ (hybridizing po sition referring to E. coli 16S rRNA gene sequence numbering 8-27 and 1541-1522 respectively). PCR amplified 16S rDNAs were purified using the NucleoFast® 96 PCR Clean-up Kit (Macherey-Nagel, Düren, Germany). Sequencing reactions were performed using the BigDye® Terminator Cycle Sequencing Kit and purified using the BigDye® XTerminatorT Purification Kit Sequencing was per- formed using an ABI Prism® 3130XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The following forward and reverse primers were used to get a partial overlap of sequences, ensuring highly reliable assembled data: 16F358 (Gamma) 5’CTC CTA CGG GAG GCA GCA GT 3’, 16R339 (Gamma) 5’ACT GCT GCC TCC CGT AGG AG 3’, 16R519 (BKL1) 5’ GTA TTA CCG CGG CTG CTG GCA 3’ (hybridizing position referring to E. coli 16S rRNA gene sequence numbering 339-358, 358-339 and 536-516). Sequence assembly was performed by using the program Autoas- sembler™ (Applied Biosystems, Foster City, CA, USA). A similarity matrix was created using the software package Bio Numerics (Applied Maths, Belgium) by homology calculation with a gap penalty of 0% and after discarding unknown bases and is based on pairwise ali- gnment using an open gap penalty of 100% and a unit gap penalty of 0%. Phylogenetic analysis was performed using the soft- ware package Bio Numerics after including the consen- sus sequence in an alignment of small ribosomal subunit sequences collected from the international nucleotide sequence library EMBL. A resulting tree was constructed using the neighbor-joining method. PCR-denaturing gradient gel electrophoresis (PCR- DGGE): PCR-DGGE analysis was carried out as de- scribed in Van Hoorde et al., 2008 [12]. Physiological and molecular characterization of yeast. Each sample of freezer-died power of MC after addi- tion of an equal amount of peptone water were cultured on dextrose yeast extract peptone agar (DYPA) with 0.02% of chloramphenicol. The physiological and mor- phological profiles of the yeast isolates were determined with the automated microplate method Allev/Biolomics (BioAware SA, Hannut, Belgium) [13], and using yeast characterization system based on standard taxonomic criteria [14,15]. The identification molecular was real- ized according to Kutzman and Robnett 1998 [16] and Barnett et al., 2000 [17]. For DNA sequence analyses, the primer pair LR0R- LR6 was used to amplify the D1/D2 region of the large subunit (LSU) of the ribosomal gene complex [18]. Suc- cessful PCR amplifications resulted in a single band ob- served on a 0.8% (w/v) agarose gel, corresponding to approximately 600 bp. PCR produ cts were cleaned using the QIAquick PCR purification kit, following the manu- facturer’s protocol. Sequencing reactions were