E. J. MULLANEY ET AL. 397
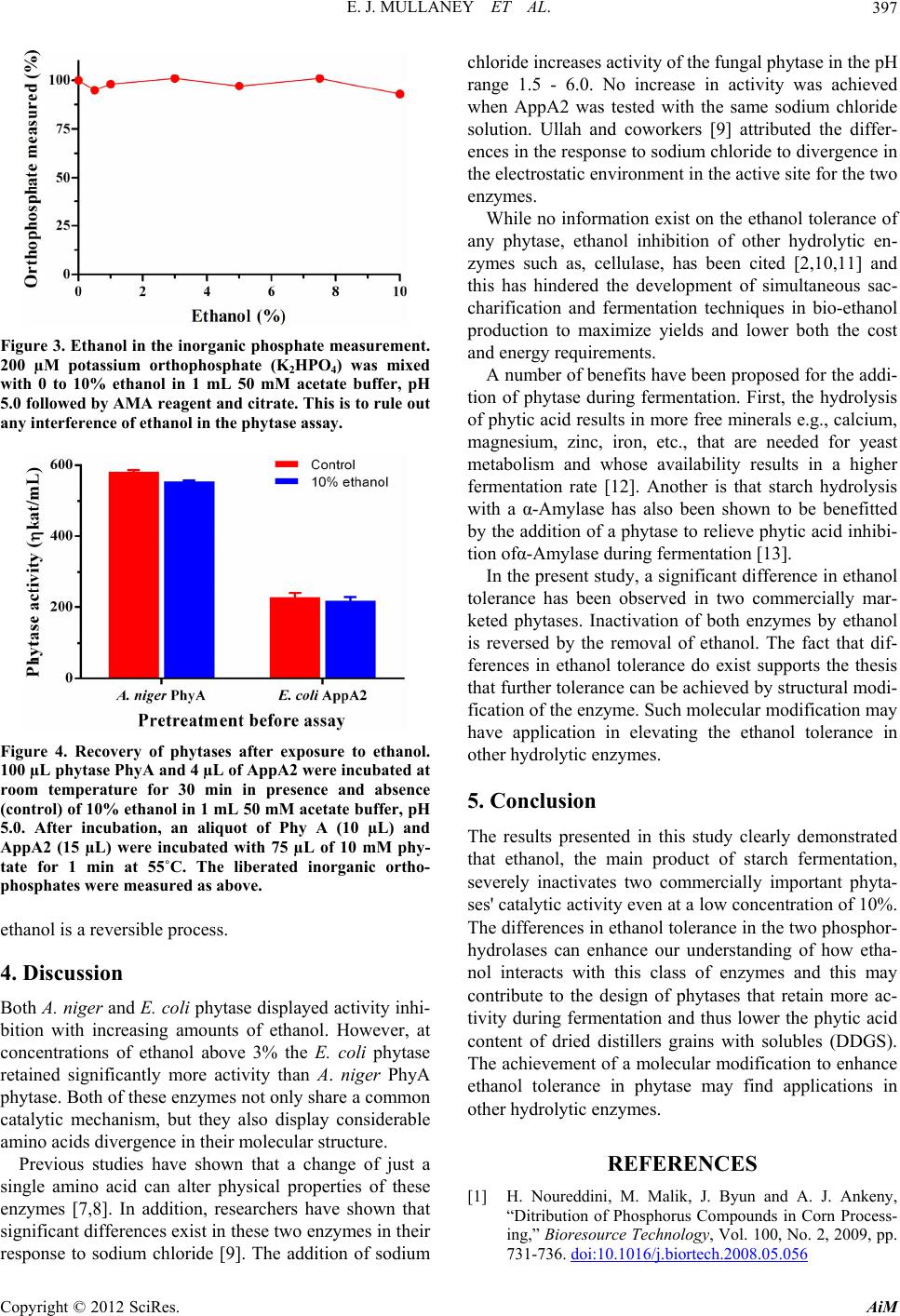
Figure 3. Ethanol in the inorganic phosphate measurement.
