 American Journal of Plant Sciences, 2012, 3, 1241-1251 http://dx.doi.org/10.4236/ajps.2012.39150 Published Online September 2012 (http://www.SciRP.org/journal/ajps) 1241 Assessments of Biodiversity Based on Molecular Markers and Morphological Traits among West-Bank, Palestine Fig Genotypes (Ficus carica L.) Rezq Basheer-Salimia1*, Murad Awad1, Joy Ward2 1Department of Plant Production and Protection, Faculty of Agriculture, Hebron University, West-Bank, Palestine; 2Department of Ecology & Evolutionary Biology, University of Kansas, Lawrence, USA. Email: *rezqbasheer@hebron.edu Received July 10th, 2012; revised August 6th, 2012; accepted August 17th, 2012 ABSTRACT Both morphological characters and PCR-based RAPD approaches were used to determine the genetic diversity and re- latedness among nine fig genotypes grown at the northern region of the West-Bank, Palestine. Although we tested 28 primers for the RAPD technique, only 9 produced reasonable amplification products. A total of 57 DNA loci were de- tected in which 70.2% were polymorphic. DNA fragments presented a minimum of 3 and a maximum of 9 polymorphic bands using primers OPT-10 and OPA-18, respectively. Primers exhibited collective resolving power values (Rp) of 18.826. The Mwazi genotype showed the highest genetic distances among all of the other genotypes. Morphologically, considerable variations were found using 41 quantitative and qualitative traits. Adloni could be a very promising geno- type for fresh consumption due to its very late maturation period, extended harvesting period, variable fruit size, and easy skin peeling. In addition, 7 genotypes presented firm fruits, which are a very important criterion for exporting purposes. Dendrogram constructed by UPGMA based on RAPD banding patterns appear somewhat contradictory to the morphological descriptors particularly with Swadi and Biadi genotypes (closed genetically and distanced morphologi- cally), which might be attributed to the phenotypic modifications caused by environmental differences across regions. These preliminary results will make a fundamental contribution to further genetic improvement of fig crops for the re- gion. Keywords: Cluster Analysis; Ficus carica L.; Genetic Variability; Random Amplified Polymorphic DNA; Morphological Descriptors 1. Introduction Fig (Ficus carica L., Moraceae) is one of the oldest cul- tivated fruit crops grown in the Mediterranean region. Because of their nutritional, medicinal, and ornamental values [1], figs have recently attracted a great deal of attention for culinary purposes and therefore, are wide- spread throughout the world. According to FAO statistics [2], the world produces over one million metric tons of figs yearly, of which 82% are produced in Mediterranean countries. Middle East countries have been the most im- portant center of fig production across several millennia [3]. The discovery of carbonized figs in an early Neoli- thic site in the Jordan Valley (between Jordan and Pale- stine), dating back 11,400 - 11,200 years ago, suggests that figs were first domesticated during the early Neoli- thic Revolution preceding cereal domestication [4]. From there, fig cultivation spread to neighboring western Asia and other Middle-East regions, and subsequently across the rest of the World [5]. In Palestine, fig trees are grown historically all over the country and are mostly located on marginal lands, in combination with other fruit trees (mainly olive and grape), or are scattered at the periphery of orchards and in home gardens. The long history of fig growth in Pale- stine and the wide range of geographical and climatical conditions under which it is grown, have combined to produce a complex picture in which fig landraces and genotypes are either misidentified or called by different names in different areas. Additionally, fig names were mainly given based on fruit skin color, internal color, local geographic origin, maturity dates, or the name of the orchard owner [3,6]. Therefore, it is crucial for discri- mination between these landraces both for conservation of plant genetic resources and for purposes of crop im- provement [7,8]. Detection and analyzing of genetic variation could be *Corresponding author. Copyright © 2012 SciRes. AJPS  Assessments of Biodiversity Based on Molecular Markers and Morphological Traits among West-Bank, Palestine Fig Genotypes (Ficus carica L.) 1242 achieved either by morphological and/or DNA molecular markers [9]. Here we make major advances in under- standing the history and genetic variation of fig by com- bining both morphological and DNA marker approaches for fig trees located throughout the West-Bank, Palestine. Morphological markers have been used for many years for identification and characterization of genotypes. In fig, several reports demonstrated the usefulness of these markers in documenting variability among genotypes [6, 9-11]. However, morphological characters can often yield ambiguous results due to high plasticity for many traits, as well as phenotypic modifications caused by environmental differences [12]. The limitations of