Paper Menu >>
Journal Menu >>
 Journal of Cancer Therapy, 2012, 3, 452-459 http://dx.doi.org/10.4236/jct.2012.324058 Published Online September 2012 (http://www.SciRP.org/journal/jct) Expression Profile of Epithelial Protein Lost in Neoplasm-Alpha (EPLIN-α) in Human Pulmonary Cancer and Its Impact on SKMES-1 Cells in Vitro Yinan Liu1,2,3, Andrew J. Sanders1,2, Lijian Zhang1,3*, Wen G. Jiang1,2* 1Cardiff University-Peking University School of Oncology Joint Institute, Cardiff, UK; 2Metastasis and Angiogenesis Research Group, Cardiff University School of Medicine, Cardiff, UK; 3Key Laboratory of Carcinogenesis and Translational Research (Minis- try of Education), Department of Thoracic Surgery II, Peking University Cancer Hospital and Institute, Beijing, China. Email: *lijzhang@yeah.net, *jiangw@cf.ac.uk Received July 27th, 2012; revised August 31st, 2012; accepted September 14th, 2012 ABSTRACT Epithelial Protein Lost in Neop lasm (EPLIN) is a cytoskeletal associated protein implicated in regulatin g actin dynamo- ics and cellular motility and whose expression is frequently downregu lated in a number of human cancers. The current study examined the expression levels of EPLIN-α in a pulmonary cancer cohort and its association with clinical patho- logical factors using quantitative polymerase chain reaction. Additionally, EPLIN-α was over-expressed in the SKMES- 1 pulmonary cancer cell line through transfection with a plasmid containing the expression sequence for EPLIN-α. The role of EPLIN-α was subsequently examined using a variety of in vitro functional assays. Decreased levels of EPLIN-α were seen in cancerous tissues compared to normal background tissue. Lower levels of EPLIN-α were also associated with higher TNM stage and nodal involvement. In vitro over-expression of EPLIN-α inhibited SKMES-1 growth rates (p = 0.05 vs plasmid control) and motility (p = 0.002 vs plasmid co ntrol), thou gh did not hav e any sign ificant effects on cell-matrix adhesion or cell invasion. Taken together, the current study indicates that lower levels of EPLIN-α may be associated with poorer prognosis and more advanced pulmonary cancer, where this molecule appears to play a suppres- sive role on cell growth and migration. Keywords: EPLIN-α; Pulmonary Cancer; Cell Migration; ECIS 1. Introduction Pulmonary cancer is one of the most fatal cancers, with distant metastasis being a key factor in the poor progno- sis associated with this cancer. Epithelial protein lost in neoplasm (EPLIN) has previously been found to localise with actin stress fibres and play important roles in regu- lating actin dynamics and linking the catenin-cadherin complex to F-actin [1-3], suggesting a key role for this molecule in regulating cellular motility. Since the discovery of EPLIN, as a molecule different- tially expressed between normal oral epithelial cells and HPV-immortalised oral epithelial cell lines, scientific in- terest has focused on the role of this molecule in cancer, with a number of studies reporting aberrant expression of EPLIN in cancerous cells and tissues of various cancer types including, breast, prostate and oesophageal can cers [4-8]. EPLIN, also known as Lima-1 (LIM domain and actin binding-1), contains a centrally located LIM do- main and exists as two isoforms, known as EPLIN-α and EPLIN-β, with the EPLIN-β isoform containing an addi- tional 160 amino acids at the N-terminus. The human gene for EPLIN consists of 11 exons, spanning more than 100 kb, with different transcriptional starting points across this gene accounting for the EPLIN-α (90 kDa) and EPLIN-β (110 kDa) isoforms [8,9 ]. Previous studies have suggested roles for EPLIN-α in cytokinesis, where loss of EPLIN-α in cancerous cells results in the mis-locali- sation of cytokinesis proteins resulting in enhanced ge- nomic instability [10]. Numerous studies have also im- plicated the loss of EPLIN-α as contributing to the en- hanced motility and invasiveness of cancer cells [1-3,5,6, 11,12]. EPLIN-α can bind actin monomers through the NH2 and COOH protein ends and can also bind F-actin, demonstrating a role in actin stabilisation [2]. Extracel- lular signal regulated kinase (ERK) phosphorylation of EPLIN has also been shown to reduce the C-terminal protein affinity for F-actin, suggesting that ERK phos- phorylation may be key to EPLINs role in regulating actin dynamics [12]. EPLIN-α has also demonstrated the capacity to link the cadherin-catenin complex to F-actin *Corresponding authors. Copyright © 2012 SciRes. JCT  Expression Profile of Epithelial Protein Lost in Neoplasm-Alpha (EPLIN-α) in Human Pulmonary Cancer and Its Impact on SKMES-1 Cells in Vitro 453 through its ability to bind α-catenin [3]. Thus, EPLIN-α appear to play key roles in regulating actin dynamics and motility in normal cells. The loss of this molecule in can - cerous cells likely impacts on these key functions, lead- ing to a more invasive phenotype. Clinical studies have further highlighted the importance of EPLIN-α in cancer progression with lower levels of this molecule being as- sociated with poor prognosis predictive factors such as TNM stage, grade and patient survival rate in numerous cancers including breast, prostate and oesophageal can- cers [5,6,11]. In support of clinical observations, addi- tional work has demonstrated that the forced expression of EPLIN-α in prostate, breast and oesophageal cancer cells can impact on aggressive traits in vitro and in vivo [5-7]. Taken together all these studies indicate that EPLIN-α may act as a potential prognostic indicator and that the molecule may act as a protective factor in cancer progression. Whilst the importance and role of EPLIN-α in a num- ber of cancers is beginning to become apparent, its role in pulmonary cancer is currently unstudied. In the present study, the expression of EPLIN-α was examined in a cohort of pulmonary cancer samples and cell lines. Pul- monary cancer cells were forced to express this molecule, though transfection with a mammalian expression plas- mid containing th e full sequence of EPLIN-α, in order to establish the functional role of EPLIN-α in pulmonary cancer cells in vitro. 2. Materials and Methods 2.1. Cell Lines and Maintenance Human pulmonary cancer cell lines, SKMES-1 and A549 were purchased from ECACC (European Collection of Animal Cell Culture, Salisbury, England, UK) and main- tained in Dubecco’s Modified Eagle Medium (DMEM) (Sigma, Dorset, UK) supplemented with penicillin, stre- ptomycin and 10% foetal calf serum (Sigma, Dorset, UK). The cells were incubated at 37˚C, 5% CO2 and 95% humidity. 2.2. Pulmonary Tissue Sample Collection Paired normal and tumour pulmonary tissues (n = 83 pairs) were collected immediately after surgery and stored at –80˚C until use. Ethical approval and informed consents were obtained from the local ethics committee and patients respectively. The clinical follow-up was routinely performed following surgery and the median follow-up period was 120 months. Details of histology were obtained from pathology reports and confirmed by a consultant pathologist. 