Paper Menu >>
Journal Menu >>
 American Journal of Analytical Chemistry, 2012, 3, 669-674 http://dx.doi.org/10.4236/ajac.2012.39088 Published Online September 2012 (http://www.SciRP.org/journal/ajac) Separation of Amino Acids Based on Thin-Layer Chromatography by a Novel Quinazoline Based Anti-Microbial Agent Supriti Sen*, Sandipan Sarkar*, Pijush Kundu, Subrata Laskar Department of Chemistry, The University of Burdwan, Golapbag, India Email: *sssensupriti@gmail.com Received March 14, 2012; revised May 30, 2012; accepted June 9, 2012 ABSTRACT A newly designed quinazoline based compound, 6-pyridin-2-yl-5,6-dihydro-benzo[4,5]imidazo[1,2-c] quinazoline (PDBIQ) has shown the ability for the easy detection of nineteen amino acids on thin-layer chromatography plates as a spray reagent. This new reagent enabled to produce various distinguishable colors with amino acids with different RF values. The detection limits and the binding ability of PDBIQ with amino acids have been calculated. PDBIQ is also able to detect aminoacids from hydrolised seed protein. The title compound also exhibited profound inhibitory action against some gm (+ve) and gm (−ve) bacterial organisms. This paper deals with synthesis, spectroscopic application and biological evaluation of the organic moity. Keywords: Thin-Layer Chromatography; Amino Acid; Ninhydrin; Binding Constant; Antimicrobial Property 1. Introduction The chemistry of Quinazolines class compounds are very promising because it shows wide spectram of biological activity like analgesic and anti-inflammatoryanti, antim- icrobial, antihypertensive, anticancer [1-4] etc. activities. Because of such enriched chemistry we are interested in evaluation and application of quinazoline compound. The detection or identification of amino acids is extremely important in biomedical and biochemical analysis for the evaluation of protein structure as the amino acids are the monomeric units of proteins; these amino acids are used by cells for protein biosynthesis, and also exist in the free state in numerous natural products (seeds and leaves) and as the C-terminal determination of degraded proteins. Sev- eral specific and non-specific reagents have been reported on thin-layer chromatography (TLC) plates [5-10]. Such identification is the most well-known reagent is ninhy- drin which is widely used for its remarkable high sensi- tivity. But, it produces same purple/violet color with all amino acids except proline and hydroxyproline. An attempt has been established to overcome this color problem us- ing 6-pyridin-2-yl-5,6-dihydro-benzo[4,5]imidazo[1,2-c] quinazoline (PDBIQ)-ninhydrine as a new reagent which