phenotype-based genetic markers led to the development of more general and now wide- spread use of DNA-based markers [13], which proved to be powerful tools to estimate genetic diversity of species, as well as genotype identity. In fact, molecular markers offer numerous advantages over conventional morpholo- gical based approaches, since they are stable and detec- table in all tissues regardless of growth, differentiation, development, or defense status of the cell. In addition, DNA markers are not confounded by the environment, pleiotropic, and epistatic effects [13]. In figs, assessment of genetic relatedness and diversity has been investigated by using RFLP, AFLP, SSR, ISSR, and RAPD methods [7,14-22]. Compared with other molecular techniques, RAPD is based on random amplification of bases from short primers. Interestingly, RAPD is a simple, fast, efficient, and inexpensive method. Further, it does not require prior knowledge of the sequences of the markers and can produce abundant polymorphic fragments [22, 23]. Therefore, RAPD has become a powerful and accu- rate tool for analyzing the genetic relatedness and diver- sity in figs. Combinations of morphological as well as molecular markers prove to be essential since morphological mar- kers continue to be a highly recommended first step for the description and classification of any germplasm [24], as well as useful tools for screening the accessions of any collection [25]. The present study is the first attempt to characterize and detect similarities among some fig geno- types grown in the northern region of Palestine using both the powerful combination of morphological and molecular markers. 2. Materials and Methods 2.1. Molecular Analysis 2.1.1. G en etic Markers This study was carried out during the growing season of 2011. A total of nine fig accessions including: Khorto- mani, Enaki, Hmadi, Hmari, Khdari, Biadi, Mwazi, Swa- di, and Adloni were surveyed throughout the northern region of West-Bank, Palestine. The climate of the re- gion is an atypical Mediterranean climate, with mild tem- peratures (18˚C - 25˚C), rainy weather (580 - 800 mm/year) in autumn and winter, and hot, dry summers. Generally, all fig trees are cultivated under rain-fed conditions. 2.1.2. D NA Extracti on Genomic DNA was extracted from fresh leaves of single adult trees using the DNeasy Plant Mini Kit (Qiagen Inc.). 2.1.3. RAPD Primers and PCR Reactions A total of 28 RAPD primers (Sigma-Aldrich) were used for the amplification of random DNA banding patterns (Tables 1, 2). PCR reactions were repeated twice and carried out in a 25 ml volume mixture containing: 5 µl of total DNA (30 ng), 2 µl primer (5 µM), 2 µl dNTPs (200 mM) (Fermentas), 2.5 µl Taq buffer (10×), 2 µl magne- sium chloride (25 mM) and 1.5 U of Taq DNA poly- merase (Hy Labs). Consequently, DNA was amplified by PCR on a Peltier Thermal Cycler-200 (MJ Research. Inc, Watertown, MA) and the PCR program was: 1 cycle, 94˚C (3 min); 35 cycles, 94˚C (1 min), 35˚C (1 min), 72 (1; 30 min) 1 cycle, 72˚C (5 min), followed by storage at 4˚C. Amplified products (25 µl) were mixed with 5 µl of orange gel loading buffer and analyzed by electrophore- sis in 2% agarose gels (Hy Labs) in 1× TAE buffer at 4 volt/cm for 4h as well as detected by staining with ethi- dium bromide (Sigma). A 100 bp DNA ladder was used as standard marker (Fermentas). Consequently, ampli- cons were visualized with a UV transilluminator (Ima- geMaster®VDS). 2.1.4. D ata Analysi s of RAPD Mar k ers For each primer, three independent researchers calculated the total number of bands and the polymorphic bands in order to avoid subject bias. The ability of the most in- formative primers to differentiate between genotypes was assessed by the estimation of their resolving power (Rp) [26]. The Rp has been described to correlate strongly with the ability to distinguish between genotypes ac- cording to the following formula: Rp = ∑Ib, where Ib = 1 – (2 × |0.5 – p|) where p is the proportion of the nine geno- types possessing the I band [27]. Banding profile data were scored as present (1) or absent (0) for each sample. Afterwards, RAPD bands were transformed into a binary matrix. Next, a genetic distance matrix was estimated (Table 3) based on Jaccard’s similarity coefficient using the multilocus fingerprinting data sets containing missing data (FAMD) software version 1.108 beta. Then a cluster analysis was made using the un-weighted pair-group Copyright © 2012 SciRes. AJPS  Assessments of Biodiversity Based on Molecular Markers and Morphological Traits among West-Bank, Palestine Fig Genotypes (Ficus carica L.) Copyright © 2012 SciRes. AJPS 1243 Table 1. List of the selected primers along with their sequences, number of banding patterns, resolving pow er, total and po- lymorphic bands gener ate d that are resultant from all tested fig genotypes. Resolving Power (Rp) Frequency Number of Patterns Percentage of Polymorphic Markers Mono- morphic Bands Poly- Morphic Bands RAPD Total Bands Sequence 5'-3'Primer Code # 1.554 3/9 2/9 1/9 1 2 2 80 1 4 5 TCCCATGCTGZ-5 1. 3.166 3/9 1/9 1 6 88 1 7 8 GGGTGGGTAAZ-8 2. 