performed using a CEQ DTCS Quick Start Kit (Beckman Coulter, Fullerton, California, USA), according to the manufac- turer’s recommendations, with the primers LR0R and LR3. Nucleotide sequences were determined using a CEQ 2000 XL capillary automated sequencer. The se- quences were assembled and edited in Sequencher 4.8 (Gene Codes Corp., Ann Arbor, Michigan, USA). After initial BLAST searches for the most similar sequences, alignments were performed to compare type strain se- quences with the query sequences. The number of sub- stitutions and potential insertions or deletions (indels) was determined from these pairwise comparisons using BioEdit 7.0.5.3 [19]. The sequences were deposited in the EMBL databank (http://www.ebi.ac.uk/EMBL; Hinx- ton, UK) with accession numbers DIV/08 182 A, 182 B, 182 C, 182 D. High-molecular-mass DNA for PCR-fingerprint ana- lyses was extracted using the Invisorb® Spin Plant Mini Kit (Invitek GmbH, Berlin, Germany) as described by Groenewald et al., 2008 [20]. PCR-based fingerprinting with the single primer M13, a ubiquitous microsatellite sequence [21], was applied as described before by Gro- enewald et al., 2008 [20]. The resulting M13-based PCR- fingerprint profiles were compared visually with refer- ence strains. “In vitro” evaluation of the MC probiotic capacity. An aliquot of MC, was inoculated into acidified MRS broth to pH 2.5, 3 and 4 with HCl (1N) and non-acidified (pH 6.8) and i n cubat e d f or 3 h at 37˚C. Copyright © 2012 SciRes. OJVM  E. N. ESTEBAN ET AL. 107 After incubation, serial dilutions in peptone water were spread onto MRS agar plates. Plates were incubated anaerobically for 48 h at 37˚C. Percentage of resistance to each analyzed pH was cal- culated by the equation: %RpH = [(UFC/ml)MRS pH... × 100]/(UFC/ml) MRS pH 6.5 [22]. An aliquot of MC was inoculated into acidified MRS broth with 0.03, 0.05 and 0.1 concentrations of bile salt and within bile salts and incubated for 3 h at 37˚C. After incubation, serial dilutions in peptone water were spread onto MRS agar plat es at 1h and 3 h p ost i ncubati on. Plates were incubated anaerobicall y for 48 h at 37 ˚C. Percentage of resistance to each analyzed bile salt concentration was calculated by the equation: %RpH = [(UFC/ml)MRS pH... × 100]/(UFC/ml) MRS pH6.5 [22]. 3. Result 3.1. MC Development In optimum anaerobic culture conditions, the biomass production in creased to 6 ± 0.5 g/l and pH fell to 3.6 ± 0.1 after 24 h culture. Periodic microscopic examination of the MC showed a homogeneous proportion in the quantity of yeast and lactic acid bacteria. The viability after MC rehydration was 3.2 × 109 lac- tobacilli plus 1.5 × 106 yeast CFU per gram lyophilized compact biomass. 