2.3. Generation of SKMES-1 Cells Over-Expressing EPLIN-α A plasmid containing the expression sequence for EPLIN-α has previously been generated and used within our laboratories [5-7,13]. In brief, the full length se- quence for EPLIN-α was generated from mammary tis- sue cDNA using a combination of the following amplify- cation primers, 5’ATGGAAAATTGTCTAGGAGA’3 (EPLINaExF1), 5’ATGGAAAATTGTCTAGGAGAA’3 (EPLINaExF2) and 5’TCACTCTTCATCCT CATCCTC’3 (EPLINaExR1), together with a high fidelity PCR mas- termix (AbGene, Epsom, UK). The product size was verified before being T-A cloned into the pEF6/V5-His mammalian expression vector (Invitrogen, Paisley, UK). Plasmids were then inserted into chemically competent TOP10 bacteria, amplified and plated under ampicillin (100 µg/ml) selection. Colonies which grew were sub- jected to orientation analysis and bacteria containing plasmids with correctly orientated inserts were selected, amplified and harvested for plasmid extraction (GenElute, Sigma, Dorset, UK). This resource was used to transfect the human pulmonary SKMES-1 cancer cell line, which demonstrated minimal expression levels of EPLIN-α. SKMES-1 cells were transfected with either the EPLIN-α expression plasmid or a closed pEF6 plasmid and sub- jected to a period of selection (5 µg/ml blasticidin) be- fore subsequent maintenance of the cells at 0.5 µg/ml blasticidin. Following the selective pe riod, the expression of EPLIN-α was examined in the wild type and trans- fected cells, at the transcriptional level, using RT-PCR. Those cells containing the expression plasmid and dis- playing enhanced EPLIN-α expression were designated SKMES-1EPLIN-α exp, those containing the pEF6 control plasmid were designated SKMES-1pEF6 and unaltered wild type cells were designated SKMES-1WT. 2.4. RNA Extraction, Reverse Transcription-PCR, and Quantitative PCR RNA isolation was performed on either a 25 cm2 tissue culture flask or homogenised tissue samples using TRI Reagent (Sigma, Dorset, UK). RNA was subsequently quantified using a spectrophotometer (WPA UV 1101, Biotech Photometer, Cambridge, UK) before standardis- ing all samples to a concentration of 500 ng for use in reverse transcription (iScript™ Reverse Transcription Supermix kit, Bio-Rad Laboratories, Hemel Hempstead, UK). Routine RT-PCR was carried out using specific primers for EPLIN-α (Table 1). Amplification conditions were as follows: 94˚C for 5 minutes, 32 cycles of 94˚C for 40 seconds, 55˚C for 40 seconds and 72˚C for 60 seconds, followed by a final extension of 72˚C for 10 minutes and 4˚C hold. PCR products were separa ted on a Copyright © 2012 SciRes. JCT  Expression Profile of Epithelial Protein Lost in Neoplasm-Alpha (EPLIN-α) in Human Pulmonary Cancer and Its Impact on SKMES-1 Cells in Vitro Copyright © 2012 SciRes. JCT 454 Table. 1. Primer sequences used in PCR and Q-PCR. Primer set Sense Anti-sense GAPDH probe ATGATATCGCCGCGCTCA CGCTCGGTGAGGATCTTCA EPLIN probe/Q-PCR AAGCAAAAATGAAAACGAAG ACTGAACCTGACCGTACAGACACCCACCTTAGCAA GAPDH Q-PCR CTGAGTACGTCGTGGAGTC ACTGAACCTGACCGTACACAGAGATGATGACCCTTTTG Z-sequence on Q-PCR primers is 5’ ACTGAACCTGACCGTACA 3’. 