1.999 2/9 1/9 1 7 60 2 3 5 CTCAGTCGCAZ-11 3. 2.443 2/9 1/9 2 7 63 3 5 8 CAATCGCCGTOPA-11 4. 2.722 2/9 1/9 1 7 63 3 5 8 CAGCACCCACOPA-13 5. 1.777 4/9 2/9 1/9 1 1 3 56 4 5 9 AGGTGACCGTOPA-18 6. 1.388 4/9 3/9 1/9 1 1 2 60 2 3 5 TCGGACGTGAOPH-02 7. 3.111 1/9 9 83 1 5 6 CTGACCAGCCOPH-19 8. 0.666 7/9 1/9 1 2 100 0 3 3 CCTTCGGAAGOPT-10 9. 2.092 4.44 6.33 Mean 18.826 70.2% 17 40 57 Total Table 2. Summary of amplification presents gener ated by the random primers tested in this study. Description Number/Frequency Total number of primers screened with all the eleven fig genotypes19 Number of primers that produced polymorphic bands9 Total number of bands amplified by the primers that generated polymorphic bands57 Average number of bands per primer6.33 Total number of polymorphic bands40 Percentage of polymorphic bands70.2 Average number of polymorphic bands per primer4.44 Total number of primers that produced more than 75% polymorphic bands4 Total number of bands produced by these 4 primers23 Number of polymorphic bands produced by these 4 primers19 Percentage of polymorphic bands82.6 Average number of polymorphic bands per primer4.75 Average size of the fragments amplified190 bp - 1300 bp  Assessments of Biodiversity Based on Molecular Markers and Morphological Traits among West-Bank, Palestine Fig Genotypes (Ficus carica L.) 1244 Table 3. Jaccard’s distance index generated for the 9 Palestinian fig genotypes based on RAP D mar kers. Genotypes Swadi Biadi Hmadi Mwazi Khdari Adloni Enaki Khorto mani Biadi 0.342 Hmadi 0.378 0.372 Mwazi 0.457 0.419 0.267 Khdari 0.333 0.368 0.286 0.444 Adloni 0.410 0.361 0.357 0.442 0.306 Enaki 0.368 0.405 0.357 0.477 0.306 0.432 Khorto mani 0.419 0.452 0.256 0.302 0.325 0.400 0.270 Hmari 0.366 0.316 0.238 0.400 0.263 0.342 0.342 0.317 method with arithmetic averages (UPGMA) [28] and the Tree view software (Win32, version 1.6.6). 2.2. Morphological and Pomological Analysis 2.2.1. P lant Mater ials and Descriptors From the nine studied genotypes, random samples of 20 adult leaves and 20 mature fruits were collected from three adult trees per genotype. 15 leaf morphological and 26 pomological traits (Tables 4, 5, respectively) were determined according to the fig descriptors prepared by IPGRI and CIHEAM [29], as well as Aljane and Ferchi- chi [3], with some minor modifications that showed high discrimination values. 2.2.2. Data Analysi s o f Mo rpholog i cal and Pomological Traits Relatedness as well as cluster analysis among genotypes was established following the same method of RAPD data analysis, in which each morphological descriptor was scored in a dominant manner and transformed into either a 1 (present) or 0 (absent). 3. Results 3.1. Molecular Results 3.1.1. Ge n etic Pol ymorphism and RAPD Patter n s Examined primers revealed various banding patterns. Among the 28 tested primers used for common fig genotypes grown at the northern region West-Bank, Pal- estine, only 9 primers produced reasonable amplification products with high intensity and pattern stability (Table 1). The remaining 19 primers exhibited ambiguous, light, and non-clear complex amplification products, and there- fore were excluded from our analysis. A total of 57 DNA fragments (loci) separated by electrophoresis on agarose gels, were detected (Table 1), ranging in size from 190 to 1300 bp. Of these fragments, 40 (70.2%) were poly- merphic and 17 (29.8%) were monomorphic. Our results also revealed an average of 6.33 loci per primer (Tables 1, 2). A minimum of three and a maximum of nine DNA fragments were obtained using OPT-10 and OPA-18 primers, respectively (Figure 1). The maximum percent- age of polymorphic markers was 100.0 (OPT-10). 3.1.2. Resolvin g P ower (Rp) The tested primers exhibited a collective Rp value of 18.826, and varied from 0.666 for the (OPT-12) primer to 3.166 for the (Z-8) with a mean of 2.092 (Table 2). 3.1.3. G e netic Di stances The data matrix size analysed was 513 entries, 300 of which were for present loci (1) and 213 for absent loci (0). Accordingly, the Jaccard coefficient was calculated and presented in (Table 3). The genetic distance matrix showed an average distance range from 0.238 to 0.477 with a mean of 0.358. The maximum genetic distance value of 0.477 was registered between Mwazi and Enaki genotypes, whereas the lowest genetic distance of 0.238 (the highest similarities of 0.762) was exhibited between the Hmadi and Hmari genotypes (Table 3). 3.1.4. D endrogram of Genetic Relationship (Similarity Matrix and Cluster Analysis) The average genetic relatedness among the genotypes is illustrated in Figure 2. RAPD UPGMA dendrogram analysis divided the genotypes studied into two main clusters. The first (I) is made up of Biadi and Swadi genotypes. The second cluster (II) is divided into two sub clusters. The first sub cluster labelled (II.a) is made up of (Khortomani and Enaki), as well as (Hmari and Hmadi) related to Khdari. The second sub-cluster (II.b) is com- posed of only genotype Adloni. However, Mwazi geno- type is separated and identified as a distant genotype. 