 American Journal of Plant Sciences, 2012, 3, 1294-1303 http://dx.doi.org/10.4236/ajps.2012.39156 Published Online September 2012 (http://www.SciRP.org/journal/ajps) Assessment of Genetic Diversity of Moroccan Cultivated Almond (Prunus dulcis Mill. DA Webb) in Its Area of Extreme Diffusion, Using Nuclear Microsatellites Abdelali Elhamzaoui1,2, Ahmed Oukabli1, Jamal Charafi1, Mohiéddine Moumni2 1Research Unit of Plant Breeding and Genetic Resources, Regional Agricultural Research Centre of Meknes, National Agronomic Research Institute, Meknes, Morocco; 2Moulay Ismaïl University, Faculty of Science, Department of Biology, Zitoune, Meknès, Morocco. Email: oukabli2001@yahoo.fr Received May 8th, 2012; revised June 5th, 2012; accepted June 18th, 2012 ABSTRACT Assessment of genetic diversity of Moroccan cultivated almond (Prunus dulcis Mill.) grown from seed and cultivated at four eco-geographical regions was performed using 16 nuclear SSRs. 238 alleles were detected with an average of 14.88 alleles per locus, ranging from 4 (locus BPPCT027) to 24 (locus CPSCT018). The size of alleles ranged from 84 bp (locus UDP96-003) to 253 bp (locus UDP96-018). A high genetic diversity of the local almonds is apparent and structured into three major clusters (Oasis cluster, High and Anti Atlas cluster, and Middle Atlas cluster). Compared to the Mediterranean genetic pools, from the East to West, the genetic diversity tends to be limited in Morocco which is the area of its extreme diffusion. Keywords: Almond; Genetic Diversity; Polymorphism; Spatial Genetic Structure; Prunus dulcis; Microsatellites; SSR 1. Introduction The almond [Prunus dulcis (Miller) DA Webb, syn. Prunusamygdalus Batsch] is a widely grown fruit tree that is commercially important th rougho ut the world . It is native to mountainous regions of Central Asia [1,2] and is probably the oldest domesticated fruit tree in the third millennium BC [3]. The almond tree was spread from its origin through the Mediterranean by the Phoenicians, Greeks and Romans in three main dispersion routes: the north route, the southern route and the route through the seas [4,5]. The cultivated almond tree was introduced in the Me- diterranean region during the second millennium BC [6,7] with a broad exchange of almond in the fourth century BC [8]. It led to the differentiation between two groups, the Mediterranean species and species of Central Asia [2]. It evolved slowly by seeding to the nineteenth century [1] and its culture, in the region, is often associated with seedling populations with selection of local varieties in some countries [9]. This mode of propagation by seed- ing generated a great variability in local genotypes. Therefore, the Mediterranean region is regarded as a se- cond source of domestication of the almond [5,10,11]. Morocco is an area of extreme diffusion of the almond tree. It was cultured by the Carthaginians in the fourth century [12] as well as by the Arabs in the sixth century [10]. Almond culture is currently about 146,000 ha [13] of which less than half (about 4 to 5 million trees) con- sists of populations grown from seed, localized mainly in the south [14]. This sexual propagation has led to high genetic diversity and the country is now considered as a secondary center of almond diversity [15]. Several works on collection and morphological characterization were performed on these populations [16-19]. This traditional plant material, resulting from many centuries of adapta- tion, may provide a basis for an almond breeding pro- gram. A collection that grouped individuals from differ- ent regions of Morocco was installed at the experimental field of INRA in Aïn Taoujdate [19] for evaluation ef- forts. This collection possesses a genetic basis necessary for any breeding program. Genetic characterization of plant material is necessary for the id entification of poten- tial genitors and their value in a breeding program. For the optimization of crossing schemes, the molecular characterization of this plant material is essential. Morphological characters were used in phenotyp ic ob- servations to characterize the genetic div ersity of almond species, but their interactions with the environment and the small number of characters [20-22] prompted the use of other more discriminating techniques. Currently, DNA markers are widely used in studies of genetic diversity Copyright © 2012 SciRes. AJPS 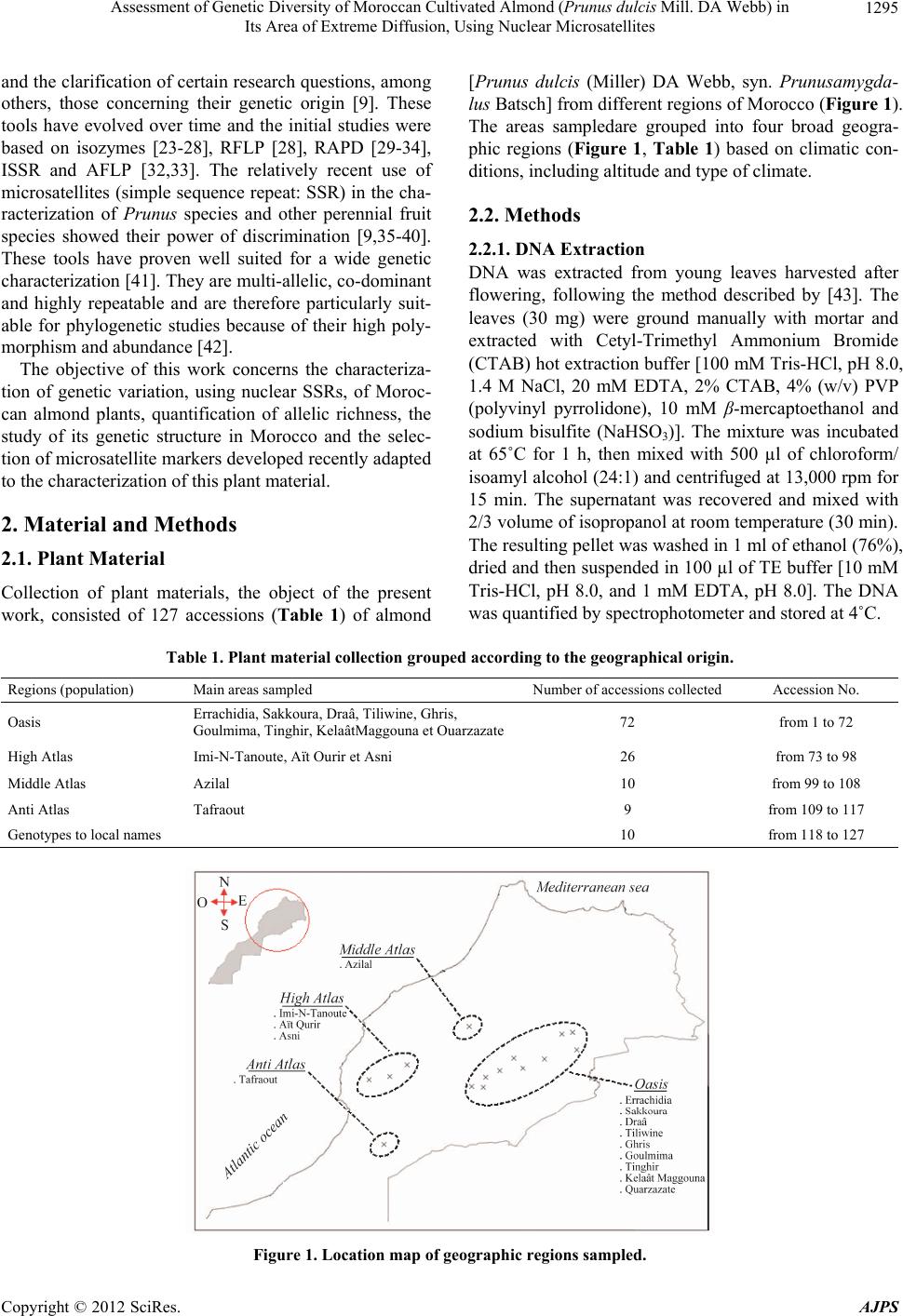 Assessment of Genetic Diversity of Moroccan Cultivated Almond (Prunus dulcis Mill. DA Webb) in Its Area of Extreme Diffusion, Using Nuclear Microsatellites 1295 and the clarification of certain research questions, among others, those concerning their genetic origin [9]. These tools have evolved over time and the initial studies were based on isozymes [23-28], RFLP [28], RAPD [29-34], ISSR and AFLP [32,33]. The relatively recent use of microsatellites (simple sequence repeat: SSR) in the cha- racterization of Prunus species and other perennial fruit species showed their power of discrimination [9,35-40]. These tools have proven well suited for a wide genetic characterization [41]. They are multi-allelic, co -dominant and highly repeatable and are therefore particularly suit- able for phylogenetic studies because of their high poly- morphism and abundance [42]. The objective of this work concerns the characteriza- tion of genetic variation, using nuclear SSRs, of Moroc- can almond plants, quantification of allelic richness, the study of its genetic structure in Morocco and the selec- tion of microsatellite markers develop ed recently adapted to the characterization of th is plant material. 2. Material and Methods 2.1. Plant Material Collection of plant materials, the object of the present work, consisted of 127 accessions (Table 1) of almond [Prunus dulcis (Miller) DA Webb, syn. Prunusamygda- lus Batsch] from different regions of Morocco (Figure 1). The areas sampledare grouped into four broad geogra- phic regions (Figure 1, Table 1) based on climatic con- ditions, including altitude and type of climate. 2.2. Methods 2.2.1. DNA Extracti on DNA was extracted from young leaves harvested after flowering, following the method described by [43]. The leaves (30 mg) were ground manually with mortar and extracted with Cetyl-Trimethyl Ammonium Bromide (CTAB) hot extraction buffer [100 mM Tris-HCl, pH 8.0, 1.4 M NaCl, 20 mM EDTA, 2% CTAB, 4% (w/v) PVP (polyvinyl pyrrolidone), 10 mM β-mercaptoethanol and sodium bisulfite (NaHSO3)]. The mixture was incubated at 65˚C for 1 h, then mixed with 500 µl of chloroform/ isoamyl alcohol (24:1) and centrifuged at 13,000 rpm for 15 min. The supernatant was recovered and mixed with 2/3 volume of isopropanol at room temperature (30 min). The resulting pellet was washed in 1 ml of ethanol (76%), dried and then suspended in 100 µl of TE buffer [10 mM Tris-HCl, pH 8.0, and 1 mM EDTA, pH 8.0]. The DNA was quantified by spectro p hotometer and stored at 4˚C. Table 1. Plant material collection grouped according to the geographical origin. Regions (population) Main areas sampled Number of accessions collected Accession No. Oasis Errachidia, Sakkoura, Draâ, Tiliwine, Ghris, Goulmima, Tinghir, KelaâtMaggouna et Ouarzazate 72 from 1 to 72 High Atlas Imi-N-Tanoute, Aït Ourir et Asni 26 from 73 to 98 Middle Atlas Azilal 10 from 99 to 108 Anti Atlas Tafraout 9 from 109 to 117 Genotypes to local names 10 from 118 to 127 Figure 1. Location map of geographic regions sampled. Copyright © 2012 SciRes. AJPS  Assessment of Genetic Diversity of Moroccan Cultivated Almond (Prunus dulcis Mill. DA Webb) in Its Area of Extreme Diffusion, Using Nuclear Microsatellites 1296 2.2.2. PCR Am pl i fi cation and Electroph o resi s The extracted DNA was amplified by PCR. The sixteen- pairs of primers flanking SSR sequences used in this work were cloned and sequenced in peach [44-48]. The multiplex PCR reactions were carried out with the Type I Microsatellite PCR Kit® (Qiagen) in a final volume of 10 µl, containing 1× of Qiagen Master Mix, 2 µM of each primer and 2 ng/µl of template DNA. The PCR pro- gram included: an initial denaturation at 95˚C for 5 min, 35 cycles of 30 sec at 95˚C, 1 min at the annealing tem- perature and 1 min 72˚C, followed by a terminal phase of 7 min at 72˚C. PCR reactions were carried out in an Ep- pendrof Mastercycler Gradient thermocycler. Samples were prepared by 3 µl diluted PCR products to 14.803 µl formamide and 0.197 GeneScan™ 500 LIZ®Size Stan- dard (Applied Biosystems, USA). The PCR products were detected by ABI 3130 XL 16-capillary sequencer (ABI Prism Applied Biosystems, Foster City, CA, USA). 2.2.3. Data Analysi s Reading the sizes of alleles (bp) was accomplished using Gene Mapper 4.0 software (Applied Biosystems). The number of alleles per locus (Na), observed heterozygos- ity (Ho) and expected heterozygosity (He) were calcu- lated using the Genetix 4.03 software [49]. The level of polymorphism was estimated by calculating the polymor- phism information content (PIC) described by [50] and modified by [51] using the formula 2 i PIC 1p , where pi is the frequency of the ith allele, by the powerof discrimination (PD, [52]), according to the formula above, for which the allele frequency was replaced by the frequency of the genotype and the probability of identity (, where pi and pj are the 2 2 i PI1p2p p ij frequency of the ith and jth alleles, respectively)for which two individuals sharing the same genetic profile by chance [53], calculated using the identity 4.0 [54]. Genetic differentiation was assessed by calculation of Wright’s fixation index (Fis) according to [55] and Fst values between each pair of populations using the Gene- pop 4.1 software [56]. The genetic relationships among genotypes based on the similarity matrix using the pro- portions of alleles [57] were studied. A dendrogram was prepared based on the unweighted pair group method with arithmetic averages (UPGMA) using the Ntsys-pc 2.02i program [58] (Rholf, 1998). A factorial correspon- dence analysis (FCA) was carried out also in our work using Ge netix 4.03 so ft w are [49]. 