affords distinguishable colors with twenty two protein ami- no acids and, enables convenient and easy detection of such compounds on silica gel “G” for TLC with very good sensi- tivity (detection limit between 0.1 - 0.5 μg at cold condi- tion and 0.05 - 0.2 μg after heating). Herein we report an account on the systematic applica- tion of a newly designed quinazoline based spraying re- agent (PDBIQ) for the detection of amino acids at trace level along with the equilibrium binding constant (k) with different amino acids and bio-activity test against some gm (+ve) and gm (−ve) bacterial organisms. 2. Experimental 2.1. Apparatus and Materials Used Thin layer chromatography plates (20 × 20 cm, thickness 0.1 mm) were prepared using silica gel “G” (Merck, In- dia) and a Unoplan coating apparatus (Shaudon, London, UK). Sample solutions were spotted on to the plates by means of a graduated micropipette (5.0 μL). Electronic absorption spectra were recorded on a JASCO UV-Vis/NIR spectrophotometer model V-570. Pyridine-2-carboxylaldehyde and 2-(2-aminophenyl) benzimidazole for the synthesis of the title compound (PDBIQ) were purchased from Aldrich. Standard amino acids and ninhydrin were procured from Sigma (USA) and n-propanol from Merck (India). All other chemicals and solvents were used as received. The spraying reagent, 6-pyridin-2-yl-5,6-dihydro-benzo[4,5]imidazo[1,2-c]quina- zoline (PDBIQ) was synthesized in our laboratory as de- scribed below. *Corresponding authors. C opyright © 2012 SciRes. AJAC  S. SEN ET AL. 670 2.2. Synthesis of 6-Pyridin-2-yl-5, 6-dihydro-benzo[4,5]imidazo[1,2-c] quinazoline (PDBIQ) An ethanolic solution of 2-(2-aminophenyl)benzimida- zole, (2.09 g, 10.0 mmol) was added to pyridine-2-carbo- xylaldehyde (1.07 g, 10.0 mmol) in ethanol (25.0 mL) at room temperature. Then this mixture was allowed to re- flux for 4.0 h. The white colored crystalline precipitate of the compound (PBBIQ) was obtained from the yellow col- ored solution through slow evaporation of the solvent in few days. The single crystals of L suitable for X-ray crys- tallography were also obtained from the methanolic solu- tion of the white colored product on slow evaporation at room temperature. These single crystals have been used in the experiments. C19H14N4: Anal. Found: C, 76.56; H, 4.75; N, 18.49; Calc.: C, 76.48; H, 4.73; N, 18.78. m.p. 231˚C ± 1˚C, MS: [M + H]+, m/z, 299.34; IR (KBr, cm−1): ν N-H, 2950, ν C=N, 1477;. 1H NMR (δ, ppm in dmso-d6): 8.437 (d, 1H, j = 3.9); 7.906 (d, 1H, j = 7.2); 7.768 - 7.697 (m, 2H); 7.631 (d, 1H, j = 7.2); 7.351 - 7.096 (m, 7H); 6.853 - 6.769 (m, 2H); Yield: 90%. 