2% agarose gel and documented, following ethidium bro- mide staining, using a digital camera mounted over a UV transluminator. The level of EPLIN-α transcript present in the clinical samples was determined using real-time quantitative PCR, based on the Amplifluor technology and modified from a method reported previously [5]. Briefly, primers were designed in a similar way as to those used in con- ventional PCR. To one of the primer pairs, an additional sequence was added, known as the Z sequence, which is complementary to the universal Z probe (Intergen Inc., Oxford, England, UK). The reaction was carried out us- ing the following: Hot-start Q-master mix (Abgene, Ep- som, UK), 10 pmol of specific forward primer, 1 pmol reverse primer (containing the Z sequence), 10 pmol of FAM-tagged probe (Intergen), and cDNA from approxi- mately 50 ng RNA. The reaction was carried out using an IcyclerIQ™ (Bio-Rad, surrey, UK). The following con- ditions were used in the reaction: 94˚C for 12 minutes, 60 cycles of 94˚C for 15 seconds, 55˚C for 40 seconds and 72˚C for 20 seconds. The levels of the EPLIN-α trans- cripts are presented as transcript copies per 50 ng RNA and values are calculated based on an internal standard that was simultaneously amplified during the same quan- titative real-time PCR. 2.5. In Vitro Cell Growth Assay Cells were seeded into triplicate 96-well plates at a seed- ing density of 3000 cells per well. The plates were incu- bated for differing periods (overnight, 3 and 5 days). Following incubation for the appropriate time, plates were fixed in 4% formalin (v/v) and stained with 0.5% (w/v) crystal violet. Crystal violet stain was extracted using 10% (v/v) acetic acid and the absorbance was de- tected at a wavelength of 540 nm using a spectropho- tometer. 2.6. In Vitro Invasion Assay This was undertaken following a previously described method [5,6]. Transwell inserts with 8.0 µm pores were coated with 50 µg Matrigel (BD Bioscience, Oxford, UK) and air dried. The Matrigel was then rehydrated before seeding 30,000 cells to each insert. After 72 hours, any cells that had invaded to the underside of the insert were fixed in 4% formalin and stained in crystal violet. Cell invasion was quantified by assessing the number of stained cells that had invaded to the underside of the in- sert in random microscopic fields. 2.7. In Vitro Matrix Adhesion Assay Wells of a 96 well plate were pre-coated with 5 µg of Matrigel and air dried. The Matrigel layer was then re- hydrated before seeding 45,000 cells onto the matrix layer and incubating for 40 minutes. Following this in- cubation, non-adherent cells were removed through nu- merous washes with BSS before fixing adherent cells in formalin and staining with crystal violet. Cell-matrix adhesion was then quantified under the microscope by assessing the number of adherent cells in random micro- scopic fields. 2.8. ECIS Based Cellular Motility Assays Cell motility was assessed using the Electric Cell-sub- strate Impedance Sensing (ECIS) system (Applied Bio- physics Inc, NJ, US) and 96W1E arrays. One hundred and twenty thousand cells (SKMES-1pEF6 or SKMES- 1EPLIN-α exp) were seeded into each well. These cells were allowed to settle and attach to the array wells to form a confluent monolayer. This monolayer was subsequently wounded, through the application of an electrical charge to the electrode. The change in resistance of the cell layer was recorded for a number of hours as the surrounding cells recovered and migrated onto the electrode to close the wound. Additional analysis was performed to assess the importance of PLCγ signalling in EPLIN-α regulated migration using a PLCγ inhibitor (CalBiochem, Notting- ham, England, UK). To accomplish this, similar experi- ments were set up to examine the motility rates of SKMES-1pEF6 and SKMES-1EPLIN-α exp cells, both in the presence and absence of 100nM PLCγ inhibitor. 