3.2. Isolation and Biochemical Characterization of the Lactic Acid Bacteria Most of the random isolates from MC (95%) expressed general characteristics corresponding to lactic acid bacte- ria (Gram-positive, catalase-negative, non-motile rods). From each resulting cluster, one to four strains were selected for further analysis so twelve lactobacilli were characterized by fermentation profiles. Phenotypic characterization of the microbial commu- nity indicated the following species and strains: Lactoba- cillus helveticus (ID11609, ID11610, ID11618), Lacto- bacillus delbrueckii (ID11611), Lactobacillus paracasei (11614), Lactobacillus fermentum (ID11613, ID11616, ID11619), Lactobacillus sp (ID11612, ID11615, ID11617), and Lactobacillus buc h neri (ID11620). 3.3. Molecular Characterization of Lactic Acid Bacteria The results of the iden tification using AFLP and 16S and PCR-DGGE are given. Two colony types were i s ol ated from ID11615: Smooth (t1) and rough (t2) col o ny. The Lactobacillus identificated were: L. helveticus (ID11609, ID11610, ID11611 and ID11612), L. fermentum (ID11613, ID11615 t1 , ID11616), L. paracasei ID11614, ID11618), L. casei (ID11617), L. parabuchneri (ID11620) as are shown in the dendo- grams of the cluster analyses of AFLPTM profiles (Fig- ures 1-4). For the rough colony and for ID11619, no profile could be obtained. Parcial 16S rDNA sequence analysis was carried out. The percentage 16S rDNA sequence similarity was Lactobacillus gasseri DSM 20243T (100) and Lactobacill us pa ni s DSM 6035T (99.8) respectively. Similar DGGE results were obtained for both gradients (35% - 60% and 35% - 70%). The results are shown in Figure 5. 3.4. Physiological and Molecular Characterization of Yeast The results obtained are shown in Table 1. 3.5. “In Vitro” Evaluation of the MC Probiotic Capacity Obtained percentage of resistance pH and bile salts are shown in Table 2. 4. Discussion and Conclusions The Kefir is a complex probiotic. Its lactobacilli and yeast components were selected over thousands of years towards milk fermentation [23]. In a similar manner, we took from Kefir the most adaptable members to grow in whey. Other lactobacilli and yeast found in our environ- ment, and which improved whey fermentation, were also selected and incorporated to the developing consortium. Based on a consistent fermentation in successive whey baths, determined by pH modification and sensorial ex- aminations like smell and taste, we considered the con- stituted community as a unit. This consistent fermenta- tion suggested a symbiotic relationship and adaptation capacity of the MC. This MC is composed by at least seven lactic acid bacteria and two yeast species, comprising agents from separated dominium like Eukaria and Bacteria. The