2.3. Detection of Amino Acids on TLC Plates Standard solutions (1 mg/ml) of amino acids were pre- pared in 0.01 M phosphate buffer (pH 8.0) and spotted on the TLC plates. Spotting volume was always 1 µL; the solutions were diluted approximately when necessary. Pla- tes were air-dried and subjected to TLC using n-propanol- water, 70 + 30 (v/v) as mobile phase. After development plates were dried and sprayed with 0.01% PDBIQ in ethyl alcohol (Reagent 1) and again dried in air for complete evaporation of solvent. The plates were then sprayed with 0.25% ninhydrin in acetone (Reagent 2), dried in air and colors were noted Table 1. The plates were then heated at 110˚C for 10 min in an oven and the colors were recorded again. Colors were always observed visually. Detection limits for the amino acids [11] after use of ninhydrin alone are also given in Table 1. Table 1. Formation of color by amino acids using PDBIQ and ninhydrin reagents, de tection limits for these reagents and for ninhydrin alone on silica gel with n-pr opanol-w a te r 70:30 as mobile phase. Cold condition After final heating Amino acids Observed colors Detection limit (μg)Observed colors Detection limit (μg) Detection limit for ninhydrin (μg) (RF) Glycine Deep orange 0.5 Deep pink 0.1 0.01 (0.03) Alanine Pinkish violet 0.1 Light pink/milky pink0.1 0.009 (0.22) Valine Reddish pink 0.1 Reddish pink 0.05 0.01 (0.14) Leucine Bluish violet 0.5 Violetish pink 0.1 0.01 (0.09) Isoleucine Very light violet 1.0 Light violet 0.1 0.20 (0.32) Serine Deep pink 0.1 Deep bluish pink 0.1 0.008 (0.38) Threonine Yellowish orange/ivory 0.5 Yellowish pink/candy0.1 0.05 (0.28) Aspartic acid Yellowish violet 0.2 Greyish violet 0.1 0.10 (0.12) Aspargine Light yellow/pale cream 1.0 Greyish yellow 0.1 0.10 (0.45) Glutamic acid Light violet 0.5 Light violet 0.1 0.04 (0.33) Glutamine Light violet 0.5 Light violet 0.2 0.10 (0.38) Lysine Reddish violet 0.2 Brick red 0.1 0.005 (0.42) Histidine Yellowish violet 0.1 Yellowish pink/petal0.1 0.05 (0.18) Arginine Light pink/mauve 0.5 Pink 0.1 0.01 (0.05) Phenyl alanine Orangish violet 1.0 Greyish pink 0.2 0.05 (0.58) Tyrosine Light violet 1.0 Light pink 0.1 0.03 (0.51) Tryptophan Greyish violet 0.5 Pinkish violet 0.1 0.05 (0.55) Cysteine Yellowish violet 2.0 Pinkish violet 1.0 0.02 (0.41) Cystine Very light pink 2.0 Light pink 1.0 0.01 (0.35) Methionine Lilac/bluish violet 0.5 Bluish violet 0.2 0.01 (0.48) Proline Light yellow/off white 1.0 Grey/beige 0.2 0.10 (0.22) Hydroxy proline Pinkish violet 0.2 Yellowish brown 0.1 0.05 (0.34) Copyright © 2012 SciRes. AJAC 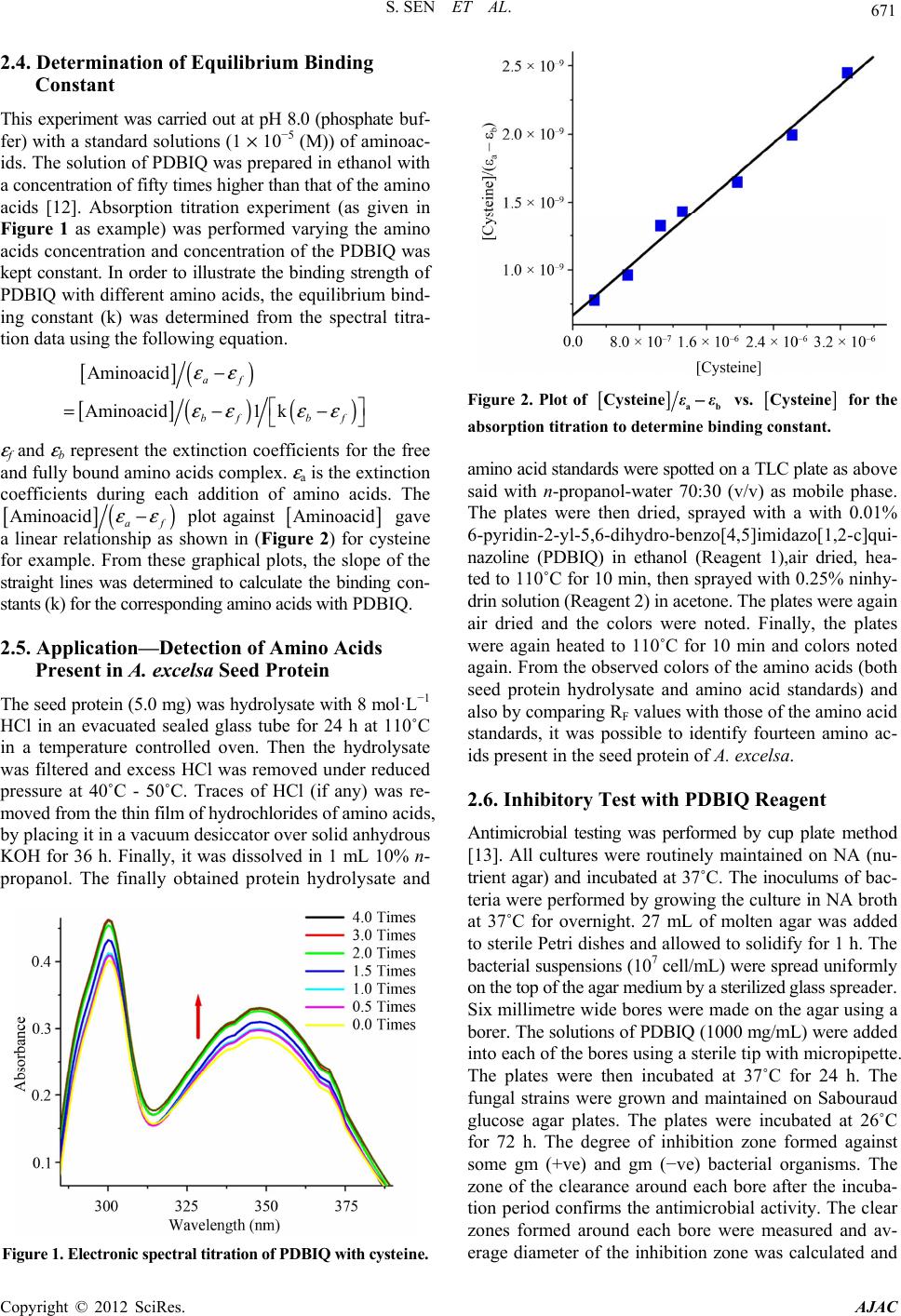 S. SEN ET AL. 671 2.4. Determination of Equilibrium Binding Constant This