3. Results The use of 16 SSR loci in the almond trees revealed a total of 238 different alleles, ranging from 4 to 24 alleles per locus. The average number of alleles per locus was 14.88. The size of these alleles varied from 84bp to 253 bp. The observed heterozygosity (Ho) varied between 0.045 (Locus BPPCT027) and 0.916 (Locus CPSCT018) with an average of 0.596. The expected heterozygosity (He) ranged from 0.043 (Locus BPPCT027) to 0.884 (Locus CPSCT018) with an average of 0.699. The calcu - lated values of the probability of identity (PI) and the power of discrimination (PD) showed that the locus CPSCT018 is the most informative with values of 0.012 and 0.979, respectively. This locus has the highest value of polymorphism information content PIC (0.921) rela- tive to other loci. Thus, the least informative locus is BPPCT027 with (PI = 0.817, PD = 0.162 and PIC = 0.098). The averages of PI, PD and PIC for all loci were, respectively, 0.119, 0.868 and 0.763 (Table 2). The comparison of almonds belonging to 4 majo r geo- graphic regions showed that the number of alleles ob- served differ from one geographic area to another. The almond trees in the oasis region is characterized by the highest number of alleles (192 alleles) while the lowest number characterized almond trees native to the Anti- Atlas. Thus, the number of alleles per locus (NA) accord- ing to geographical origin, follows the same order with the highest value obtained at the oasis (NA = 12) and the smallest value in the Anti Atlas (NA = 5.38). The ob- served heterozygosity (Table 3) is similar in all four geo- graphic areas studied (Ho = 0.600). The comparison of pairwise Fst values of populations shows that th e values vary b etween 0.00726 and 0 .04354 (Table 4). Genetic distances are low for the almond trees that come from three geographic regions of the Atlas (High, Middle and Anti Atlas). Fst values of these are not significant. However, the difference is significant be- tween the Oasis almonds and those of the Atlas. The dendrogram was constructed using the UPGMA method and is based on similarity data of 127 genotypes, reveal- ing the existence of a very significant level of genetic diversity among genotypes. Thus, positioning arbitrarily at a level of 27% similarity, three homogeneous groups are distinguished (Figure 2). The first group consists of accessions from the region of the Oasis and most of the accessions of the Middle Atlas, the second group is com- posed mainly of those from the regions of High and Anti Atlas and the third group includes, in addition to geno- types to local names, a mixture of genotypes from re- gions of the Oasis and High Atlas. A more advanced structure was obtained by three-dimensional factorial correspondence analysis (FCA). The three axes explain, respectively, 48.49%, 32.22% and 19.29% of the vari- ance and allow the distinction of three homogeneous clusters (Figure 3). Cluster A contains mostly the acces- sions of Oasis, cluster B consists of genotypes of High and Anti Atlas and the last cluster C is composed only of Copyright © 2012 SciRes. AJPS 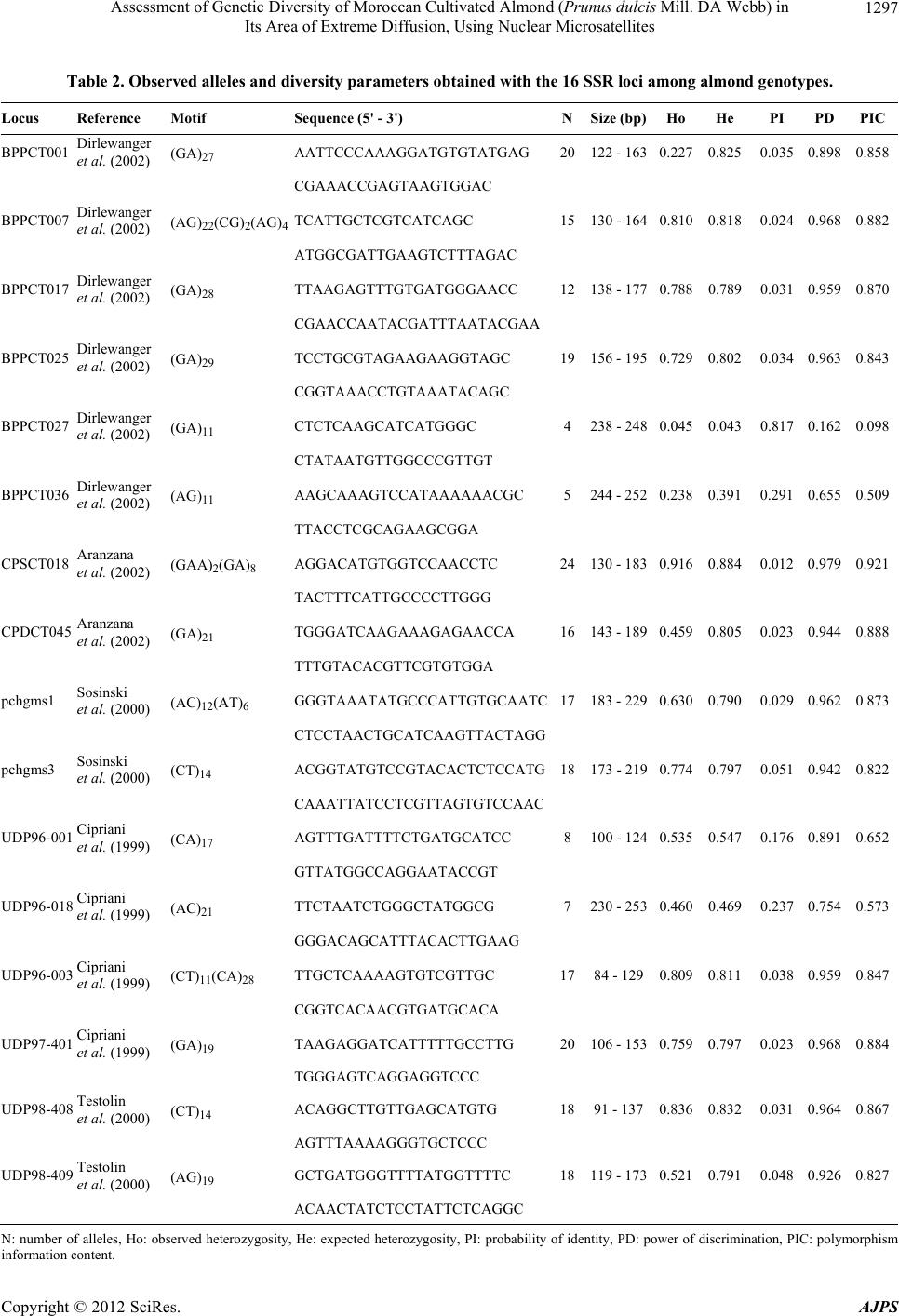 Assessment of Genetic Diversity of Moroccan Cultivated Almond (Prunus dulcis Mill. DA Webb) in Its Area of Extreme Diffusion, Using Nuclear Microsatellites 1297 Table 2. Observed alleles and diversity parameters obtained with the 16 SSR loci among almond genotypes. Locus Reference Motif Sequence (5' - 3') NSize (bp)Ho He PI PDPIC BPPCT001 Dirlewanger et al. (2002) (GA)27 AATTCCCAAAGGATGTGTATGAG 20122 - 1630.227 0.825 0.035 0.8980.858 CGAAACCGAGTAAGTGGAC BPPCT007 Dirlewanger et al. (2002) (AG)22(CG)2(AG)4 TCATTGCTCGTCATCAGC 15130 - 1640.810 0.818 0.024 0.9680.882 ATGGCGATTGAAGTCTTTAGAC BPPCT017 Dirlewanger et al. (2002) (GA)28 TTAAGAGTTTGTGATGGGAACC 12138 - 1770.788 0.789 0.031 0.9590.870 CGAACCAATACGATTTAATACGAA BPPCT025 Dirlewanger et al. (2002) (GA)29 TCCTGCGTAGAAGAAGGTAGC 19156 - 1950.729 0.802 0.034 0.9630.843 CGGTAAACCTGTAAATACAGC BPPCT027 Dirlewanger et al. (2002) (GA)11 CTCTCAAGCATCATGGGC 4238 - 2480.045 0.043 0.817 0.1620.098 CTATAATGTTGGCCCGTTGT BPPCT036 Dirlewanger et al. (2002) (AG)11 AAGCAAAGTCCATAAAAAACGC 5244 - 2520.238 0.391 0.291 0.6550.509 TTACCTCGCAGAAGCGGA CPSCT018 Aranzana et al. (2002) (GAA)2(GA)8 AGGACATGTGGTCCAACCTC 24130 - 1830.916 0.884 0.012 0.9790.921 TACTTTCATTGCCCCTTGGG CPDCT045 Aranzana et al. (2002) (GA)21 