ap- plication of molecular biology tools to better understand the integrated process of this complex consortium would allow us, in the near future, to use specific community members, or specifically tailored consortia of members, to target desired interactions, resulting in health benefits and food production impr ovement [24,25]. Selected lactic acid bacteria and yeast are multispecies and also multistrain assuring high biodiversity. The ad- vantage of multistra in and multispecies probiotics is that a number of favorable characteristics of individual strains are combined in a single preparation [26]. It has been clearly shown that multispecies preparations have advan- tages when compared to monostrain probiotics or, to a lesser extent, multistrain probiotics [27]. Well-designed Copyright © 2012 SciRes. OJVM 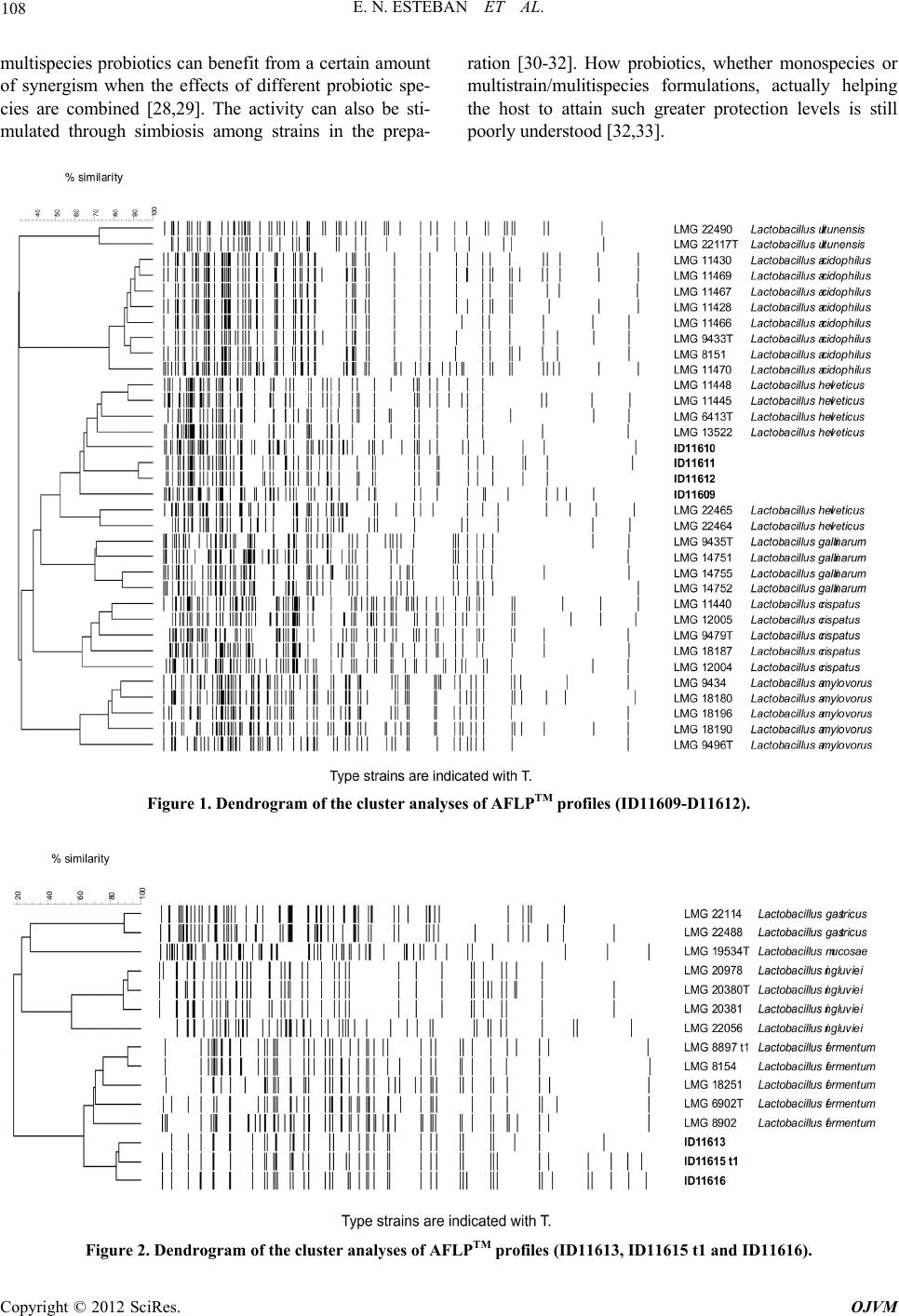 E. N. ESTEBAN ET AL. Copyright © 2012 SciRes. OJVM 108 multispecies probiotics can benefit from a certain amount of synergism when the effects of different probiotic spe- cies are combined [28,29]. The activity can also be sti- mulated through simbiosis among strains in the prepa- ration [30-32]. How probiotics, whether monospecies or multistrain/mulitispecies formulations, actually helping the host to attain such greater protection levels is still poorly underst ood [32,33]. Figure 1. Dendrogram of the cluster analyses of AFLPTM profiles (ID11609-D11612). Figure 2. Dendrogram of the cluster analyses of AFLPTM profiles (ID11613, ID11615 t1 and ID11616). 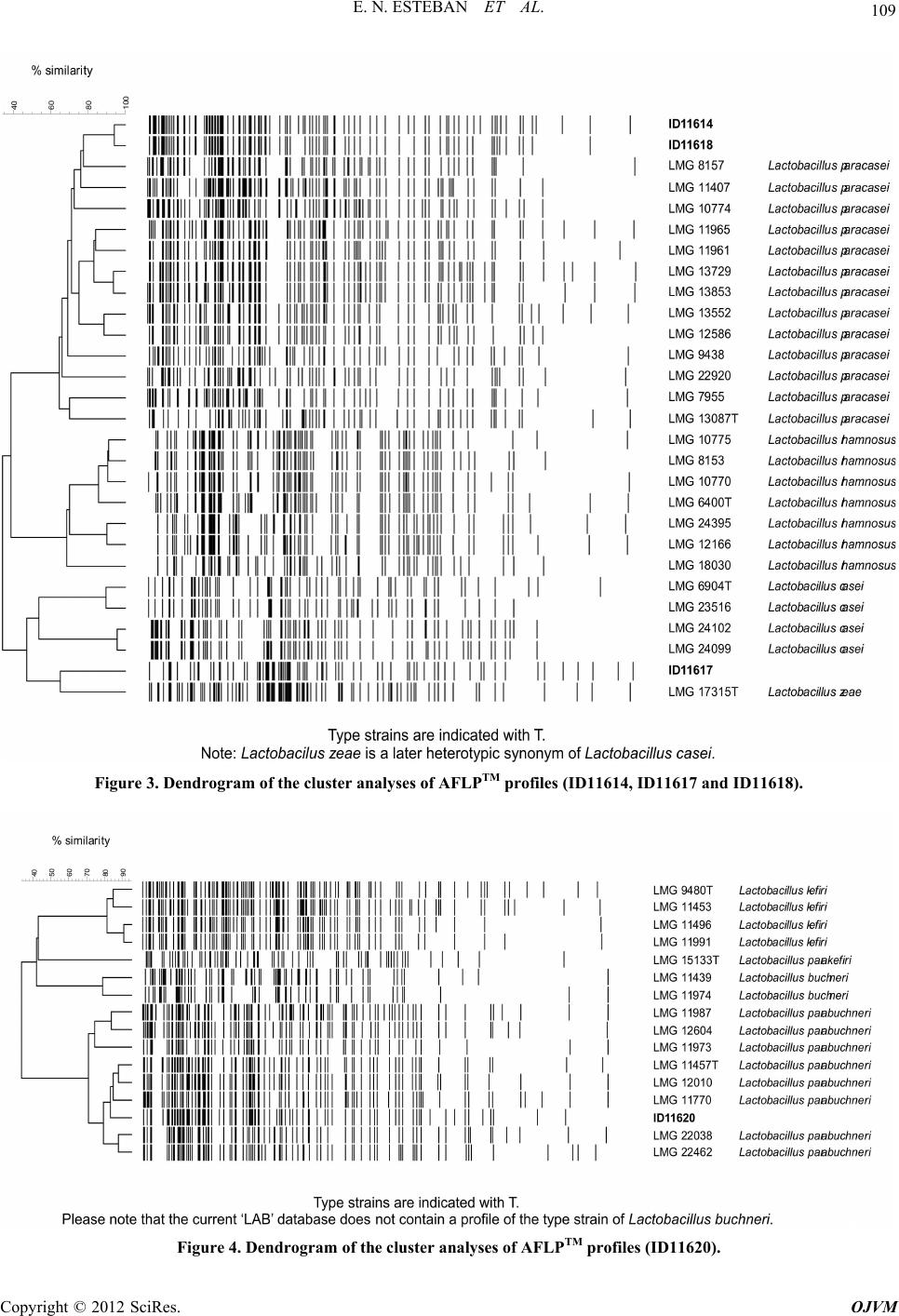 E. N. ESTEBAN ET AL. 109 Figure 3. Dendrogram of the cluster analyses of AFLPTM profiles (ID11614, ID11617 and ID11618). Figure 4. Dendrogram of the cluster analyses of AFLPTM profiles (ID11620). Copyright © 2012 SciRes. OJVM 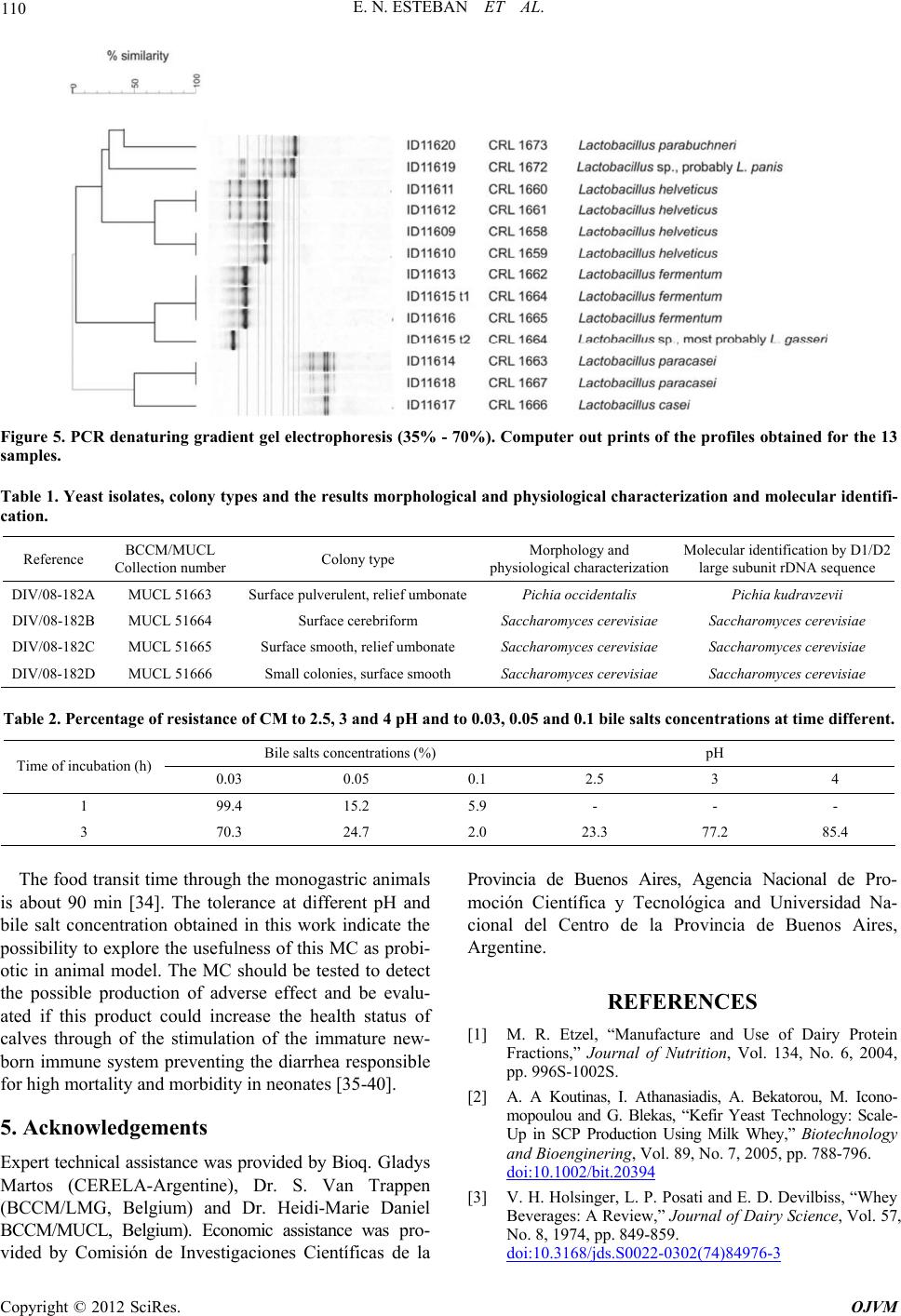 E. N. ESTEBAN ET AL. Copyright © 2012 SciRes. OJVM 110 Figure 5. PCR denaturing gradient gel electrophoresis (35% - 70%). Computer out prints of the profiles obtained for the 13 samples. Table 1. Yeast isolates, colony types and the results morphological and physiological characterization and molecular identifi- cation. Reference BCCM/MUCL Collection numberColony type Morphology and physiological characterizationMolecular identification by D1/D2 large subunit rDNA sequence DIV/08-182A MUCL 51663 Surface pulverulent, relief umbonatePichia occidentalis Pichia kudravzevii DIV/08-182B MUCL 51664 Surface cerebriform Saccharomyces cerevisiae Saccharomyces cerevisiae DIV/08-182C MUCL 51665 Surface smooth, relief umbonate Saccharomyces cerevisiae Saccharomyces cerevisiae DIV/08-182D MUCL 51666 Small colonies, surface smooth Saccharomyces cerevisiae Saccharomyces cerevisiae Table 2. Percentage of resistance of