experiment was carried out at pH 8.0 (phosphate buf- fer) with a standard solutions (1 × 10−5 (M)) of aminoac- ids. The solution of PDBIQ was prepared in ethanol with a concentration of fifty times higher than that of the amino acids [12]. Absorption titration experiment (as given in Figure 1 as example) was performed varying the amino acids concentration and concentration of the PDBIQ was kept constant. In order to illustrate the binding strength of PDBIQ with different amino acids, the equilibrium bind- ing constant (k) was determined from the spectral titra- tion data using the following equation. [ ] () [] ()() Aminoacid Aminoacid 1 af bf εε εε − =−kbf εε − ε f and ε b represent the extinction coefficients for the free and fully bound amino acids complex. ε a is the extinction coefficients during each addition of amino acids. The [ ] () Aminoacid εε − af plot against [ ] Aminoacid gave a linear relationship as shown in (Figure 2) for cysteine for example. From these graphical plots, the slope of the straight lines was determined to calculate the binding con- stants (k) for the corresponding amino acids with PDBIQ. 2.5. Application—Detection of Amino Acids Present in A. excelsa Seed Protein The seed protein (5.0 mg) was hydrolysate with 8 mol·L−1 HCl in an evacuated sealed glass tube for 24 h at 110˚C in a temperature controlled oven. Then the hydrolysate was filtered and excess HCl was removed under reduced pressure at 40˚C - 50˚C. Traces of HCl (if any) was re- moved from the thin film of hydrochlorides of amino acids, by placing it in a vacuum desiccator over solid anhydrous KOH for 36 h. Finally, it was dissolved in 1 mL 10% n- propanol. The finally obtained protein hydrolysate and Figure 1. Electronic spectral titration of PDBIQ with cysteine. [ ] Figure 2. Plot of ab Cysteine −εε vs. [ ] Cysteine for the absorption titration to deter mine binding constant. amino acid standards were spotted on a TLC plate as above said with n-propanol-water 70:30 (v/v) as mobile phase. The plates were then dried, sprayed with a with 0.01% 6-pyridin-2-yl-5,6-dihydro-benzo[4,5]imidazo[1,2-c]qui- nazoline (PDBIQ) in ethanol (Reagent 1),air dried, hea- ted to 110˚C for 10 min, then sprayed with 0.25% ninhy- drin solution (Reagent 2) in acetone. The plates were again air dried and the colors were noted. Finally, the plates were again heated to 110˚C for 10 min and colors noted again. From the observed colors of the amino acids (both seed protein hydrolysate and amino acid standards) and also by comparing RF values with those of the amino acid standards, it was possible to identify fourteen amino ac- ids present in the seed protein of A. excelsa. 