TGGGATCAAGAAAGAGAACCA 16143 - 1890.459 0.805 0.023 0.9440.888 TTTGTACACGTTCGTGTGGA pchgms1 Sosinski et al. (2000) (AC)12(AT)6 GGGTAAATATGCCCATTGTGCAATC17183 - 2290.630 0.790 0.029 0.9620.873 CTCCTAACTGCATCAAGTTACTAGG pchgms3 Sosinski et al. (2000) (CT)14 ACGGTATGTCCGTACACTCTC CATG18173 - 2190.774 0.797 0.051 0.9420.822 CAAATTATCCTCGTTAGTGTCCAAC UDP96-001 Cipriani et al. (1999) (CA)17 AGTTTGATTTTCTGATGCATCC 8100 - 1240.535 0.547 0.176 0.8910.652 GTTATGGCCAGGAATACCGT UDP96-018 Cipriani et al. (1999) (AC)21 TTCTAAT CTGGG CTATGGCG 7230 - 2530.460 0.469 0.237 0.7540.573 GGGACAGCATTTACACTTGAAG UDP96-003 Cipriani et al. (1999) (CT)11(CA)28 TTGCTCAAAAGTGTCGTTGC 1784 - 1290.809 0.811 0.038 0.9590.847 CGGTCACAACGTGATGCACA UDP97-401 Cipriani et al. (1999) (GA)19 TAAGAGGATCATTTTTGCCTTG 20106 - 1530.759 0.797 0.023 0.9680.884 TGGGAGTCAGGAGGTCCC UDP98-408 Testolin et al. (2000) (CT)14 ACAGGCTTGTTGAGCATGTG 1891 - 1370.836 0.832 0.031 0.9640.867 AGTTTAAAAGGGTGCTCCC UDP98-409 Testolin et al. (2000) (AG)19 GCTGATGGGTTTT AT GGTTTTC 18119 - 1730.521 0.791 0.048 0.9260.827 ACAACTATCTCCTATTCTCAGGC N: number of alleles, Ho: observed heterozygosity, He: expected heterozygosity, PI: probability of identity, PD: power of discrimination, PIC: polymorphism informat i o n content. Copyright © 2012 SciRes. AJPS  Assessment of Genetic Diversity of Moroccan Cultivated Almond (Prunus dulcis Mill. DA Webb) in Its Area of Extreme Diffusion, Using Nuclear Microsatellites 1298 Table 3. Genetic diversity in the geographical areas. Population Genotypes N NA Ho He Fis Oasis 72 192 12.00 0.597 0.742 0.180 High Atlas 26 136 08.50 0.599 0.711 0.174 Middle Atlas 10 107 06.69 0.598 0.683 0.169 Anti Atlas 09 086 05.38 0.590 0.661 0.167 N: total number of alleles detected in each zone, NA: average number of alleles per locus, Ho: observed heterozygosity, He: expected heterozygosity, Fis: fixa- tion index in tra-popul ation. Table 4. Fst pairwise values between different geographical origins. Pop Oasis High Atlas Middle Atlas High Atlas 0.03899** Middle Atlas 0.03843* 0.03239ns Anti Atlas 0.04354** 0.00726ns 0.03823ns ns: non-si gnificant; *P < 0.05; **P < 0.001. genotypes from the Middle Atlas. 4. Discussion The present work is the first study which characterizes the diversity of almond grown in Morocco using mi- crosatellite markers as molecular tools. The plant mate- rial analyzed comes from seedlings carrying no specific name but it is known locally, by farmers, under the “Louzbeldi” name. Farmers continue to maintain these almond accessions and to further exploit the germplasm by traditional planting of seedlings to establish new or- chards. The SSR loci used in this study were selected, from a set of primer pairs developed in peach, on the basis of their rate of allelic polymorphism [44-48]. The results obtained in our study are consistent with Sosinski et al. [45] and Xie et al. [59] data, which confirmed the inter- specific usefulness of microsatellite markers in Prunus species. The three genetic parameters (PI, PD and PIC) have shown therefore that all SSR loci may be recom- mended for future studies of genetic diversity of almond except BPPCT027locus, which is not very discriminat- ing. The number of alleles obtained for each locus is high in general but with differences between sub-populations located in the regions. The differences in the number of alleles detected in the oasis population (192 alleles) and that of the Anti-Atlas (86 alleles) could be due to the number of genotypes analyzed at each site. The level of genetic diversity is quite importan t because of traditional multiplication mode (by seeds) for a crop formerly intro- duced in Morocco [10,12]. A spatial genetic structure appears to be demonstrated by the parameters of Fst. This differentiation is most notable between the Oasis genotypeson the one hand and the provenances from the High Atlas, Middle Atlas and Anti Atlas on the other hand. These latter are genetically similar and Fst values are not significant (Table 4). The three homogeneous clusters, emerged by FCA, maybe the cause of an exchange of plant material as seeds between regions of High Atlas and those of Middle and Anti Atlas which constitute a geographical contin- uum. This exchange seems to be limited (or non-existent) with the Oasis probably because of the remoteness and geographical isolation of the Oasis zones. By comparing the Moroccan population with other genetic pools, since the center of origin of this species in the east to west through the different distribution centers around the Mediterranean, the average number of alleles per locus (14.88) found at the national level is high. This number is higher than that of the almond trees from Iran; countries belonging to the center of origin of the species since the values reported by Shiran et al. [37] and by