CM to 2.5, 3 and 4 pH and to 0.03, 0.05 and 0.1 bile salts concentrations at time different. Bile salts concentrations (%) pH Time of incubation (h) 0.03 0.05 0.1 2.5 3 4 1 99.4 15.2 5.9 - - - 3 70.3 24.7 2.0 23.3 77.2 85.4 The food transit time through th e monogastric animals is about 90 min [34]. The tolerance at different pH and bile salt concentration obtained in this work indicate the possibility to exp lore the usefulness of this MC as probi- otic in animal model. The MC should be tested to detect the possible production of adverse effect and be evalu- ated if this product could increase the health status of calves through of the stimulation of the immature new- born immune system preventing the diarrhea responsible for high mortality and morbidity in neonates [35-40]. 5. Acknowledgements Expert technical assistance was provided by Bioq. Glad ys Martos (CERELA-Argentine), Dr. S. Van Trappen (BCCM/LMG, Belgium) and Dr. Heidi-Marie Daniel BCCM/MUCL, Belgium). Economic assistance was pro- vided by Comisión de Investigaciones Científicas de la Provincia de Buenos Aires, Agencia Nacional de Pro- moción Científica y Tecnológica and Universidad Na- cional del Centro de la Provincia de Buenos Aires, Argentine. REFERENCES [1] M. R. Etzel, “Manufacture and Use of Dairy Protein Fractions,” Journal of Nutrition, Vol. 134, No. 6, 2004, pp. 996S-1002S. [2] A. A Koutinas, I. Athanasiadis, A. Bekatorou, M. Icono- mopoulou and G. Blekas, “Kefir Yeast Technology: Scale- Up in SCP Production Using Milk Whey,” Biotechnology and Bioenginering, Vol. 89, No. 7, 2005, pp. 788-796. doi:10.1002/bit.20394 [3] V. H. Holsinger, L. P. Posati and E. D. Devilbiss, “Whey Beverages: A Review,” Journal of Dairy Science, Vol. 57, No. 8, 1974, pp. 849-859. doi:10.3168/jds.S0022-0302(74)84976-3  E. N. ESTEBAN ET AL. 111 [4] M. Chartrain, L. Bhatnagar and J. G. Zeikus, “Microbial Ecophysiology of Whey Biomethanation: Comparison of Carbon Transformation Parameters, Species Composition, and Starter Culture Performance in Continuous Culture,” Applied Environmental Microbiology, Vol. 53, No. 5, 1987, pp. 1147-1156. [5] R. Fuller, “Probiotics in Man and Animals, a Review,” Journal of Applied Microbiology, Vol. 66, No. 5, 1989, pp. 365-378. doi:10.1111/j.1365-2672.1989.tb05105.x [6] X. Chen, J. Xu, J. Shuai, J. Chen, Z. Zhang and W. Fang, “The S-Layer Proteins of Lactobacillus crispatus Strain ZJ001 Is Responsible for Competitive Exclusion against Escherichia coli O157: H7 and Salmonella typhimurium,” International Journal of Food Microbiology, Vol. 115, No. 3, 2007, pp. 307-312. doi:10.1016/j.ijfoodmicro.2006.11.007 [7] R. D. Rolfe, “The Role of Probiotic Cultures in the Con- trol of Gastrointestinal Health,” The Journal of Nutrition, Vol. 