2.9. Statistical Analysis The Minitab 14 statistical software package was used to carry out statistical analysis. Normally distributed data was analysed using two-sample, two-tailed t-tests and non-normally distributed data was analysed using the Mann-Whitney test. Two Way ANOVA analysis of mi- 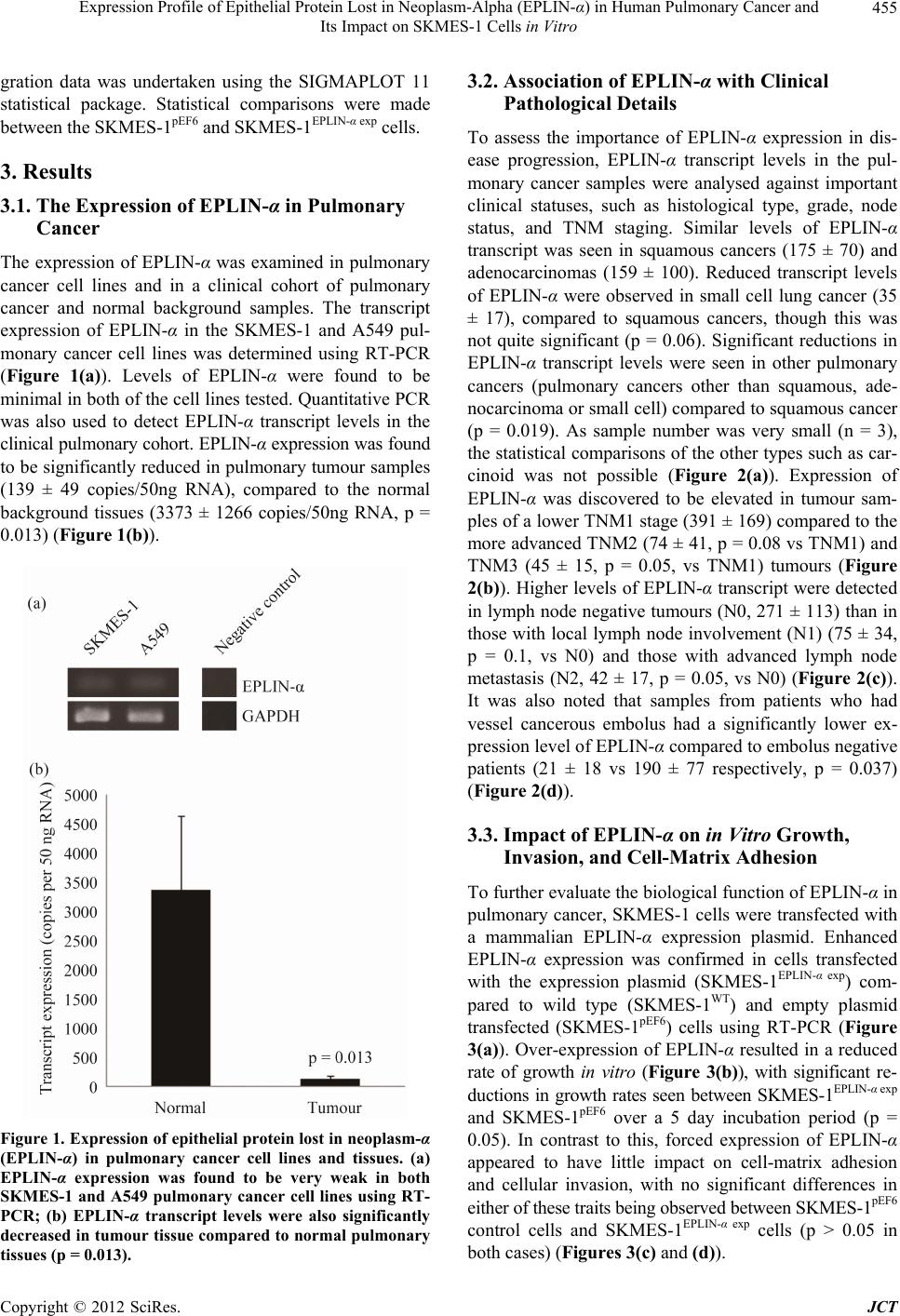 Expression Profile of Epithelial Protein Lost in Neoplasm-Alpha (EPLIN-α) in Human Pulmonary Cancer and Its Impact on SKMES-1 Cells in Vitro 455 gration data was undertaken using the SIGMAPLOT 11 statistical package. Statistical comparisons were made between the SKMES-1pEF6 and SKMES-1EPLIN-α exp cells. 3. Results 3.1. The Expression of EPLIN-α in Pulmonary Cancer The expression of EPLIN-α was examined in pulmonary cancer cell lines and in a clinical cohort of pulmonary cancer and normal background samples. The transcript expression of EPLIN-α in the SKMES-1 and A549 pul- monary cancer cell lines was determined using RT-PCR (Figure 1(a)). Levels of EPLIN-α were found to be minimal in both of the cell lines tested . Quantitative PCR was also used to detect EPLIN-α transcript levels in the clinical pulmonary cohort. EPLIN-α expression was found to be significantly reduced in pu lmonary tumour samples (139 ± 49 copies/50ng RNA), compared to the normal background tissues (3373 ± 1266 copies/50ng RNA, p = 0.013) (Figure 1(b)). Figure 1. Expression of epithelial protein lost in neoplasm-α (EPLIN-α) in pulmonary cancer cell lines and tissues. (a) EPLIN-α expression was found to be very weak in both SKMES-1 and A549 pulmonary cancer cell lines using RT- PCR; (b) EPLIN-α transcript levels were also significantly decreased in tumour tissue compared to normal pulmonary tissues (p = 0.013). 3.2. Association of EPLIN-α with Clinical Pathological Details To assess the importance of EPLIN-α expression in dis- ease progression, EPLIN-α transcript levels in the pul- monary cancer samples were analysed against important clinical statuses, such as histological type, grade, node status, and TNM staging. Similar levels of EPLIN-α transcript was seen in squamous cancers (175 ± 70) and adenocarcinomas (159 ± 100). Reduced transcript levels of EPLIN-α were observed in small cell lung cancer (35 ± 17), compared to squamous cancers, though this was not quite significant (p = 0.06). Significant reductions in EPLIN-α transcript levels were seen in other pulmonary cancers (pulmonary cancers other than squamous, ade- nocarcinoma or small cell) compared to squamous cancer (p = 0.019). As sample number was very small (n = 3), the statistical comparisons of the other types such as car- cinoid was not possible (Figure 2(a)). Expression of EPLIN-α was discovered to be elevated in tumour sam- ples of a lower TNM1 stage (391 ± 169) compared to the more advanced TNM2 (74 ± 41, p = 0.08 vs TNM1) and TNM3 (45 ± 15, p = 0.05, vs TNM1) tumours (Figure 2(b)). Higher levels of EPLIN-α transcript were detected in lymph node negative tumours (N0, 271 ± 113) than in those with local lymph node involvement (N1) (75 ± 34, p = 0.1, vs N0) and those with advanced lymph node metastasis (N2, 42 ± 17, p = 0.05, vs N0) (Figure 2(c)). It was also noted that samples from patients who had vessel cancerous embolus had a significantly lower ex- pression level of EPLIN-α compared to embolus negative patients (21 ± 18 vs 190 ± 77 respectively, p = 0.037) (Figure 2(d)). 