Kadkhodaei et al. [60] are 6.64 and 12.86, respectively. For those authors who have characterized a small number of known cultivars (39 and 53, respectively), the average value of observed heterozygosity (0.50) and (0.54) re- mains low compared to the Moroccan population. This result is due to the bursting of the species diversity fol- lowing to its old propagation by seeds and the self-in- compatibility characterization of the almond compared to the other Prunus species. These high values may also be due to differences of SSR primer pairs used, the large number of genotypes analyzed in our study and that any work on pre-selected genotypes (case of works of Shiran et al. [37] and Kadkhodaei et al. [60]) may limit the ge- netic diversity. The average value of the polymorphism information content PIC (0.76), which provides an estimate of the Copyright © 2012 SciRes. AJPS 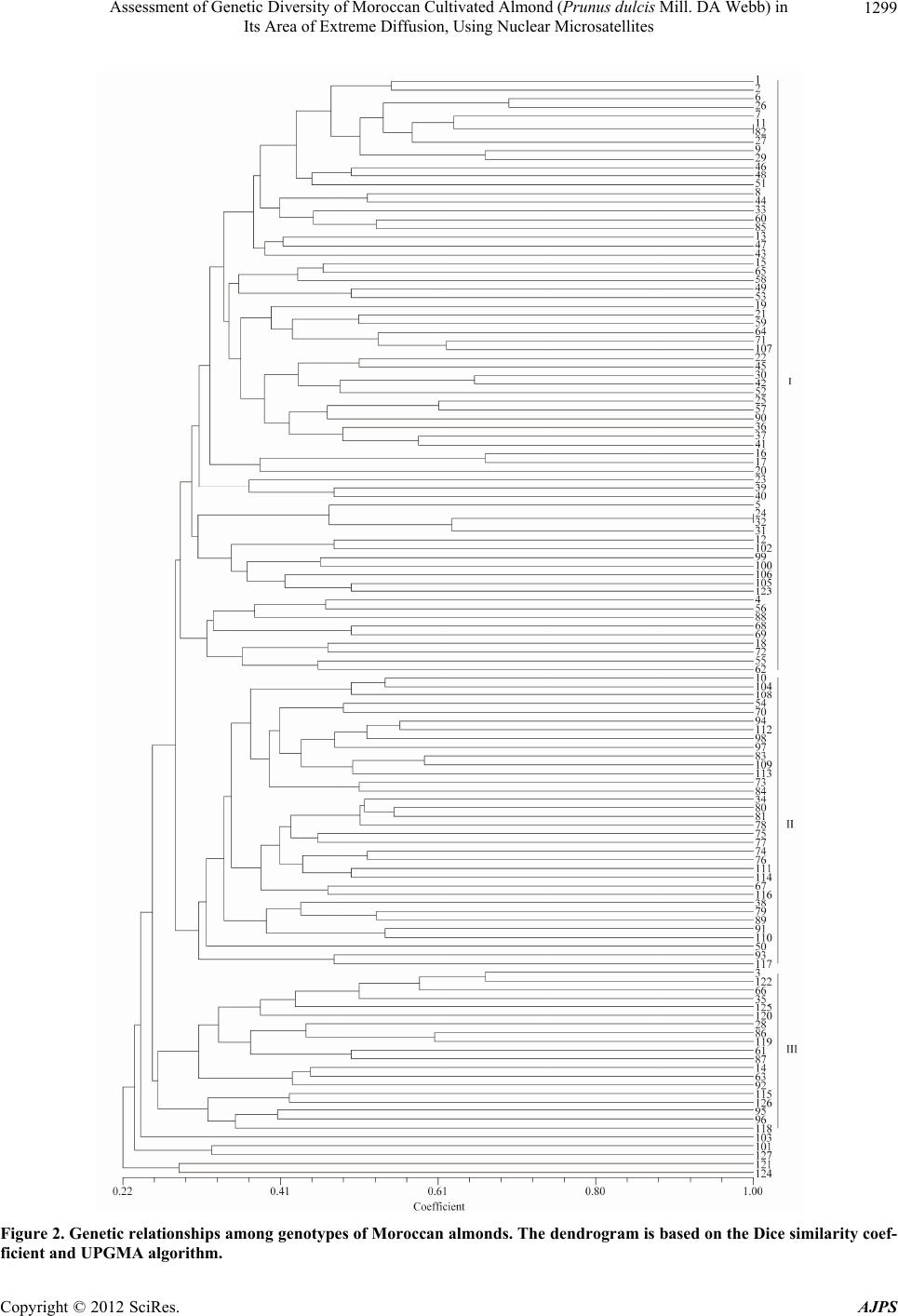 Assessment of Genetic Diversity of Moroccan Cultivated Almond (Prunus dulcis Mill. DA Webb) in Its Area of Extreme Diffusion, Using Nuclear Microsatellites 1299 Figure 2. Genetic relationships among genotypes of Moroccan almonds. The dendrogram is based on the Dice similar ity coef- ficient and UPGMA algorithm. Copyright © 2012 SciRes. AJPS 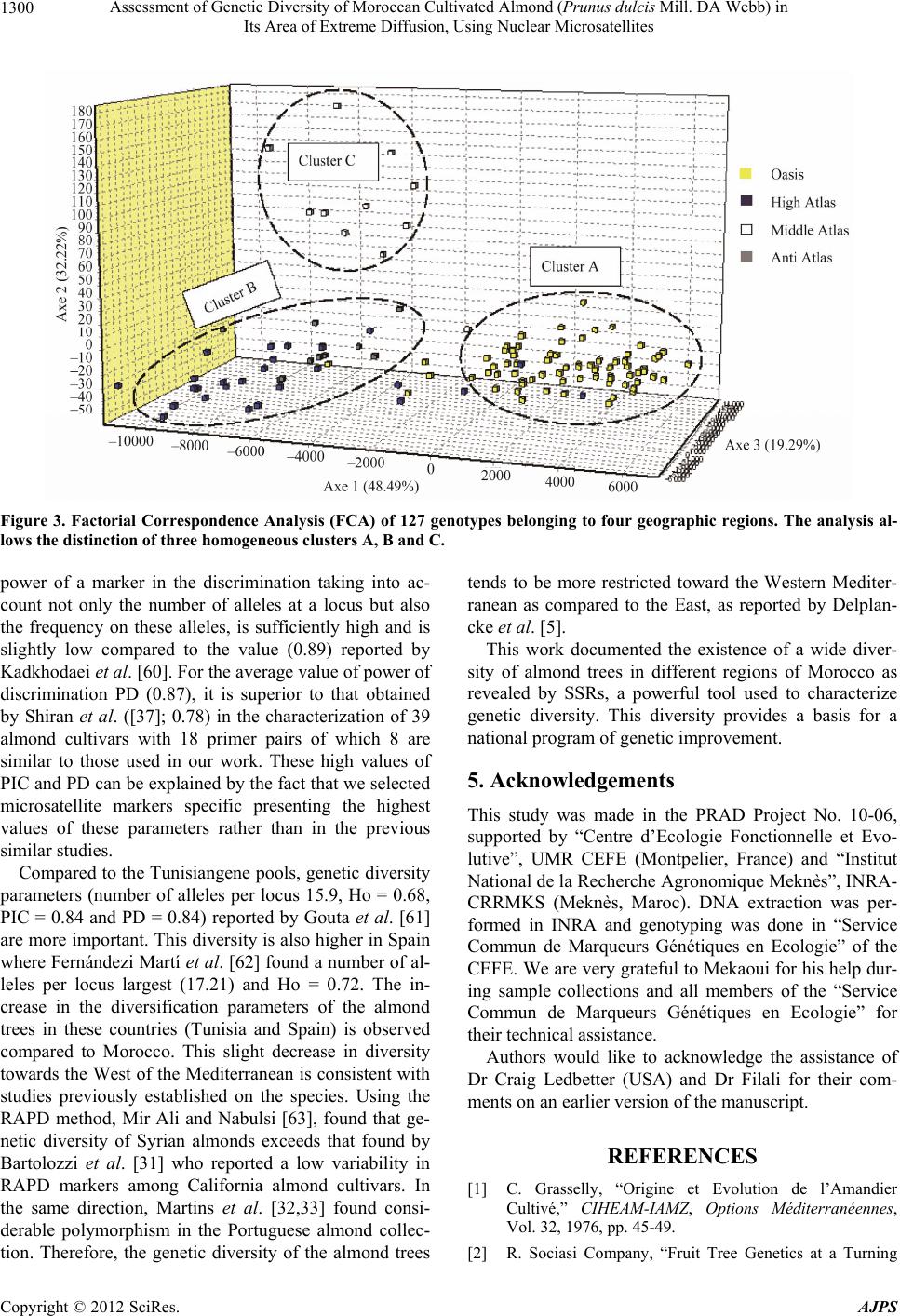 Assessment of Genetic Diversity of Moroccan Cultivated Almond (Prunus dulcis Mill. DA Webb) in Its Area of Extreme Diffusion, Using Nuclear Microsatellites 1300 Figure 3. Factorial Correspondence Analysis (FCA) of 127 genotypes belonging to four geographic regions. The analysis al- lows the distinction of three homogeneous c l usters A, B and C. power of a marker in the discrimination taking into ac- count not only the number of alleles at a locus but also the frequency on these alleles, is sufficiently high and is slightly low compared to the value (0.89) reported by Kadkhodaei et al. [60]. For the average value of power of discrimination PD (0.87), it is superior to that obtained by Shiran et al. ([37]; 0.78) in the characterization of 39 almond cultivars with 18 primer pairs of which 8 are similar to those used in our work. These high values of PIC and PD can be explained by the fact that we selected microsatellite markers specific presenting the highest values of these parameters rather than in the previous similar studies. Compared to the Tunisiangene pools, genetic diversity parameters (number of alleles per locus 15.9, Ho = 0.68, PIC = 0.84 and PD = 0.84) reported by Gouta et al. [61] are more important. This diversity is also higher in Spain where Fernándezi Martí et al. [62] found a number of al- leles per locus largest (17.21) and Ho = 0.72. The in- crease in the diversification parameters of the almond trees in these countries (Tunisia and Spain) is observed compared to Morocco. This slight decrease in diversity towards the West of the Mediterranean is co nsistent with studies previously established on the species. Using the RAPD method, Mir Ali and Nabulsi [63], found that ge- netic diversity of Syrian almonds exceeds that found by Bartolozzi et al. [31] who reported a low variability in RAPD markers among California almond cultivars. In the same direction, Martins et al. [32,33] found consi- derable polymorphism in the Portuguese almond collec- tion. Therefore, the genetic diversity of the almond trees tends to be more restricted toward the Western Mediter- ranean as compared to the East, as reported by Delplan- cke et al. [5]. This work documented the existence of a wide diver- sity of almond trees in different regions of Morocco as revealed by SSRs, a powerful tool used to characterize genetic diversity. This diversity provides a basis for a national program of genetic improvement. 5. Acknowledgements This study was made in the PRAD Project No. 10-06, supported by “Centre d’Ecologie Fonctionnelle et Evo- lutive”, UMR CEFE (Montpelier, France) and “Institut National de la Recherche Agronomique Meknès”, INRA- CRRMKS (Meknès, Maroc). DNA extraction was per- formed in INRA and genotyping was done in “Service Commun de Marqueurs Génétiques en Ecologie” of the CEFE. We are very grateful to Mekaoui for his help dur- ing sample collections and all members of the “Service Commun de Marqueurs Génétiques en Ecologie” for their technical assistance. Authors would like to acknowledge the assistance of Dr Craig Ledbetter (USA) and Dr Filali for their com- ments on an earlier version of the manuscript. REFERENCES [1] C. Grasselly, “Origine et Evolution de l’Amandier Cultivé,” CIHEAM-IAMZ, Options Méditerranéennes, Vol. 32, 1976, pp. 45-49. [2] R. Sociasi Company, “Fruit Tree Genetics at a Turning Copyright © 2012 SciRes. AJPS  Assessment of Genetic Diversity of Moroccan Cultivated Almond (Prunus dulcis Mill. DA Webb) in Its Area of Extreme Diffusion, Using Nuclear Microsatellites 