130, 2000, pp. 396S-402S. [8] J. C. de Man, M. Rogosa and M. E. Sharpe, “A Medium for the Cultivation of Lactobacilli,” Journal of Appied Bacteriology, Vol. 23, No. 1, 1960, pp. 130-135. doi:10.1111/j.1365-2672.1960.tb00188.x [9] P. Raibaud, M. Caulet, J. V. Galpín and G. Mocquot, “Studies on the Bacterial Flora of the Alimentary Tract of Pigs, II Streptococci: Selective Enumeration of the Dominant Groups,” Journal of Applied Microbiology, Vol. 24, No. 3, 1961, pp. 285-306. doi:10.1111/j.1365-2672.1961.tb00262.x [10] P. Vos, R. Hogers, M. Ble eker, M. Reijans, T. van de Lee, M. Hornes, A. Friters, J. Pot, J. Paleman, M. Kuiper and M. Zabeau, “AFLP: A New Technique for DNA Fingerprint- ing,” Nucleic Acids Research, Vol. 2, No. 21, 1995, pp. 4407-4414. doi:10.1093/nar/23.21.4407 [11] S. Gevers, G. Huys and J. Swings, “Applicability of Rep-PCR Fingerprinting for Differentiation of Lactoba- cillus Species,” FEMS Microbiology Letters, Vol. 205, No. 1, 2001, pp. 31-36. doi:10.1111/j.1574-6968.2001.tb10921.x [12] K. Van Hoorde, T. Verstraete, P. Vandamme and G. Huys, “Diversity of Lactic Acid Bacteria in Two Flemish Arti- san Raw Milk Gouda-Ty pe Cheeses,” Food Microbiology, Vol. 25, No. 7, 2008, pp. 929-935. doi:10.1016/j.fm.2008.06.006 [13] V. Robert, P. Evrard and G. L. Hennebert, “BCCM- TM/Allev 2.00 an Automated System for the Identifica- tion of Yeast,” Mycotaxon, Vol. 64, 1997, pp. 455- 463. [14] N. J. W. Kreger-Van Rij, “Classification of Yeast,” In: A. H. Rose and J. S. Harrisonn, Eds., The Yeast, Academic Press Inc., London, 1987, pp. 5-61. [15] J. P. Van der Walt and D. Yarrow, “Methods for de Isola- tion, Maintenance, Classification and Identification of Yeast,” In: N. J. W. Kreger-Van Rij, Ed., The Yeast, a Taxonomic Study, B. V. Elsevier Science Publishers, Am- sterdam, 1984, pp. 45-104. [16] C. P. Kurtzman and C. J. Robnett, “Identification and Phylogeny of Ascomycetous Yeasts from Analysis of Nuclear Large Subunit (26S) Ribosomal DNA Partial Sequences,” Antonie Van Leeuwenhoek, Vol. 73, No. 4, 1998, pp. 331-371. doi:10.1023/A:1001761008817 [17] J. A. Barnett, R. W. Payne and D. Yarrow, “Yeasts: Characteristics and Identification,” 3rd Edition, Cam- bridge University Press, Cambridge, 2000. [18] T. J. White, T. Bruns, S. Lee and J. W. Taylor, “Con- served Primer Sequences for PCR Amplification and Se- quencing from Nuclear Ribosomal RNA,” 1990. www.biology.duke.edu/fungi/mycolab/primers.htm [19] T. A. Hall, “BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT,” Nucleic Acids Symposium Series, Vol. 41, No. 41, 1999, pp. 95-98. [20] M. Groenewald, H. M. Daniel, V. Robert, G. A. Poot and M. T. Smith, “Polyphasic Re-Examination of Debaryo- myces hansenii Strains and Reinstatement of