3.3. Impact of EPLIN-α on in Vitro Growth, Invasion, and Cell-Matrix Adhesion To further evaluate the biological function of EPLIN-α in pulmonary cancer, SKMES-1 cells were transfected with a mammalian EPLIN-α expression plasmid. Enhanced EPLIN-α expression was confirmed in cells transfected with the expression plasmid (SKMES-1EPLIN-α exp) com- pared to wild type (SKMES-1WT) and empty plasmid transfected (SKMES-1pEF6) cells using RT-PCR (Figure 3(a)). Over-expression of EPLIN-α resulted in a reduced rate of growth in vitro (Figure 3(b)), with significant re- ductions in growth rates seen between SKMES-1EPLIN-α exp and SKMES-1pEF6 over a 5 day incubation period (p = 0.05). In contrast to this, forced expression of EPLIN-α appeared to have little impact on cell-matrix adhesion and cellular invasion, with no significant differences in either of these traits being observed between SKMES-1pEF6 control cells and SKMES-1EPLIN-α exp cells (p > 0.05 in both cases) (Figures 3(c) and (d)). Copyright © 2012 SciRes. JCT 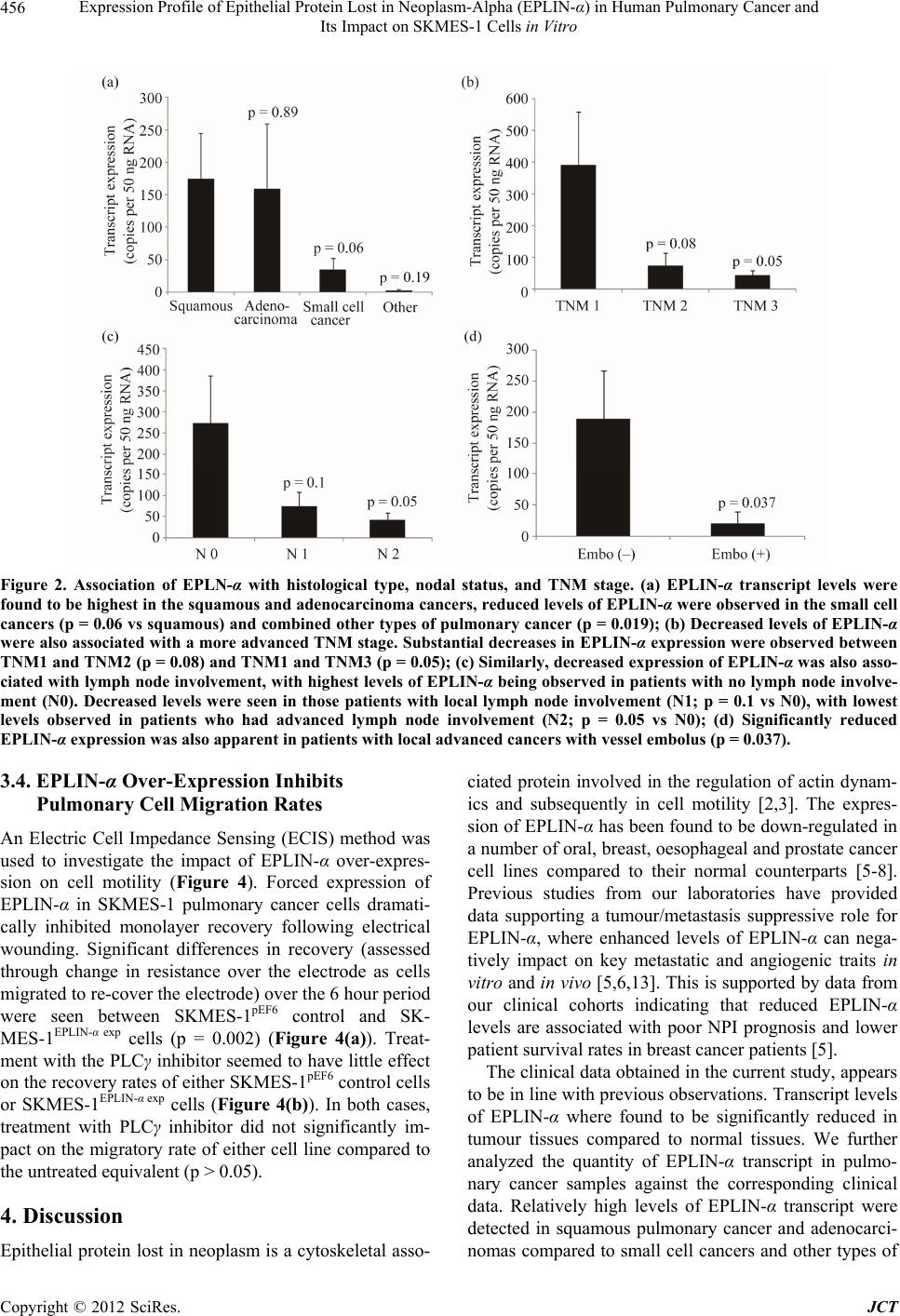 Expression Profile of Epithelial Protein Lost in Neoplasm-Alpha (EPLIN-α) in Human Pulmonary Cancer and Its Impact on SKMES-1 Cells in Vitro Copyright © 2012 SciRes. JCT 456 Figure 2. Association of EPLN-α with histological type, nodal status, and TNM stage. (a) EPLIN-α transcript levels were found to be highest in the squamous and adenocarcinoma cancers, reduced levels of EPLIN-α were observed in the small cell cancers (p = 0.06 vs squamous) and combined other types of pulmonary cancer (p = 0.019); (b) Decreased levels of EPLIN-α were also associated with a more advanced TNM stage. Substantial decreases in EPLIN-α expression were observed between TNM1 and TNM2 (p = 0.08) and TNM1 and TNM3 (p = 0.05); (c) Similarly, decreased expression of EPLIN-α was also asso- ciated with lymph node involvement, with highest levels of EPLIN-α being observed in patients with no lymph node involve- ment (N0). Decreased levels were seen in those patients with local lymph node involvement (N1; p = 0.1 vs N0), with lowest levels observed in patients who had advanced lymph node involvement (N2; p = 0.05 vs N0); (d) Significantly reduced EPLIN-α expression was also apparent in patients with local advanced cancers with vessel embolus (p = 0.037). 3.4. EPLIN-α Over-Expression Inhibits Pulmonary Cell Migration Rates An Electric Cell Impedance Sensing (ECIS) method was used to investigate the impact of EPLIN-α over-expres- sion on cell motility (Figure 4). Forced expression of EPLIN-α in SKMES-1 pulmonary cancer cells dramati- cally inhibited monolayer recovery following electrical wounding. Significant differences in recovery (assessed through change in resistance over the electrode as cells migrated to re-cover the electrode) over the 6 hour period were seen between SKMES-1pEF6 control and SK- MES-1EPLIN-α exp cells (p = 0.002) (Figure 4(a)). Treat- ment with the PLCγ inhibitor seemed to h ave little effect on the recov ery rate s of either SK MES-1 pEF6 control cells or SKMES-1EPLIN-α exp cells (Figure 4(b)). In both cases, treatment with PLCγ inhibitor did not significantly im- pact on the migratory rate of either cell line compared to the untreated equivalent (p > 0.05). 4. Discussion Epithelial protein lost in neoplasm is a cytoskeletal asso- ciated protein involved in the regulation of actin dynam- ics and subsequently in cell motility [2,3]. The expres- sion of EPLIN-α has been found to be down-regulated in a number of oral, breast, oesophageal and prostate cancer cell lines compared to their normal counterparts [5-8]. Previous studies from our laboratories have provided data supporting a tumour/metastasis suppressive role for EPLIN-α, where enhanced levels of EPLIN-α can nega- tively impact on key metastatic and angiogenic traits in vitro and in vivo [5,6,13]. This is supported by data from our clinical cohorts indicating