1301 Point: The Almond Example,” Theoretical and Applied Genetics, Vol. 96, No. 5, 1998, pp. 588-601. doi:10.1007/s001220050777 [3] P. Spiegel-Roy, “Domestication of Fruit Trees,” In: C. Barigozzi, Ed., The Origin and Domestication of Culti- vated Plants, Elsevier, Amsterdam, 1986, pp. 201-211. [4] C. Grasselly and P. Crossa-Raynaud, “L’Amandier. Techniques Agricoles et Productions Méditerranéennes,” G. P. Maison Neuve et Larose, Paris, XII, 1980. [5] M. Delplancke, “Histoire Evolutive de l’Amandier Cultivé (Prunus dulcis) en Méditerranée: Regards Croisés sur la Domestication, Dialogue Entre la Biologie et l’Ethnobiologie, Biologie des Populations,” Université Montpellier 2, Montpellier, 2011. [6] N. I. Vavilov, “Wild Progenitors of the Fruit Trees of Turkestan and the Caucasus and the Problem of the Ori- gin of Fruit Trees,” Proceedings of the 9th International Horticultural Congress, London, 1930, pp. 271-286. [7] D. Zohary and M. Hopf, “Domestication of Plants in the Old World,” Clarendon Press, Oxford, 1993. [8] D. Cerdá, “Economía Antigua de Mallorca,” In: J. Mascaró Pasarius, Ed., Historia de Mallorca, Vol. I, Pasarius, Palma de Mallorca, Spain, 1973, pp. 417-448. [9] M. Zeinalabedini, M. Khayam-Nekoui, V. Grigorian, T. M. Gradziel and P. Martínez-Gómezd, “The Origin and Dissemination of the Cultivated Almond as Determined by Nuclear and Chloroplast SSR Marker Analysis,” Sci- entia Horticulturae, Vol. 125, No. 4, 2010, pp. 593-601. doi:10.1016/j.scienta.2010.05.007 [10] D. E. Kester, T. M. Gradziel and C. Grasselly, “Almonds (Prunus),” In: J. N. Moore and H. J. Ballington, Eds., Genetic Resources of Temperate Fruit and Nut Crops, International Society for Horticultural Science, The Netherlands, 1991, pp. 701-758. [11] A. J. Felipe,“Variedades de Almendro,” In: Integrum, Ed., El Almendro, Zaragoza, Spain, 2000, pp. 204-279. [12] N. El-Khatib-Boujibar, “Le Maroc et Carthage,” Le Mémorial du Maroc (I), Nord organizationEdition, 1983, p. 140. [13] DDFP, “Bilan Annuel des Rosacées Fruitières,” Direction de Développement des Filières de Productions, Ministre de l’Agriculture et de la Pêche Maritime, Maroc, 2011. [14] A. Oukabli, A. Mamouni, M. Laghezali, A.Chahbar, A. Mekkaoui, M. Lahlou and A. Bari, “Caractérisation de la Diversité Genétique des Populations Locales d’Amandier Cultivé [Prunus dulcis (Miller) D. A. Webb] au Maroc,” Proceeding IV Journées Nationales de Biodiversité, Tetouan, Maroc, 2008. [15] N. I. Vavilov, “Studies on the Origin of Cultivated Plants,” Leningrad, 1926, p. 248. [16] G. Barbeau and A. Elbouami, “Les Hybrides Amandier x Pêcher Naturels du sud Marocain,” Fruits, Vol. 35, No. 3, 1980, pp. 171-176. [17] M. Laghezali, “L ’Amandier au Maroc,” Options Méditer- ranéennes, Edition IAMZ, Vol. 85, No. 1, 1985, pp. 91-95. [18] A. Lansari, A. F. Iezzoni and D. E. Kester, “Morphologi- cal Variation within Collections of Moroccan Almond Clones and Mediterranean and North American Culti- vars,” Euphytica, Vol. 78, No. 1-2, 1994, pp. 27-41. [19] A. Oukabli, “Almond Breeding in Morocco: A Chrono- logical Perspective,” NUCIS FAO-CIHEAM, Vol. 15, 2011, pp. 4-7. [20] K. Sorkheh, B. Shiran, T. M. Gradziel, B. K. Eppe rson, P. Martínez-Gómez and E. Asadi, “Amplified Fragment Length Polymorphism as a Tool for Molecular Charac- terization of Almond Germplasm: Genetic Diversity among Cultivated Genotypes and Related Wild Species of Almond, and Its Relationships with Agronomic Traits,” Euphytica, Vol. 156, No. 3, 2007, pp. 327-344. doi:10.1007/s10681-007-9382-x [21] K. Sorkheh, B. Shiran,V. Rouhi, E. Asadi, H. Jahanbazi, H. Moradi, T. M. Gradziel and P. Martínez-Gómez, “Phenotypic Diversity within Native Iranian Almond Species and Their Breeding Potential,” Genetic Resources and Crop Evolution, Vol. 56, No. 7, 2009, pp. 947-96. doi:10.1007/s10722-009-9413-7 [22] M. Zeinalabedini, K. Majourhat, V. Grigorian, M. Torchi, F. Dicenta, P. Martínez-Gómez, “Comparison of the Use of Morphological, Protein and DNA Markers in the Ge- netic Characterization of Iranian Wild Prunus Species,” Scientia Horticulturae, Vol. 116, No. 1, 2008, pp. 80-88. doi:10.1016/j.scienta.2007.10.022 [23] S. Arulsekar, D. E. Parfitt and D. E. Kester, “Comparison of Isozyme Variability in Peach and Almond Cultivars,” Journal of Heredity, Vol. 77, No. 4, 1986, pp. 272-274. [24] R. Hauagge, D. E. Kester, S. Arulsekar, D. E. Parfitt and L. Liu, “Isozyme Variation among California almond Cultivars. II. Cultivar Characterization and Origins,” Journal of the American Society for Horticultural Science, Vol. 112, 1987, pp. 693-698. [25] M. Cerezo, R. Sociasi Company and F. Vargas, “Identifi- cation of almond Cultivars by Pollen Isoenzymes,” Jour- nal of the American Society for Horticultural Science, Vol. 114, 1989, pp. 164-169. [26] J. F. Jackson and G. R. Clarke, “Gene Flow in an Almond