2.6. Inhibitory Test with PDBIQ Reagent Antimicrobial testing was performed by cup plate method [13]. All cultures were routinely maintained on NA (nu- trient agar) and incubated at 37˚C. The inoculums of bac- teria were performed by growing the culture in NA broth at 37˚C for overnight. 27 mL of molten agar was added to sterile Petri dishes and allowed to solidify for 1 h. The bacterial suspensions (107 cell/mL) were spread uniformly on the top of the agar medium by a sterilized glass spreader. Six millimetre wide bores were made on the agar using a borer. The solutions of PDBIQ (1000 mg/mL) were added into each of the bores using a sterile tip with micropipette. The plates were then incubated at 37˚C for 24 h. The fungal strains were grown and maintained on Sabouraud glucose agar plates. The plates were incubated at 26˚C for 72 h. The degree of inhibition zone formed against some gm (+ve) and gm (−ve) bacterial organisms. The zone of the clearance around each bore after the incuba- tion period confirms the antimicrobial activity. The clear zones formed around each bore were measured and av- erage diameter of the inhibition zone was calculated and Copyright © 2012 SciRes. AJAC  S. SEN ET AL. 672 expressed in millimeter. 3. Results and Discussion 3.1. Synthesis and Characterization of PDBIQ The spraying reagent, 6-pyridin-2-yl-5,6-dihydro-benzo [4,5]imidazo[1,2-c]quinazoline (PDBIQ) was synthesiz- ed by refluxing 2-(2-aminophenyl)-benzimidazol and pyridine-2-carboxaldehyde in equi-molar ratio in metha- nol viz. Scheme 1. The rearranged white product (PDBIQ) was obtained as the end-product. The structural analysis by spectroscopic tools and the X-ray crystallographic tools has confirmed this cyclic rearranged product. The solid state structure of PDBIQ has already been reported in the literature [9], and for that reason we are not describing the structure here, and the solid state structure of PDBIQ has been included in supplementary file as Figure S1. 3.2. Detection of Amino Acids It is observed from Table 1 that detection limits obtained after uses of PDBIQ are very low in both cases before heating (0.1 - 2.0 μg) and after heating (0.05 - 1.0 μg) and various distinguishable colors were produced. Sometimes the detection limit is same before and after heating and in other cases it is somewhat different. It should be noted that identification of amino acids by ninhydrin is in prac- tice difficult, in spite of the high sensitivity of ninhydrin. So the new spray reagent unable to differentiate the amino acids by color reaction. The mechanism of the color for- mation is still uncertain, but we may assume that carbox- ylic group of aminoacids first condensed with PDBIQ (hea- ting at 90˚C for 10 min) to form a carbamide type inter- mediates which form charge transfer complexes with nin- hydrin. 