that reduced EPLIN-α levels are associated with poor NPI prognosis and lower patient surviv al rates in breast cancer patients [5]. The clinical data obtained in the cu rrent study, appears to be in line with previous observations. Transcript levels of EPLIN-α where found to be significantly reduced in tumour tissues compared to normal tissues. We further analyzed the quantity of EPLIN-α transcript in pulmo- nary cancer samples against the corresponding clinical data. Relatively high levels of EPLIN-α transcript were detected in squamous pulmonary cancer and adenocarci- nomas compared to small cell cancers and other types of  Expression Profile of Epithelial Protein Lost in Neoplasm-Alpha (EPLIN-α) in Human Pulmonary Cancer and Its Impact on SKMES-1 Cells in Vitro 457 Figure 3. Impact of EPLIN-α over-expression on SKMES-1 pulmonary cancer cells. (a) Successful over-expression of EPLIN- α in SKMES-1 cells was seen in cells containing the expression plasmid (SKMES-1EPLIN-α exp) compared to wild type (SKMES-1WT) and plasmid control (SKMES-1pEF6) cells using RT-PCR; (b) Over-expression of EPLIN-α resulted in a de- creased rate of growth, in comparison to the plasmid control line, over a 5 day incubation period (p = 0.05). No significant effect on SKMES-1 invasiveness; (c) Or cell matrix-adhesion; (d) was observed following over-expression of EPLIN-α (p > 0.05). Mean values +/− SEM are shown. Figure 4. Impact of EPLIN-α over-expression on SKMES-1 cell motility. (a) Over-expression of EPLIN-α significantly inhi- bited cell motility in SKMES-1 cells, with significant differences being seen between SKMES-1pEF6 and SKMES-1EPLIN-α exp cells over time (p = 0.002); (b) Inhibition of PLCγ signalling by treatment of cells with a PLCγ inhibitor did not have any sig- nificant effects on the motility rate of either cell line (vs untreated control, p > 0.05). Representative data is shown. Copyright © 2012 SciRes. JCT  Expression Profile of Epithelial Protein Lost in Neoplasm-Alpha (EPLIN-α) in Human Pulmonary Cancer and Its Impact on SKMES-1 Cells in Vitro 458 pulmonary cancers, such as carcinoid. Expression levels of EPLIN-α appeared to associate with the TNM stage of the cancer, with highest levels being seen in the lower TNM1 stage and levels reducing at TNM2 and TNM3 stages with close to significant and significant diffe- rences observed between TNM1 vs TNM2 and TNM1 vs TNM3 respectively. This observation is in line with ob- servations from the clinical breast cohort [5], where lower level of EPLIN-α were seen in the higher TNM stages and similarly indicates that a loss of EPLIN-α ex- pression occurs during the advancement of pulmonary cancer. Decreased expression levels of EPLIN-α were also found to associate with lymphatic involvement. Highest levels of EPLIN-α transcripts were observed in cancers with no lymphatic involvement (N0) and large reductions in EPLIN-α expression, in comparison to no lymphatic involvement, were seen in tissues where local lymph node involvement was apparent (N1), with further reductions in EPLIN-α expression being apparent in the tissues from patients with advanced lymph node in- volvement (N2). A recent study has also highlighted re- duced expression of EPLIN in lymph node metastasis in a number of epithelial cancers, including prostate, colo- rectal, breast and squamous cell carcinoma of the head and neck [11]. The data presented here further suggests that EPLIN-α may be a key contributing factor in deter- mining the likelihood of cancer cells metastasising through the lymphatic route in a variety