Orchard,” Theoretical and Applied Genetics, Vol. 82, No. 2, 1991, pp. 169-173. doi:10.1007/BF00226208 [27] P. Arús, C. Olarte, M. Romero and F. Vargas, “Linkage Analysis of Ten Isozyme Genes in F1 Segregating Al- mond Progenies,” Journal of the American Society for Horticultural Science, Vol. 119, 1994, pp. 339-34. [28] M. A. Viruel, R. Messeguer, M. C. de Vicente, J. Gar- cia-Mas, P. Puigdomenech, F. J. Vargas and P. Arus, “A Linkage Map with RFLP and Isozyme Markers for Al- mond,” Theoretical and Applied Genetics, Vol. 91, No. 6-7, 1995, pp. 964-971. doi:10.1007/BF00223907 [29] P. Resta, M. G. Corona, G. Faniz za, M. Palasciano and A. Godini, “Random Amplified DNA Polymorphisms in Amygdaluscommunis L., A. Webbii Spach,” Acta Hor- ticulturae, Vol. 470, 1997, pp. 82-90. [30] T. Joobeur, M. A. Viruel, M. C. de Vicente, B. Jauregui, J. Bellester, M. T. Dettori, I. Verde, M. J. Truco, R. Messe- guer, J. Balester, R. Quarta, E. Dirlewanger and P. Arús, Copyright © 2012 SciRes. AJPS  Assessment of Genetic Diversity of Moroccan Cultivated Almond (Prunus dulcis Mill. DA Webb) in Its Area of Extreme Diffusion, Using Nuclear Microsatellites 1302 “Construction of a Saturated Linkage Map for Prunus Using an Almond × Peach F2 Progeny,” Theoretical and Applied Genetics, Vol. 97, No. 7, 1998, pp. 1034-1041. doi:10.1007/s001220050988 [31] F. Bartolozzi, M. L. Warburton, S. Arulsekar and T. M. Gradziel, “Genetic Characterization and Relatedness among California Almond Cultivars and Breeding Lines Detected by Randomly Amplified Polymorphic DNA (RAPD) Analysis,” Journal of the American Society for Horticultural Science, Vol. 123, 1998, pp. 381-387. [32] M. Martins, A. Farinha, E. Ferreira, V. Cordeiro, A. Monteiro, R. Tenreiro and M. M. Oliveira, “Molecular Analysis of the Genetic Variability of Portuguese Almond Collections,” Acta Horticulturae, Vol. 546, 2001, pp. 449-452. [33] M. Martins, R. Tenreiro and M. M. Oliveira, “Genetic Relatedness of Portuguese Almond Cultivars Assessed by RAPD and ISSR Markers,” Plant Cell Reports, Vol. 22, No. 1, 2003, pp. 71-78. doi:10.1007/s00299-003-0659-9 [34] F. J. Ryan, C. A. Ledbetter, D. W. Ramming, D. Palm- quist, D. E. Bell and S. J. Peterson, “Challenges in De- veloping Molecular Markers for Almond (Prunus) and Grape (Vitis Species),”Acta Horticulturae, Vol. 546, 2001, pp. 629-639. [35] P. Martínez-Gómez, S. Arulsekar, D. Potter and T. M. Gradziel, “Relationships among Peach, Almond and Re- lated Species as Detected by Simple Sequence Repeat Markers,” Journal of the American Society for Horticul- tural Science, Vol. 128, 2003, pp. 667-671. [36] B. Khadari, J. Charafi, A. Moukhli and M. Ater, “Sub- stantial Genetic Diversity in Cultivated Moroccan Olive Despite a Single Major Cultivar: A Paradoxical Situation Evidenced by the Use of SSR Loci,” Tree Genetics & Genomes, Vol. 4, No. 2, 2007, pp. 213-221. doi:10.1007/s11295-007-0102-4 [37] B. Shiran, N. Amirbakhtiar, S. Kiani, Sh. Mohammadi, B. E. Sayed-Tabatabaei and H. Moradi, “Molecular Charac- terization and Genetic Relationship among Almond Cul- tivars Assessed by RAPD and SSR Markers,” Scientia Horticulturae, Vol. 111, No. 3, 2007, pp. 280-292. doi:10.1016/j.scienta.2006.10.024 [38] A. Fathi, B. Ghareyazi, A. Haghnazari, M. R. Ghaffari, S. M. Pirseyedi, S. Kadkhodaei, M. R. Naghavi and M. Mardi, “Assessment of the Genetic Diversity of Almond (Prunus dulcis) Using Microsatellite Markers and Mor- phological Traits,” Iranian Journal of Biotechnology, Vol. 6, No. 2, 2008, pp. 98-106. [39] H. Achtak, A. Oukabli, M. Ater, S. Santoni, F. Kjellberg and B. Khadari, “Microsatellite Markers as Reliable Tools for Fig Cultivar Identification,” Journal of the American Society for Horticultural Science, Vol. 134, 2009, pp. 624-631. [40] H. Achtak, M. Ater, A. Oukabli, S. Santoni, F. Kjellberg and B. Khadar, “Traditional Agroecosystems as Conser- vatories and Incubators of Cultivated Plant Varietal Di- versity: The Case of Fig (Ficuscarica L.) in Morocco,” BMC Plant Biology, Vol. 10, 2010, pp. 1471-2229. doi:10.1186/1471-2229-10-28 [41] P. Martínez-Gómez, R. Sánchez-Pérez, F. Dicenta, W. Howad, P. Arus and T. M. Gradziel, “Almonds,” In: C. R. Kole, Ed., Genome Mapping and Molecular Breeding, Fruits & Nuts, Springer, Heidelberg, Berlin, New York, Tokyo, Vol. 4, 2007, pp. 229-242. [42] P. K. Gupta, H. S. Balyan, P. C. Sharma and B. Ramesh, “Microsatellites in Plants: A New Class of Molecular Markers,” Current Science, Vol. 70, 1996, pp. 45-54. [43] J. J. Doyle and J. L. Doyle, “A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue,” Phytochemical Bulletin, Vol. 19, 1987, pp. 11-15. [44] G. Cipriani, G. Lot, H. G. Huang, M. T. Marrazzo, E. Peterlunger and R. Testolin, “AC/GT and AG/CT Mi- crosatellite Repeats in Peach (Prunuspersica (L) Basch): Isolation, Characterization and Cross-Species Amplifica- tion