3.3. Determination of Equilibrium Binding Constant The equilibrium binding constant of PDBIQ with differ- ent amino acids was given in Table 2. The values are quite high. This high value indicates that there are some inter- action between PDBIQ and amino acids through charge transfer transition. The Job’s plot Figure 3 shows the maximum 1:1 adduct formation which confirms the ratio of adduct formation between ligand and amino acids dur- ing charge transfer transition. NNH NH2N OHC MeOH N N H N N +Reflux Rearr angement PDBIQ Scheme 1. Synthetic strategy of the reage nt PDBIQ. Table 2. Equilibrium bindi ng constants (k) of the molecular complexes between organic reagent and amino acids at pH 8.0 (phosphate buffer) at 25˚C. Amino acids k [dm3·mole−1] L-Glycine 1.25 × 106 L-Alanine 1.32 × 106 L-Valine 0.89 × 106 L-Leucine 5.47 × 106 L-Isoleucine 3.94 × 106 L-Serine 2.56 × 106 L-Threonine 1.66 × 106 L-Aspartic acid 0.72 × 106 L-Aspargine 1.21 × 106 L-Glutamic acid 3.40 × 106 L-Glutamine 2.72 × 106 L-Lysine 1.72 × 106 L-Histidine 5.40 × 106 L-Arginine 3.34 × 106 L-Phenyl alanine 1.53 × 106 L-Tyrosine 0.86 × 106 L-Tryptophan 1.87 × 106 L-Cysteine 0.79 × 106 L-Cystine 8.52 × 106 L-Methionine 8.89 × 106 L-Proline 5.23 × 106 L-Hydroxy proline 4.04 × 106 3.4. Use of the Method for TLC Detection of the Amino Acids Present in A. excelsa Seed Protein It was found that spraying with a 0.01% solution of 6- pyridin-2-yl-5,6-dihydro-benzo[4,5]imidazo[1,2-c]quinazo- line (PDBIQ) in ethanol combined with spraying with 0.25% ninhydrin solution in acetone enabled detection of fourteen amino acids-arginine, isoleucine, glutamine, lysine, asparagine, phenylalanine, serine, alanine, glutamic acid, valine, aspartic acid, leucine, glycine, and proline even at low concentration of the amino acids. The results were also agreement with the respective RF values of the acids. 3.5. Inhibitory Test with PDBIQ Antimicrobial test of PDBIQ was checked using cup plate method and the observed results of the inhibitory test per- formed with the reagent has been tabulated in Table 3. Here, the diameter of inhibition zone for the organisms in presence of PDBIQ is significantly greater than that of the corresponding controlled replicate experiment. This study indicates that the compound has profound activity against the test organisms. Copyright © 2012 SciRes. AJAC 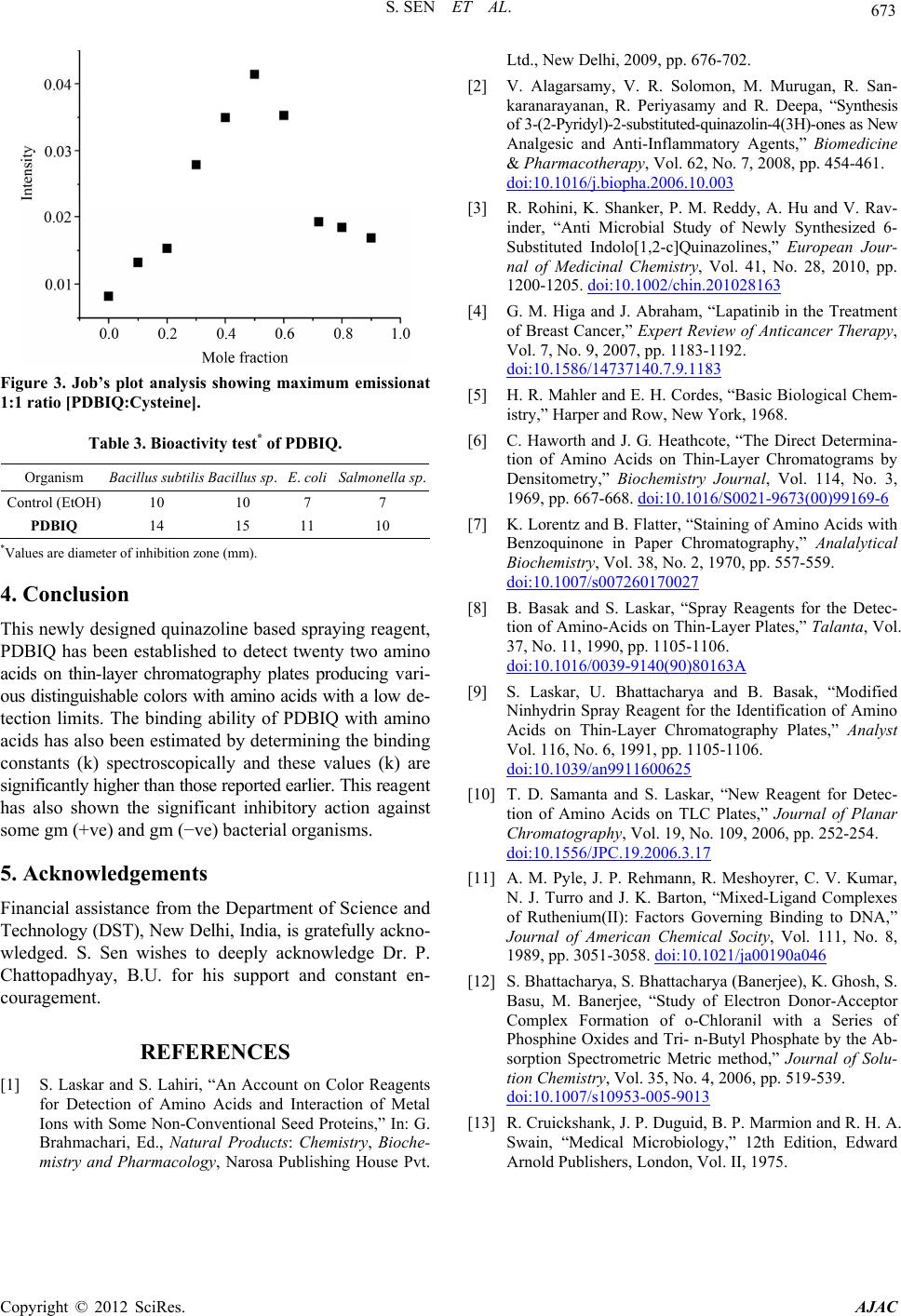 S. SEN ET AL. Copyright © 2012 SciRes. AJAC 673 Figure 3. Job’s plot analysis showing maximum emissionat 1:1 ratio [PDBIQ:Cysteine]. Table 3. Bioactivity test* of PDBIQ. Organism Bacillus subtilis Bacillus sp. E. coli Salmonella sp. Control (EtOH) 10 10 7 7 PDBIQ 14 15 11 10 *Values are diameter of inhibition zone (mm). 