3.2. Morphological Descriptors Fifteen quantitative and qualitative morphological traits Copyright © 2012 SciRes. AJPS  Assessments of Biodiversity Based on Molecular Markers and Morphological Traits among West-Bank, Palestine Fig Genotypes (Ficus carica L.) 1245 Table 4. Morphological descriptors of some fig genotypes grown in the northern region of West-Bank, Palestine. # Descriptors Khort- omani Enaki Hmari Hmadi KhdariBiadi Mwazi Swadi Adloni 1 Bud Break Mar, 15 - 30 Mar, 15 - 30 Mar, 1 - 15 Mar, 1 - 15 Mar, 1 - 15 Mar, 1 - 15 Mar, 15 - 30 Mar, 1 - 15 Mar, 15 - 30 2 Leaf Color Light green Green dark green Dark greenDark green Dark greenDark greenLight green green Green dark green Light green 3 Leaf Shape Base cordate, lobes spatulate Base cordate, lobes spatulate Base cordate, lobes spatulate Base cordate, lobes spatulate-base calcarate, lobes latate Base cordate, lobes spatulate Base cordate, lobes spatulate Base cordate, lobes spatulate Base calcarate lobes lineate Base cordate, lobes spatulate 4 Lobes Number Five Five Five Five Five Five Five Five Five 5 Leaf Venation Apparent Apparent ApparentApparent Apparent Apparent Apparent Apparent Apparent 6 Apex Shape Tri Anleobtuse Tri Angle- obtuse Obtuse rounded Triangle- obtuse TriangleTriangleTriangle Triangle Obtuserounded 7 Counter Crenate dentate Crenate dentate Crenate undulate Crenate undulate Crenate CrenateCrenate Crenate undulate Crenate 8 Leaf Roughness Fairly rough Fairly rough Fairly rough Fairly rough Fairly rough Fairly rough Fairly rough Rough Fairly rough 9 Leaf Area (cm2) Medium Large Large Large Large Large Small Medium Small 10 Limb Length (mm) Short Medium Medium Long MediumLong Short Medium Short 11 Limb Width (mm) Medium Medium Medium Medium Large Large Small Medium Small 12 Lateral Sinus Depth (mm) Medium Medium Small Medium MediumMediumMedium Long Medium 13 Petiole Length (mm) Medium Long Long Long MediumMediumMedium Medium Medium 14 Petiole Width (mm) Medium Medium Medium Medium Large Large Small Small Small 15 Beginning of Defoliation Sep-01 Sep-15 Aug-01 Aug-01 Aug-01Aug-01 Aug-20 Jul-01 Sep-15 are shown in Table 4. Bud break was observed between March 1-15 in five genotypes (Hmari, Hmadi, Khdari, Biadi and Swadi), with the remainder occurring between March 15-30. Among all genotypes tested, leaf color ranged from light green to dark green, leaf shape was always base chordate with lobes spatulate (except for Hmadi and Swadi), lobe number was five, leaf venation was appar- ent, leaf apex shape was variable, leaf serration tended to be crenate, leaf roughness was fairly rough except for the Swadi (rough) genotype. The Biadi genotype presented the greatest value for leaf area, leaf limb length and width, whereas Adloni tended to have the smallest values for each of these parameters. Lateral sinus depth was small for Hmari and long for Swadi, with the other genotypes being medium for this trait. Petiole length was long for genotypes Enaki, Hmari, and Hmadi and it was medium for the remaining genotypes. Additionally, peti- ole width was large for Biadi and Khdari genotypes, me- dium for (Khortomani, Enaki, Hmari, and Hmadi), and small for (Mwazi, Swadi, and Adloni).Defoliation was early (July 1) for the Swadi genotype, while it was latest (Sept 15) for the Enaki and Adloni genotypes; others were intermediate. 3.3. Pomological Descriptors Twenty-six quantitative and qualitative pomological traits are presented in Table 5 . In terms of fruit maturation, the nine genotypes studied were categorized very early (Hmari, Hmadi, Khdari, Biadi, and Swadi), early (Khor- tomani and Mwazi), or mid-season (Enaki and Adloni). However, no genotypes were present which would be categorized as either late or very late. The same trend was observed for full fruit maturity for each genotype, with the exception of Adloni, where maturation extended beyond October 1 (very late). For all tested genotypes, the harvesting period observed ranged from medium (Khortomani, Hmari, Hmadi, Khdari, Biadi, and Swadi), to long (Enaki), to very long (Mwazi and Adloni). Fruit Copyright © 2012 SciRes. AJPS 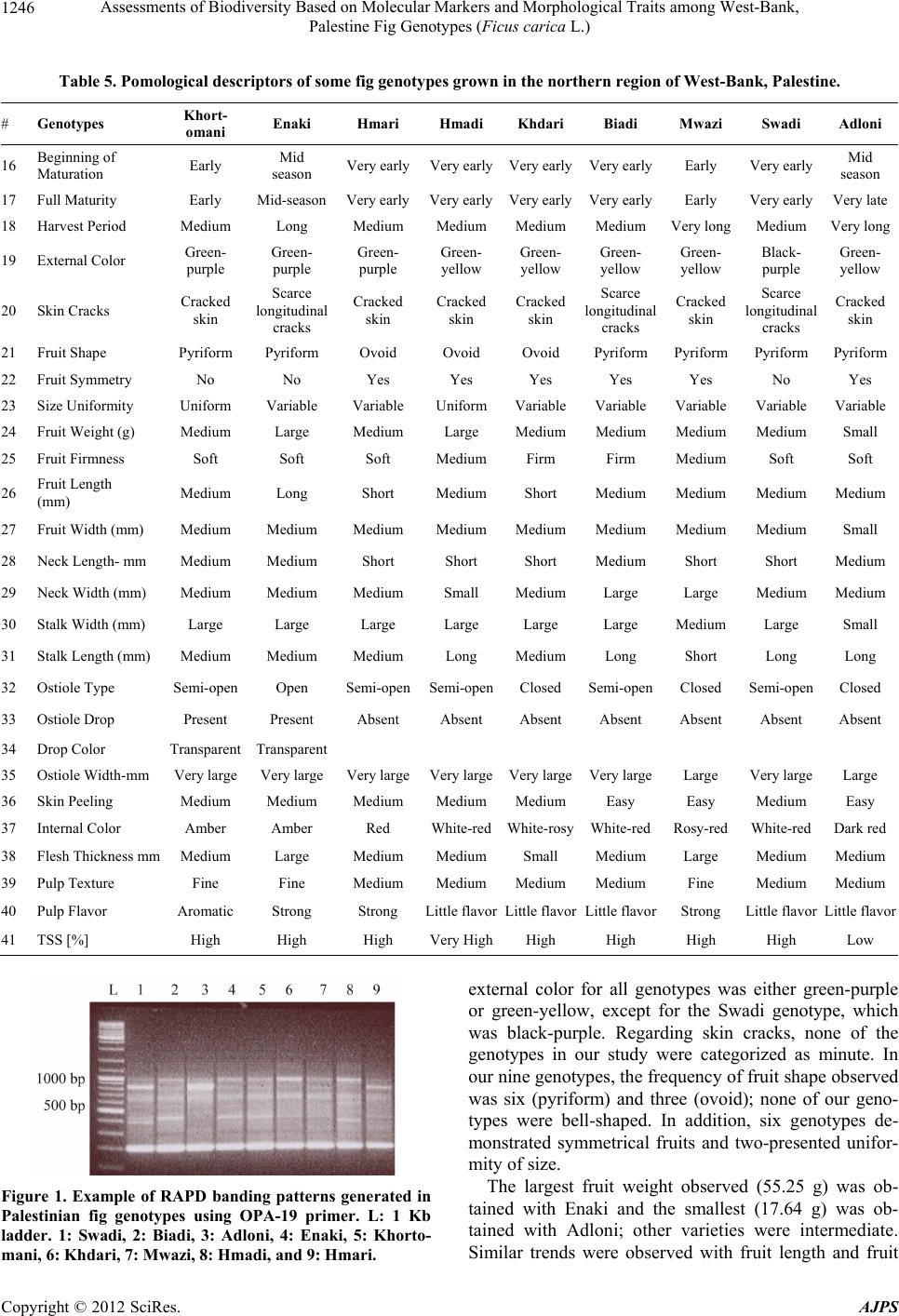 Assessments of Biodiversity Based on Molecular Markers and Morphological Traits among West-Bank, Palestine Fig Genotypes (Ficus carica L.) 1246 Table 5. Pomological descriptors of some fig genotypes grown in the northern region of West-Bank, Palestine. # Genotypes Khort- omani Enaki Hmari Hmadi Khdari Biadi Mwazi Swadi Adloni 16 Beginning of Maturation Early Mid season Very earlyVery earlyVery earlyVery earlyEarly Very earlyMid season 17 Full Maturity Early Mid-season Very earlyVery earlyVery earlyVery earlyEarly Very earlyVery late 18 Harvest Period Medium Long Medium Medium MediumMedium Very long Medium Very long 19 External Color Green- purple Green- purple Green- purple Green- yellow Green- yellow Green- yellow Green- yellow Black- purple Green- yellow 20 Skin Cracks Cracked skin Scarce longitudinal cracks Cracked skin Cracked skin Cracked skin Scarce longitudinal cracks Cracked skin Scarce longitudinal cracks Cracked skin 21 Fruit Shape Pyriform Pyriform Ovoid Ovoid Ovoid PyriformPyriform Pyriform Pyriform 22 Fruit