in Prunus,” Theoretical and Applied Genetics, Vol. 99, No. 1-2, 1999, pp. 65-72. doi:10.1007/s001220051209 [45] B. Sosinski, M. Gannavarapu, L. D. Hager, L. E. Beck, G. J. King, C. D. Ryder, S. Rajapakse, W. V. Baird, R. E. Ballard and A. G. Abbott, “Characterisation of Microsa- tellite Markers in Peach [Prunuspersica (L.) Batsch],” Theoretical and Applied Genetics, Vol. 101, No. 3, 2000, pp. 421-428. doi:10.1007/s001220051499 [46] R. Testolin, T. Marrazo, G. Cipriani, R. Quarta, I. Verde, T. Dettori, M. Pancaldi and S. Sansavini, “Microsatellite DNA in Peach (Prunuspersica L. Batsch), and Its Use in Fingerprinting and Testing the Genetic Origin of Culti- vars,” Genome, Vol. 43, 2000, pp. 512-520. [47] M. J. Aranzana, J. García-Mas, J. Carbóand P. Arús, “Development and Variability Analysis of Microsatellite Markers in Peach,” Plant Breeding, Vol. 121, No. 1, 2002, pp. 87-92. doi:10.1046/j.1439-0523.2002.00656.x [48] E. Dirlewanger, A. Crosson, P. Tavaud, M.J. Aranzana, C. Poizat, A. Zanetto, P. Arus and L. Laigret, “Development of Microsatellite Markers in Peach and Their Use in Ge- netic Diversity Analysis in Peach and Sweet Cherry,” Theoretical and Applied Genetics, Vol. 105, No. 1, 2002, pp. 127-138. doi:10.1007/s00122-002-0867-7 [49] K. Belkhir, P. Borsa, L. Chikhi, N. Raufaste and F. Bonhomme, “GENETIX 4.03, Logiciel sous Windows TM pour la Génétique des Populations,” Laboratoire Génome, Populations, Interactions CNRS UMR 5000, Université de Montpellier II, Montpellier (France), 2004. [50] D. Botstein, R. L. White, M. Skolnick and R. W. Davis, “Construction of Genetic Linkage Map in Man Using Re- striction Fragment Length Polymorphisms,” The Ameri- can Journal of Human Genetics, Vol. 32, 1980, pp. 314- 331. [51] J. A. Anderson, G. A. Churchill, J. E. Sutrique, S. D. Tanksley and M. E. Sorrels, “Optimizing Parental Elec- tion for Genetic Linkage Maps,” Genome, Vol. 36, No. 1, 1993, pp. 181-186. doi:10.1139/g93-024 [52] A. D. Kloosterman, B. Budowle and M. Daselaar, “PCR-Amplification and Detection of the Human DIS80 VNTR Locus. Amplification Conditions, Population Ge- netics and Application in Forensic Analysis,” Interna- tional Journal of Legal Medicine, Vol. 105, No. 5, 1993, Copyright © 2012 SciRes. AJPS  Assessment of Genetic Diversity of Moroccan Cultivated Almond (Prunus dulcis Mill. DA Webb) in Its Area of Extreme Diffusion, Using Nuclear Microsatellites Copyright © 2012 SciRes. AJPS 1303 pp. 257-264. doi:10.1007/BF01370382 [53] D. Paetkau, W. Calvert, I. Stirling and C. Strobeck, “Mi- crosatellite Analysis of Population Structure in Canadian Polar Bears,” Molecular Ecology, Vol. 4, No. 3, 1995, pp. 347-354. doi:10.1111/j.1365-294X.1995.tb00227.x [54] H. W. Wagner and K. M. Sefc, “IDENTITY 4.0. Centre for Applied Genetics,” University of Agricultural Sci- ences, Vienna, 1999. [55] B. S. Weir and C. C. Cockerham, “Estimating F-Statistics for the Analysis of Population Structure,” Evolution, Vol. 38, No. 6, 1984, pp. 1358-1370. doi:10.2307/2408641 [56] F. Rousset, “Genepop’007: A Complete Reimplementa- tion of the Genepop Software for Windows and Linux,” Molecular Ecology Resources, Vol. 8, No. 1, 2008, pp. 103-106. doi:10.1111/j.1471-8286.2007.01931.x [57] M. Nei and W. H. Li, “Mathematical Model for Studying Genetic Variation in Terms of Restriction Endonucle- ases,” Proceedings of the National Academy of Sciences of the USA, Vol. 76, 1979, pp. 5269-5273. doi:10.1073/pnas.76.10.5269 [58] F. J. Rohlf, “NTSYS-pc: Numerical Taxonomy and Mul- tivariate Analysis System, Version 2.02i,” Exeter Soft- ware, New York, 1998. [59] H. Xie, Y. Sui, F. Chang, Y. Xu and R. Ma, “SSR Allelic Variation in Almond (Prunus dulcis Mill.),” Theoretical and Applied Genetics, Vol. 112, No. 2, 2006, pp. 366-372. doi:10.1007/s00122-005-0138-5 [60] S. Kadkhodaei, M. Shahnazari, M. Khayyam Nekouei, M. Ghasemi, H. Etminani, A. Imani and A. B. Ariff, “A Comparative Study of Morphological and Molecular Di- versity Analysis among Cultivated Almonds (Prunus dul- cis),”Australian Journal of Crop Science, Vol. 5, No. 1, 2011, pp. 82-91. [61] H. Gouta, E. Ksia, T. Buhner, M. A. Moreno, M. Zarrouk, A. Mliki and Y. Gogorcena, “Assessment of Genetic Di- versity and Relatedness among Tunisian Almond Germ- plasm Using SSR Markers,” Hereditas, Vol. 147, No. 6, 2010, pp. 283-292. doi:10.1111/j.1601-5223.2009.02147.x [62] A. FernándeziMartí, J. M. Alonso, M. J. Espiau, Rubio- Cabetas and R. Sociasi Company, “Genetic Diversity in Spanish and Foreign Almond Germplasm Assessed by Molecular Characterization with Simple Sequence Re- peats,” Journal of the American Society for Horticultural Science, Vol. 134, No. 5, 2009, pp. 535-542. [63] N. MirAli and I. Nabulsi, “Genetic Diversity of Almond s (Prunus dulcis) Using RAPD Technique,” Scientia Hor- ticulturae, Vol. 98, 2003, pp. 461-471.
|