D. hansenii, D. fabryi and D. subglobosus,” Persoonia-Molecular Phylogeny and Evolution of Fungi, Vol. 21, No. 11, 2008, pp. 17-27. doi:10.3767/003158508X336576 [21] G. Vassart, M. Georges, R. Monsieur, H. Brogas, A. S. Lequarre and D. Christophe, “A Sequence in M13 Phage Detects Hypervariable Minisatellites in Human and Ani- mal DNA,” Science, Vol. 235, No. 4789, 1987, pp. 683- 684. doi:10.1126/science.2880398 [22] G. Kociubinski, P. Pérez and G. de Antoni, “Screening of Bile Resistance and Bile of Precipitation in Lactic Acid Bacteria and Bifidobacteria,” Journal Food Protection, Vol. 62, No. 8, 1999, pp. 905-912. [23] E. R. Famworth, “Kefir, a Complex Probiotic,” Food Science and Technology Bulletin: Functional Foods, Vol. 2, No. 1, 2005, pp. 1-17. doi:10.1616/1476-2137.13938 [24] K. B. Amor, E. E. Vaughan and W. M. de Vos, “Advanced Molecular Tools for the Identification of La ctic Aci d Ba c- teria,” The Journal of Nutrition, Vol. 137, No. 3, 2007, pp. 741S-747S. [25] C. Vael and K. Desager, “The Importance of Develop- ment of the Intestinal Microbiota in Infancy,” Current Opinion of Pediatrics, Vol. 21, No. 6, 2009, pp. 794-800. doi:10.1097/MOP.0b013e328332351b [26] M. Campieri and P. Gionchetti, “Probiotics in Inflamma- tory Bowel Disease: New Insight to Pathogenesis or a Possible Therapeutic Alternative?” Gastroenterology, Vol. 116, No. 5, 1999, pp. 1246-1249. doi:10.1016/S0016-5085(99)70029-6 [27] I. Salinas, L. Abelli, F. Bertoni, S. Picchietti, A. Roque, D. Furones, A. Cuesta, J. Meseguer and M. A. Esteban, “Mono- species and Multispecies Probiotic Formulations Produce Different Systemic and Local Immunostimulatory Effects in the Gilthead Seabream (Sparus aurata L.),” Fish & Shellfish Inmunology, Vol. 25, No. 1-2, 2008, pp. 114- 123. doi:10.1016/j.fsi.2008.03.011 [28] M. Collado, J. Meriluoto and S. Salminen, “Measurement of Aggregation Properties between Probiotics and Patho- gens: In Vitro Evaluation of Different Methods,” Journal of Microbiological Methods, Vol. 71, No. 1, 2007, pp. 71-74. doi:10.1016/j.mimet.2007.07.005 [29] H. M. Timmerman, L. E. Niers, B. U. Ridwan, C. J. Koning, L. Mulder, L. M. Akkermans, F. M. Rombouts and G. T. Rijkers, “Design of a Multispecies Probiotic Mixture to Prevent Infectious Complications in Critically Copyright © 2012 SciRes. OJVM  E. N. ESTEBAN ET AL. Copyright © 2012 SciRes. OJVM 112 Ill Patients,” Clinical Nutrition, Vol. 26, No. 4, 2007, pp. 450-459. doi:10.1016/j.clnu.2007.04.008 [30] T. Hosoi, A. Ametani, K. Kiuchi and S. Kaminogawa, “Improved Growth and Viability of Lactobacilii in the Presence of Bacillus subtilis, Catalase, or Subtilisin,” Ca- nadian Journal of Microbiology, Vol. 46, No. 10, 2000, pp. 892-897. [31] A. C. Ouwehand, E. Isolauri, P. V. Kirjavainen, S. Tö