Symmetry No No Yes Yes Yes Yes Yes No Yes 23 Size Uniformity Uniform Variable Variable UniformVariable Variable Variable Variable Variable 24 Fruit Weight (g) Medium Large Medium Large MediumMedium Medium Medium Small 25 Fruit Firmness Soft Soft Soft Medium Firm Firm Medium Soft Soft 26 Fruit Length (mm) Medium Long Short Medium Short Medium Medium Medium Medium 27 Fruit Width (mm) Medium Medium Medium Medium MediumMedium Medium Medium Small 28 Neck Length- mm Medium Medium Short Short Short Medium Short Short Medium 29 Neck Width (mm) Medium Medium Medium Small MediumLarge Large Medium Medium 30 Stalk Width (mm) Large Large Large Large Large Large Medium Large Small 31 Stalk Length (mm) Medium Medium Medium Long MediumLong Short Long Long 32 Ostiole Type Semi-openOpen Semi-openSemi-openClosed Semi-open Closed Semi-openClosed 33 Ostiole Drop Present Present Absent Absent Absent Absent Absent Absent Absent 34 Drop Color Transparent Transparent 35 Ostiole Width-mm Very largeVery large Very largeVery largeVery largeVery largeLarge Very largeLarge 36 Skin Peeling Medium Medium Medium Medium MediumEasy Easy Medium Easy 37 Internal Color Amber Amber Red White-redWhite-rosyWhite-redRosy-red White-redDark red 38 Flesh Thickness mm Medium Large Medium Medium Small Medium Large Medium Medium 39 Pulp Texture Fine Fine Medium Medium Medium Medium Fine Medium Medium 40 Pulp Flavor Aromatic Strong Strong Little flavorLittle flavorLittle flavor Strong Little flavorLittle flavor 41 TSS [%] High High High Very HighHigh High High High Low Figure 1. Example of RAPD banding patterns generated in Palestinian fig genotypes using OPA-19 primer. L: 1 Kb ladder. 1: Swadi, 2: Biadi, 3: Adloni, 4: Enaki, 5: Khorto- mani, 6: Khdari, 7: Mwazi, 8: Hmadi, and 9: Hmari. external color for all genotypes was either green-purple or green-yellow, except for the Swadi genotype, which was black-purple. Regarding skin cracks, none of the genotypes in our study were categorized as minute. In our nine genotypes, the frequency of fruit shape observed was six (pyriform) and three (ovoid); none of our geno- types were bell-shaped. In addition, six genotypes de- monstrated symmetrical fruits and two-presented unifor- mity of size. The largest fruit weight observed (55.25 g) was ob- tained with Enaki and the smallest (17.64 g) was ob- tained with Adloni; other varieties were intermediate. Similar trends were observed with fruit length and fruit Copyright © 2012 SciRes. AJPS  Assessments of Biodiversity Based on Molecular Markers and Morphological Traits among West-Bank, Palestine Fig Genotypes (Ficus carica L.) 1247 Figure 2. Dendrogram of 9 local Palestinian fig genotypes constructed by UPGM A base d on RAPD banding patterns. width. Genotypes Khdari and Biadi presented firm fruits, whereas soft fruits were observed for genotypes Khorto- mani, Enaki, Hmari, Swadi, and Adloni. Fruit neck length, neck width, stalk width, and stalk length varied among genotypes in our study. Among all tested genotypes, three (Khdari, Mwazi, and Adloni) presented closed ostiole type, five (Khorto- mani, Hmari, Hmadi, Biadi, and Swadi) presented semi- open, and one (Enaki) had open ostiole. Furthermore, transparent ostiole dew (drop) was observed in Khorto- mani and Enaki genotypes. Additionally, most of the genotypes exhibited very large ostiole width. Three ge- notypes (Biadi, Mwazi, and Adloni) presented easy skin peeling and the remainders were medium for this trait. Internal fruit color was highly variable in this study, ranging from amber to dark red. Flesh thickness was large for Enaki and Mwazi genotypes, and small for Khdari genotypes; other genotypes were medium. Pulp texture in all genotypes was either fine or me- dium, whereas the strongest pulp aromatic flavor was observed in Enaki, Hmari, and Mwazi genotypes. Total soluble solid (TSS) was either high or very high in all genotypes, with the exception of Adloni, which had low TSS. 3.4. Dendrogram of Morphological and Pomological Relationship (Similarity Matrix and Cluster Analysis) Genetic distances ranged from 0.374 to 0.654 (Table 6). “Hmari and Khdari” were the most closely related geno- types, followed by Hmari and Hmadi. In contrary, Adloni and Enaki were the most distantly related ones. UPGMA dendrogram clustered the genotypes into two main clus- ters (Figure 3). The smallest cluster, I, was composed of Adloni and Mwazi genotypes. The largest cluster, II, consisted of two sub-clusters, namely IIa, and IIb. Sub cluster (II.a) was composed of the Khortomani and Enaki genotypes, whereas the major sub cluster (II.b) included highly related Khdari and Biadi as well as the two other related genotypes Hmari and Hmadi, all of which derived from the Swadi genotype. 