4. Conclusion This newly designed quinazoline based spraying reagent, PDBIQ has been established to detect twenty two amino acids on thin-layer chromatography plates producing vari- ous distinguishable colors with amino acids with a low de- tection limits. The binding ability of PDBIQ with amino acids has also been estimated by determining the binding constants (k) spectroscopically and these values (k) are significantly higher than those reported earlier. This reagent has also shown the significant inhibitory action against some gm (+ve) and gm (−ve) bacterial organisms. 5. Acknowledgements Financial assistance from the Department of Science and Technology (DST), New Delhi, India, is gratefully ackno- wledged. S. Sen wishes to deeply acknowledge Dr. P. Chattopadhyay, B.U. for his support and constant en- couragement. REFERENCES [1] S. Laskar and S. Lahiri, “An Account on Color Reagents for Detection of Amino Acids and Interaction of Metal Ions with Some Non-Conventional Seed Proteins,” In: G. Brahmachari, Ed., Natural Products: Chemistry, Bioche- mistry and Pharmacology, Narosa Publishing House Pvt. Ltd., New Delhi, 2009, pp. 676-702. [2] V. Alagarsamy, V. R. Solomon, M. Murugan, R. San- karanarayanan, R. Periyasamy and R. Deepa, “Synthesis of 3-(2-Pyridyl)-2-substituted-quinazolin-4(3H)-ones as New Analgesic and Anti-Inflammatory Agents,” Biomedicine & Pharmacotherapy, Vol. 62, No. 7, 2008, pp. 454-461. doi:10.1016/j.biopha.2006.10.003 [3] R. Rohini, K. Shanker, P. M. Reddy, A. Hu and V. Rav- inder, “Anti Microbial Study of Newly Synthesized 6- Substituted Indolo[1,2-c]Quinazolines,” European Jour- nal of Medicinal Chemistry, Vol. 41, No. 28, 2010, pp. 1200-1205. doi:10.1002/chin.201028163 [4] G. M. Higa and J. Abraham, “Lapatinib in the Treatment of Breast Cancer,” Expert Review of Anticancer Therapy, Vol. 7, No. 9, 2007, pp. 1183-1192. doi:10.1586/14737140.7.9.1183 [5] H. R. Mahler and E. H. Cordes, “Basic Biological Chem- istry,” Harper and Row, New York, 1968. [6] C. Haworth and J. G. Heathcote, “The Direct Determina- tion of Amino Acids on Thin-Layer Chromatograms by Densitometry,” Biochemistry Journal, Vol. 114, No. 3, 1969, pp. 667-668. doi:10.1016/S0021-9673(00)99169-6 [7] K. Lorentz and B. Flatter, “Staining of Amino Acids with Benzoquinone in Paper Chromatography,” Analalytical Biochemistry, Vol. 38, No. 2, 1970, pp. 557-559. doi:10.1007/s007260170027 [8] B. Basak and S. Laskar, “Spray Reagents for the Detec- tion of Amino-Acids on Thin-Layer Plates,” Talanta, Vol. 37, No. 11, 1990, pp. 1105-1106. doi:10.1016/0039-9140(90)80163A [9] S. Laskar, U. Bhattacharya and B. Basak, “Modified Ninhydrin Spray Reagent for the Identification of Amino Acids on Thin-Layer Chromatography Plates,” Analyst Vol. 116, No. 6, 1991, pp. 1105-1106. doi:10.1039/an9911600625 [10] T. D. Samanta and S. Laskar, “New Reagent for Detec- tion of Amino Acids on TLC Plates,” Journal of Planar Chromatography, Vol. 19, No. 109, 2006, pp. 252-254. doi:10.1556/JPC.19.2006.3.17 [11] A. M. Pyle, J. P. Rehmann, R. Meshoyrer, C. V. Kumar, N. J. Turro and J. K. Barton, “Mixed-Ligand Complexes of Ruthenium(II): Factors Governing Binding to DNA,” Journal of American Chemical Socity, Vol. 111, No. 8, 1989, pp. 3051-3058. doi:10.1021/ja00190a046 [12] S. Bhattacharya, S. Bhattacharya (Banerjee), K. Ghosh, S. Basu, M. Banerjee, “Study of Electron Donor-Acceptor Complex Formation of o-Chloranil with a Series of Phosphine Oxides and Tri- n-Butyl Phosphate by the Ab- sorption Spectrometric Metric method,” Journal of Solu- tion Chemistry, Vol. 35, No. 4, 2006, pp. 519-539. doi:10.1007/s10953-005-9013 [13] R. Cruickshank, J. P. Duguid, B. P. Marmion and R. H. A. Swain, “Medical Microbiology,” 12th Edition, Edward Arnold Publishers, London, Vol. II, 1975. 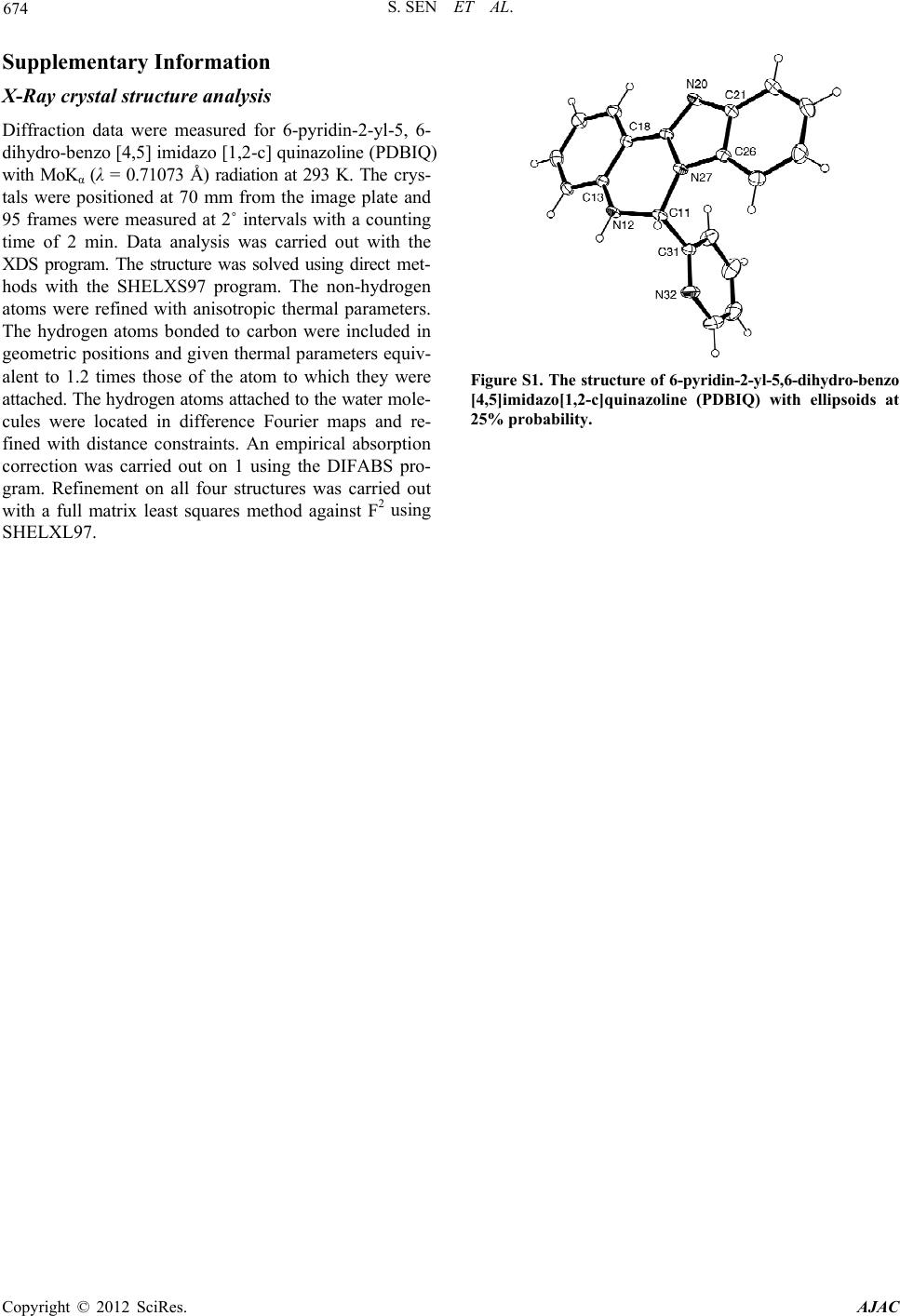 S. SEN ET AL. 674 Supplementary Information X-Ray crystal structure analysis Diffraction data were measured for 6-pyridin-2-yl-5, 6- dihydro-benzo [4,5] imidazo [1,2-c] quinazoline (PDBIQ) with MoKα (λ = 0.71073 Å) radiation at 293 K. The crys- tals were positioned at 70 mm from the image plate and 95 frames were measured at 2˚ intervals with a counting time of 2 min. Data analysis was carried out with the XDS program. The structure was solved using direct met- hods with the SHELXS97 program. The non-hydrogen atoms were refined with anisotropic thermal parameters. The hydrogen atoms bonded to carbon were included in geometric positions and given thermal parameters equiv- alent to 1.2 times those of the atom to which they were attached. The hydrogen atoms attached to the water mole- cules were located in difference Fourier maps and re- fined with distance constraints. An empirical absorption correction was carried out on 1 using the DIFABS pro- gram. Refinement on all four structures was carried out with a full matrix least squares method against F2 using SHELXL97. Figure S1. The structure of 6-pyridin-2-yl-5,6-dihydro-benzo [4,5]imidazo[1,2-c]quinazoline (PDBIQ) with ellipsoids at 25% probability. Copyright © 2012 SciRes. AJAC |