Ikkö and S. J. Salminen, “The Mucus Binding of Bifido- Bacterium Lactis Bb12 Is Enhanced in the Presence of Lactobacillus GG and Lact. Delbrüekii Subsp. Bulgari- cus,” Letters in Applied Microbiology, Vol. 30, No. 1, 2000, pp. 10-13. doi:10.1046/j.1472-765x.2000.00590.x [32] H. M. Timmerman, C. J. Koning, L. Mulder, F. M. Rom- boust and A. C. Beynen, “Monostrain, Multistrain and Multispecies Probiotics: A Comparison of Functionality and Efficacy,” International Journal of Food Microbio- logy, Vol. 96, No. 219, 2004, pp. 219-233. doi:10.1016/j.ijfoodmicro.2004.05.012 [33] I. Salinas, A. Cuesta, M. A. Esteban and J. Meseguer, “Dietary Administration of Lactobacillus delbrüeckii and Bacillus subtilis, Single or Combined, on Gilthead Seabream Cellular Innate Immune Responses,” Fish & Shellfish Inmunology, Vol. 19, No. 1, 2005, pp. 67-77. doi:10.1016/j.fsi.2004.11.007 [34] E. Renner, “Cultured Dairy Products in Human Nutri- tion,” Bulletin of the International Dairy Federation, Vol. 225, 1991, pp. 2-24. [35] E. L. Morrell, D. P. Moore, A. C. Odeón, M. A. Poso, E. Odriozola, G. Cantón, F. Paolicchi, F. Malena, M. R. Leunda, C. Marsella and C. M. Campero, “Retrospective Study of Bovine Neonatal Mortality: Cases Reported from INTA Balcarce, Argentina,” Revista Argentina de Microbiología, Vol. 40, No. 3, 2008, pp. 151-157. [36] L. S. Frizzo, E. Bertozzi, L. P. Soto, M. V. Zbrun, G. J. Sequeira, R. Dalla Santina and R. Rodriguez Armestro, “The Effect of Supplementation with Three Lactic Acid Bacteria from Bovine Origin on Growth Performance and Health Status of Group Calves,” Journal of Animal and Veterinary Advances, Vol. 7, No. 4, 2008, pp. 400-408. [37] M. R. Mokhber-Dezfouli, P. Tajik, M. Bolourchi and H. Mahmoudzadeh, “Effects of Probiotics Supplementation in Daily Milk Intake of Newborn Calves on Body Weight Gain, Body Height, Diarrhea Occurrence and Health Condition,” Journal of Biological Science, Vol. 10, No. 18, 2007, pp. 3136-3140. [38] H. M. Timmerman, L. Mulder, H. Everts, D. C. van Espen, E. van der Wal, G. Klaassen, S. M. Rouwers, R. Hartemink, F. M. Rombousts and A. C. Beynen, “Health and Growth of Veal Calves Fed Milk Replacers with or without Probiotics,” Journal of Dairy Science, Vol. 88, No. 6, 2005, pp. 2154-2165. doi:10.3168/jds.S0022-0302(05)72891-5 [39] Q. Shu and H. S. Gill, “Immune Protection Mediated by the Probiotic Lactobacillus Rhamnosus HN001 against Escherichia coli O157: H7 Infection in Mice,” FEMS Immonulogy and Medical Microbiology, Vol. 34, No. 1, 2002, pp. 59-64. [40] E. Isolauri, Y. Sütas, P. Kankaanpää, H. Arvilommi and S. Salminen, “Probiotics: Effects on Immunity,” American Journal of Clinical Nutrition, Vol. 73, No. 2, 2001, pp. 444S-450S.
|