4. Discussion Fig (Ficus carica, Moraceae) is the only species among 700 species belonging to the Ficus genus that bear edible fruits with significant commercial value. During the last 20 years, many Mediterranean countries have investi- gated the genetic diversity of this crop; however, up to now, therereally has not been substantial progress made on this issue. This study is the first attempt to charac- terize the figs grown in Palestine using a combination of the PCR-based RAPD technique and morphological characterization. At the molecular level, our results (Tables 1, 2) and comparable studies in the literature presented high poly- morphism ratio (70.2% in 9 RAPD primers) among fig genotypes grown in the Mediterranean countries which commonly ranged between 39% - 81% within the same marker (39% in 12 RAPD primers [14]; 67% in 7 RAPD primers [19]; 72% in 6 RAPD primers [10]; 70% in 13 RAPD primers [30]; 77% in 6 RAPD primers [7]; 81% in 7 RAPD primers [21]). Indeed, the high polymorphism that was observed here is indicative of more divergent genotypes, and consequently a potential for success in future selection programs [31]. Additionally, the average of 6.33 amplicons (loci) per primer presented in this study (Table 2) was sufficient to produce useful fin-ger- prints for genotype and clone discrimination [14,17]. Copyright © 2012 SciRes. AJPS 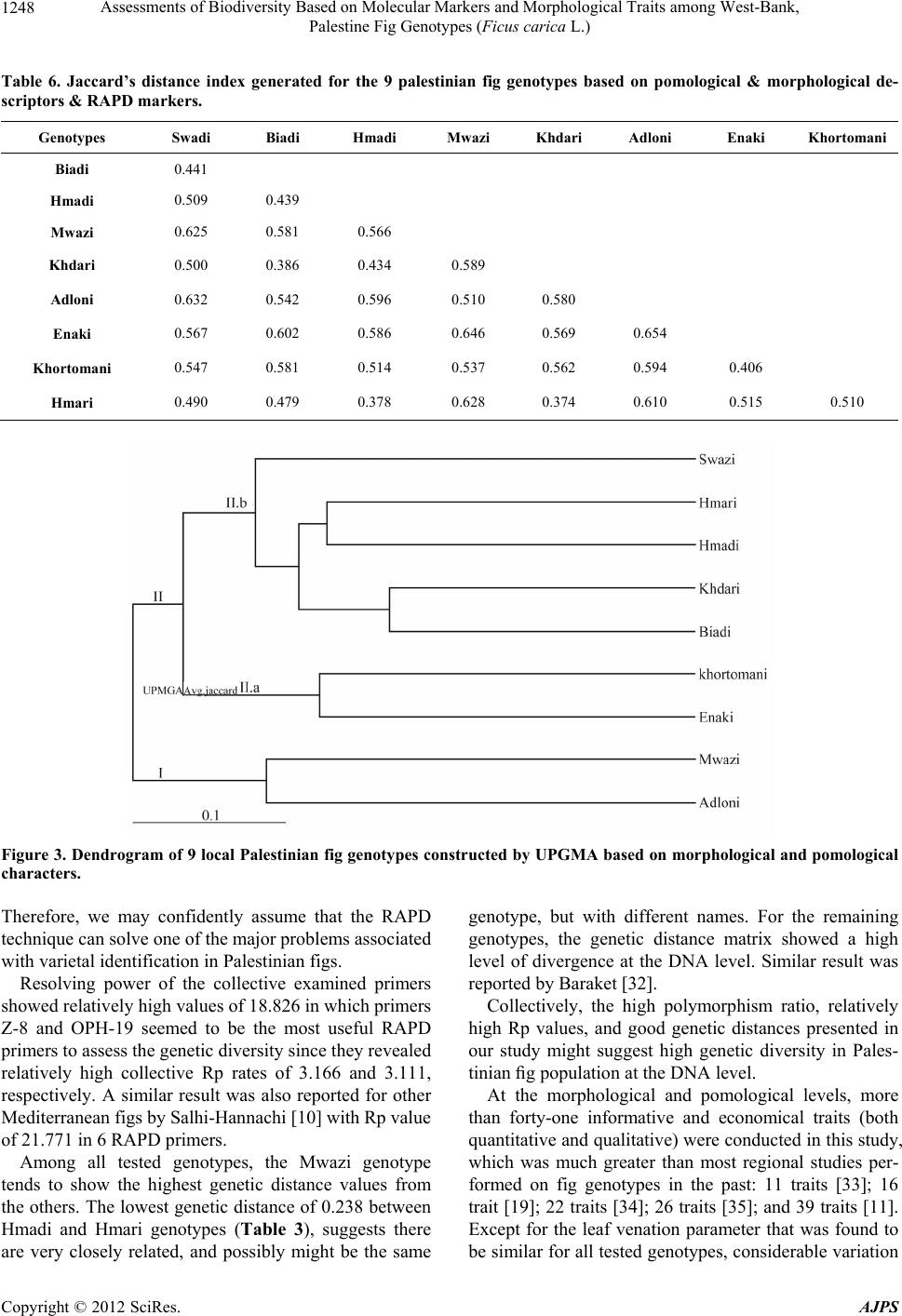 Assessments of Biodiversity Based on Molecular Markers and Morphological Traits among West-Bank, Palestine Fig Genotypes (Ficus carica L.) 1248 Table 6. Jaccard’s distance index generated for the 9 palestinian fig genotypes based on pomological & morphological de- scriptors & RAPD markers. Genotypes Swadi Biadi Hmadi Mwazi Khdari Adloni Enaki Khortomani Biadi 0.441 Hmadi 0.509 0.439 Mwazi 0.625 0.581 0.566 Khdari 0.500 0.386 0.434 0.589 Adloni 0.632 0.542 0.596 0.510 0.580 Enaki 0.567 0.602 0.586 0.646 0.569 0.654 Khortomani 0.547 0.581 0.514 0.537 0.562 0.594 0.406 Hmari 0.490 0.479 0.378 0.628 0.374 0.610 0.515 0.510 Figure 3. Dendrogram of 9 local Palestinian fig genotypes constructed by UPGMA based on morphologica l and pomological characters. Therefore, we may confidently assume that the RAPD technique can solve one of the major problems associated with varietal identification in Palestinian figs. Resolving power of the collective examined primers showed relatively high values of 18.826 in which primers Z-8 and OPH-19 seemed to be the most useful RAPD primers to assess the genetic diversity since they revealed relatively high collective Rp rates of 3.166 and 3.111, respectively. A similar result was also reported for other Mediterranean figs by Salhi-Hannachi [10] with Rp value of 21.771 in 6 RAPD primers. Among all tested genotypes, the Mwazi genotype tends to show the highest genetic distance values from the others. The lowest genetic distance of 0.238 between Hmadi and Hmari genotypes (Table 3), suggests there are very closely related, and possibly might be the same genotype, but with different names. For the remaining genotypes, the genetic distance matrix showed a high level of divergence at the DNA level. Similar result was reported by Baraket [32]. Collectively, the high polymorphism ratio, relatively high Rp values, and good genetic distances presented in our study might suggest high genetic diversity in Pales- tinian fig population at the DNA level. At the morphological and pomological levels, more than forty-one informative and economical traits (both quantitative and qualitative) were conducted in this study, which was much greater than most regional studies per- formed on fig genotypes in the past: 11 traits [33]; 16 trait [19]; 22 traits [34]; 26 traits [35]; and 39 traits [11]. Except for the leaf venation parameter that was found to be similar for all tested genotypes, considerable variation Copyright © 2012 SciRes. AJPS  Assessments of Biodiversity Based on Molecular Markers and Morphological Traits among West-Bank, Palestine Fig Genotypes (Ficus carica L.) 1249 among the genotypes was exhibited for the remaining fourteen leaf traits (Ta ble 4), which may be highly effec- tive in differentiating among fig genotypes [9,36]. A great diversity of fruit harvesting period, fruit size, fruit weight, fruit uniformity, and fruit external color ex- hibited in our examined genotypes (Table 5) could be potentially incorporated to both local and regional breed- ing programs. Such a high degree of morphological vari- ation and fruit characteristics could be utilized to interest farmers in diversifying fig genotypes [37], increasing fig production [34], and stimulating attraction of consumers for fresh fruit consumption. Additionally, the majority of our genotypes presented firm fruits, short fruit neck, and acceptable TSS (Tab le 5 ), which are very important qua- lity criterion particularly for exporting purposes. Dendrogram constructed by UPGMA based on RAPD banding patterns (Figure 2) appear contradictory to the morphological descriptors in some cases (Figure 3). This was particularly the case for the Swadi and Biadi geno- types, which were found to be close to each other ge- netically, but were morphologically distanced. However, for the remaining genotypes they were often similar. This difference could be attributed to the phenotypic modify- cations caused by the harshest prevailing weather condi- tions in the Mediterranean basin where fig can grow [7, 12], particularly since plasticity in morphology is present in fig [9]. However, RAPD is not confounded by the en- vironment, pleiotropic and epistatic effects [13], and has been found to be a useful and powerful tool to estimate genetic diversity of species and genotype identity. It is also important to point out through morphological, pomological and genetic comparisons that the Mwazi genotype was separated and identified as a distant geno- type (Table 6) indicating that its genome may differ from the other common fig genotypes [7]. 