200 µM potassium orthophosphate (K2HPO4) was mixed
with 0 to 10% ethanol in 1 mL 50 mM acetate buffer, pH
5.0 followed by AMA reagent and citrate. This is to rule out
any interference of ethanol in the phytase assay.
Figure 4. Recovery of phytases after exposure to ethanol.
100 µL phytase PhyA and 4 µL of AppA2 were incubated at
room temperature for 30 min in presence and absence
(control) of 10% ethanol in 1 mL 50 mM acetate buffer, pH
5.0. After incubation, an aliquot of Phy A (10 µL) and
AppA2 (15 µL) were incubated with 75 µL of 10 mM phy-
tate for 1 min at 55˚C. The liberated inorganic ortho-
phosphates were me asur e d as above.
ethanol is a reversible process.
4. Discussion
Both A. niger and E. coli phytase displayed activity inhi-
bition with increasing amounts of ethanol. However, at
concentrations of ethanol above 3% the E. coli phytase
retained significantly more activity than A. niger PhyA
phytase. Both of these enzymes not only share a common
catalytic mechanism, but they also display considerable
amino acids divergence in their molecular structure.
Previous studies have shown that a change of just a
single amino acid can alter physical properties of these
enzymes [7,8]. In addition, researchers have shown that
significant differences exist in these two enzymes in their
response to sodium chloride [9]. The addition of sodium
chloride increases activity of the fungal phytase in the pH
range 1.5 - 6.0. No increase in activity was achieved
when AppA2 was tested with the same sodium chloride
solution. Ullah and coworkers [9] attributed the differ-
ences in the response to sodium chloride to divergence in
the electrostatic environment in the active site for the two
enzymes.
While no information exist on the ethanol tolerance of
any phytase, ethanol inhibition of other hydrolytic en-
zymes such as, cellulase, has been cited [2,10,11] and
this has hindered the development of simultaneous sac-
charification and fermentation techniques in bio-ethanol
production to maximize yields and lower both the cost
and energy requirements.
A number of benefits have been proposed for the addi-
tion of phytase during fermentation. First, the hydrolysis
of phytic acid results in more free minerals e.g., calcium,
magnesium, zinc, iron, etc., that are needed for yeast
metabolism and whose availability results in a higher
fermentation rate [12]. Another is that starch hydrolysis
with a α-Amylase has also been shown to be benefitted
by the addition of a phytase to relieve phytic acid inhibi-
tion ofα-Amylase during fermentation [13].
In the present study, a significant difference in ethanol
tolerance has been observed in two commercially mar-
keted phytases. Inactivation of both enzymes by ethanol
is reversed by the removal of ethanol. The fact that dif-
ferences in ethanol tolerance do exist supports the thesis
that further tolerance can be achieved by structural modi-
fication of the enzyme. Such molecular modification may
have application in elevating the ethanol tolerance in
other hydrolytic enzymes.
5. Conclusion
The results presented in this study clearly demonstrated
that ethanol, the main product of starch fermentation,
severely inactivates two commercially important phyta-
ses' catalytic activity even at a low concentration of 10%.
The differences in ethanol tolerance in the two phosphor-
hydrolases can enhance our understanding of how etha-
nol interacts with this class of enzymes and this may
contribute to the design of phytases that retain more ac-
tivity during fermentation and thus lower the phytic acid
content of dried distillers grains with solubles (DDGS).
The achievement of a molecular modification to enhance
ethanol tolerance in phytase may find applications in
other hydrolytic enzymes.
REFERENCES
[1] H. Noureddini, M. Malik, J. Byun and A. J. Ankeny,
“Ditribution of Phosphorus Compounds in Corn Process-
ing,” Bioresource Technology, Vol. 100, No. 2, 2009, pp.
731-736. doi:10.1016/j.biortech.2008.05.056
Copyright © 2012 SciRes. AiM