5. Conclusion Morphological and pomological results will be very use- ful in characterizing each considered genotype and to create the first reference and catalogue of the local Pales- tinian fig genotypes. RAPD is a very useful technique in characterizing Palestinian fig genotypes. Here we com- bined morphological and genetic characters to further refine this Palestinian fig database for use by both re- searchers and farmers. 6. Acknowledgements We thank the U.S. National Academy of Sciences and the Kuwait Institute for Scientific Research for hosting the Arab-American Frontiers Program that helped make this collaboration among the authors possible. We also thank both of these agencies especially to Kathrin Hum- phrey and Dalal Najib for the seed grants to help support preparation of the manuscript. REFERENCES [1] M. Flaishman, V. Rodov and E. Stover, “The Fig: Botany, Horticulture, and Breeding,” Horticultural Reviews, Vol. 34, 2008, pp. 113-197. [2] FAO, “The FAO Statistical Database-Agriculture,” 2009. http://www.fao.org/economic/the-statistics-division-ess/p ublitions-studies/statistical-yearbook/fao-statistical-year book-2009/en/. [3] F. Aljane and A. Ferchichi, “Assessment of Genetic Di- versity among Some Southern Tunisian Fig (Ficus carica L.) Cultivars Based on Morphological Descriptors,” Jor- dan Journal of Agricultural Sciences, Vol. 5, No. 1, 2009, pp. 1-16. [4] M. E. Kislev, A. Hartmann and O. Bar-Yosef, “Early Domesticated Fig in the Jordan Valley,” Science, Vol. 312, No. 5778, 2006, pp. 1372-1374. doi:10.1126/science.1125910 [5] M. Aradhya, E. Velasco and A. Koehmstedt, “Genetic Structure and Differentiation in Cultivated Fig (Ficus carica L.),” Genetica, Vol. 138, No. 6, 2010, pp. 681-694. doi:10.1007/s10709-010-9442-3 [6] O. Caliskan and A. A. Polat, “Morphological Diversity among Fig (Ficus carica L.) Accessions Sampled from the Eastern Mediterranean Region of Turkey,” Turkish Journal of Agriculture and Forestry, Vol. 36, 2012, pp. 179-193. [7] M. T. Sadder and A. F. Atteyyeh, “Molecular Assessment of Polymorphism among Local Jordanian Genotypes of The Common Fig (Ficus carica L.),” Scientica Horticul- turae, Vol. 107, No. 4, 2006, pp. 347-351. doi:10.1016/j.scienta.2005.11.006 [8] G. R. Rout and A. Mohapatra, “Use of Molecular Mark- ers in Ornamental Plants: A Critical Reappraisal,” Euro- pean Journal of Horticultural Science, Vol. 71, No. 2, 2006, pp. 53-68. [9] O. Saddoud, G. Baraket, K. Chatti, M. Trifi, M. Marrak- chi, A. Salhi-Hannachi and M. Mars, “Morphological Variability of Fig (Ficus carica L.) Cultivars,” Interna- tional Journal of Fruit Science, Vol. 8, No. 1-2, 2008, pp. 35-51. doi:10.1080/15538360802365921 [10] A. Salhi-Hannachi, K. Chatti, O. Saddoud, M. Mars, A. Rhouma, M. Marrakchi and M. Trifi, “Genetic Diversity of Different Tunisian Fig (Ficus carica L.) Collection Revealed by RAPD Fingerprints,” Hereditas, Vol. 143, 2006, pp. 15-22. doi:10.1111/j.2005.0018-0661.01904.x [11] M. Podgornik, I. Vuk, I. Vrhovnik and D. B. Mavsar, “A Survey and Morphological Evaluation of Fig (Ficus carica L.) Genetic Resources from Slovenia,” Scientia Horticulturae, Vol. 125, No. 3, 2010, pp. 380-389. doi:10.1016/j.scienta.2010.04.030 [12] M. L. A. Lima, A. A. F. Garcia, K. M. Matsuoka, H. Ari- zono and C. L. De-Souza, “Analysis of Genetic Similarity Detected by AFLP and Coefficient of Percentage among Copyright © 2012 SciRes. AJPS  Assessments of Biodiversity Based on Molecular Markers and Morphological Traits among West-Bank, Palestine Fig Genotypes (Ficus carica L.) 1250 Genotypes of Sugar Cane (Saccharum spp.),” Theoretical and Applied Genetics, Vol. 104, No. 1, 2002, pp. 30-38. doi:10.1007/s001220200003 [13] M. Agarwal, N. Shrivastava and H. Padth, “Advances in Molecular Markers Techniques and Their Applications in Plant Sciences,” Plant Cell Reports, Vol. 27, No. 4, 2008, pp. 617-631. doi:10.1007/s00299-008-0507-z [14] B. Khadari, P. H. Lashermes and F. Kjellberg, “RAPD Fingerprints for Identification and Genetic Characteriza- tion of Fig (Ficus carica L.) Genotypes,” Journal of Ge- netics and Breeding, Vol. 49, 1995, pp. 77-86. [15] B. Khadari, I. Hochu, S. Santoni, A. Oukabli, M. Ater, J. P. Roger and F. Kjellberg, “Which Molecular Markers are Best Suited to Identify Fig Cultivars: A Comparison of RAPD, ISSR and Microsatellite Markers,” Acta Hor- ticulturae, Vol. 605, 2003, pp. 69-75. [16] P. J. Elisiario, M. C. Neto, L. F. Cabrita and J. M. Leitao, “Isoenzyme and RAPDs Characterization of a Collecion on Fig (Ficus carica L.) Traditional Varieties,” Acta Hor- ticulturae, Vol. 480, 1998, pp. 149-154. [17] U. Galderisi, M. Cipollaro, G. Di Bernardo, L. De Masi, G. Galano and A. Cascino, “Identification of the Edible Fig ‘Bianco del Cilento’ by Random Amplified Poly- morphic DNA Analysis,” HortScience, Vol. 3, 1999, pp. 1263-1265. [18] L. F. Cabrita, U. Aksoy, S. Hepaksoy and J. M. Leitao, “Suitability of Isozyme, RAPD and AFLP Markers to Assess Genetic Differences and Relatedness among Fig (Ficus carica L.) Clones,” Scientia Hortculturae, Vol. 87, No. 4, 2001, pp. 261-273. doi:10.1016/S0304-4238(00)00181-3 [19] K. Papadopoulou, C. Ehaliotis, M. Tourna, P. Kastanis, I. Karydis and G. Zervakis, “Genetic Relatedness among Dioecious Fig (Ficus carica L.) Cultivars by Random Amplified Polymorphic DNA Analysis, and Evaluation of Agronomic and Morphological Characters,” Genetica, Vol. 114, No. 2, 2002, pp. 183-194. doi:10.1023/A:1015126319534 [20] Y. Aka-Kacar, A. B. Kuden and S. Cetiner, “Identifica- tion of Varietal Polymorphism in Ficus carica L. by RAPD (Randomly Amplified Polymorphic DNA) Mark- ers,” Acta Horticulturae, Vol. 598, 2003, pp. 167-172. [21] L. De Masi, M. Cipollaro, G. Di Bernardo, U. Galderisi, G. Galano, A. Cascino, G. Grassi, E. Pavone and A. Simeone, “Clonal Selection and Molecular Characteriza- tion by RAPD Analysis of The Fig (Ficus carica L.) ‘Dottato’ and ‘Blanco del Cilento’ Cultivars in Italy,” Acta Horticulturae, Vol. 605, 2003, pp. 65-68. [22] H. Achtak, A. Oukabli, M. Ater, S. Santoni, F. Kjellberg and B. Khadari, “Microsatellite Markers as Reliable Tools for Fig Cultivar Identification,” Journal of Ameri- can Society for Horticultural Science, Vol. 134, No. 6, 2009, pp. 624-631. [23] M. Kocsis, L. Jaromi, P. Putnoky, P. Kozma and A. Borhidi, “Genetic Diversity among Twelve Grape Culti- vars Indigenous to the Carpathian Basin Revealed by RAPD Markers,” Vitis, Vol. 44, No. 2, 2005, pp. 87-91. [24] M. Hoogendijk and D. E. Williams, “Characterizing the Genetic Diversity of Home Garden Crops: Some Exam- ples from the Americas,” In: J. W. Watson and P. D. Eyzaguirre, Eds., Home Gardens and in Situ Conserva- tions of Plant Genetic Resources in Farming Systems, Proceeding of the 2nd International Home Gardens Workshop, Witzenhausen, 17-19 July 2001, pp. 34-40. [25] C. Cantini, A. Cimato and G. Sani, “Morphological Evaluation of Olive Germplasm Present in Tuscany Re- gion,” Euphytica, Vol. 109, No. 3, 1999, pp. 173-181. doi:10.1023/A:1003728800464 [26] A. Prevost and M. J. Wilkinson, “A New System of Comparing PCR Primers Applied to ISSR Fingerprinting of Potato Cultivars,” Theoretical and Applied Genetics, Vol. 98, No. 1, 1999, pp. 107-112. doi:10.1007/s001220051046 [27] J. E. Gilbert, R. V. Lewis and M. J. Wilkinson, “Devel- oping an Appropriate Strategy to Assess Genetic Vari- ability in Plant Germplasm Collections,” Theoretical and Applied Genetics, Vol. 98, No. 6-7, 1999, pp. 1125-1131. doi:10.1007/s001220051176 [28] P. M. Schluter and S. A. Harris, “Analysis of Multilocus Fingerprinting Data Sets Containing Missing Data,” Mo- lecular Ecology Notes, Vol. 6, No. 2, 2006, pp. 569-572. doi:10.1111/j.1471-8286.2006.01225.x [29] IPGRI and CIHEAM, “Descriptors for Fig (Ficus carica L.),” International Plant Genetic Resources Institute (IP- GRRI), Rome, Italy and International Center for Ad- vanced Mediterranean Agronomic Studies (CIHEAM), Paris, 2003. [30] M. Akbulut, S. Ercisli and H. Karlidag “RAPD-Based Study of Genetic Variation and Relationships among Wild Fig Genotypes in Turkey,” Genetics and Molecular Research, Vol. 8, No. 3, 2009, pp. 1109-1115. doi:10.4238/vol8-3gmr634 [31] D. Bandelj, J. Jakse and B. Javornik, “DNA Fingerprint- ing of Olive Varieties by Microsatellite Markers,” Food Technology and Biotechnology, Vol. 40, 2002, pp. 185- 190. [32] G. Baraket, K. Chatti, O. Saddoud, A. Ben-Abdelkarim, M. Mars, M. Trifi and A. Hannachi, “Comparative As- sessment of SSR and AFLP Markers for Evaluation of Genetic Diversity and Conservation of Fig, Ficus carica L., Genetic Resources in Tunisia,” Plant Molecular Biol- ogy Reporter, Vol. 29, No. 1, 2011, pp. 171-184. [33] L. Chalak, A. Chehade, E. Mattar and B. Khadari, “Mor- phological Characterization in Fig Accessions Cultivated in Lebanon,” Acta Horticulturae, Vol. 798, 2008, pp. 49- 56. [34] O. Caliskan and A. A. Polat, “Fruit Characteristics of Fig Cultivars and Genotypes Grown in Turkey,” Scientica Horticulturae, Vol. 115, No. 4, 2008, pp. 360-367. doi:10.1016/j.scienta.2007.10.017 [35] M. Simsek, “Fruit Performances of the Selected Fig Types in Turkey,” African Journal of Agricultural Re- search, Vol. 4, 2009, pp. 1260-1267. [36] F. Aljane, “Analyse de la Diversité Génétique de Culti- Copyright © 2012 SciRes. AJPS  Assessments of Biodiversity Based on Molecular Markers and Morphological Traits among West-Bank, Palestine Fig Genotypes (Ficus carica L.) Copyright © 2012 SciRes. AJPS 1251 vars Locaux du Figuier (Ficus carica L.) Cultivés dans la Chaîne de Matmata,” Revue des Régions Arides, 2004, pp. 95-104. [37] A. Oukabli, A. Mamouni, M. Laghezali and B. Khadari, “Genetic Variability in Moroccan Fig Cultivars (Ficus carica L.) Based on Morphological and Pomological Data,” Acta Horticulturae, Vol. 605, 2003, pp. 311-318.
|