of human can- cers. Finally, our clinical data indicated that reduced EPLIN-α levels were associated with patients who had local advanced cancers with vessel cancerous embolus. Thus, the clinical results indicate that, similar to other studies on different cancer types, lower levels of EPLIN- α are associated with a more aggressive pulmonary can- cer and an increased likelihood of metastatic spread. To support our clinical findings, we generated a SKMES-1 human lung cancer line over-expressing EP- LIN-α and examined the impact of this over-expression on the cellular functions of this cell line. Forced expres- sion of EPLIN-α brought about a reduction in SKMES-1 cell growth and motility co mpared to control cells. How- ever, over-expression of EPLIN-α in this cell line did not seem to influence SKMES-1 invasiveness. This data is somewhat in line with the established role of EPLIN in motility. Previous studies from our laboratory have also found the over-expression of EPLIN-α to negatively im- pact on the cellular invasiveness and motility of MDA- MB-231 breast cancer cells [5] and negatively affect the motility of HECV endothelial cells [13], though in oeso- phageal cancer cells over-expression of EPLIN-α had a greater influence on cellular invasiveness than motility [7]. Additionally, treatment with a PLCγ inhibitor seemed to have similar effects on both control and EPLIN-α over-expressing cells, suggesting that the role that EPLIN-α plays in cellular motility in this cell line may not be linked to the PLCγ signalling pathway. Thus, our data suggests that, in this lung cancer cell line, EPLIN-α may not play as great a role in cell invasion. The exact reason behind this observation, which is in contrast to other studies, is currently unknown. Further work to cla- rify this observation is required, examining the role of EPLIN-α in other pulmonary cancer cell lines using both over-expression and knockdown studies where possib le. Collectively, the present study suggests that EPLIN-α is inversely associated with the aggressiveness and clini- cal outcome of human pulmonary cancers and can influ- ence in vitro cell growth and migration of lung cancer cells. Further data examining EPLIN-α expression in larger cohorts and also using in vivo metastasis models are required to fully explore the potential importance of EPLIN-α in pulmonary cancer. 5. Acknowledgements Dr Yinan Liu is a recipient of the Albert Hung China Medical Scholarship of Cardiff University. The authors wish to thank Cancer Research Wales for their kind sup- port of this wo rk . REFERENCES [1] Y. Song, R. E. Maul, C. S. Gerbin and D. D. Chang, “In- hibition of Anchorage-Independent Growth of Trans- formed NIH3T3 Cells by Epithelial Protein Lost in Neo- plasm (EPLIN) Requireslocalization of EPLIN to Actin Cytoskeleton,” Molecular Biology of the Cell, Vol. 13, No. 4, 2002, pp. 1408-1416. doi:10.1091/mbc.01-08-0414 [2] R. S. Maul, Y. Song, K. J. Amann, S. C. Gerbin, T. D. Pollard and D. D. Chang, “EPLIN Regulates Actin Dy- namics by Cross-Linking and Stabilizing Filaments,” The Journal of Cell Biology, Vol. 160, No. 3, 2003, pp. 399- 407. doi:10.1083/jcb.200212057 [3] K. Abe and M. Takeichi, “EPLIN Mediates Linkage of the Cadherin-Catenin Complex to F-Actin and Stabilizes the Circumferential Actin Belt,” Proceedings of the Na- tional Academy of Sciences, Vol. 105, No. 1, 2008, pp.13- 19. doi:10.1073/pnas.0710504105 [4] D. D. Chang, N.-H. Park, C. T. Denny, S. F. Nelson and M. Pe, “Characterization of Transformation Related Genes in Oral Cancer Cells,” Oncogene, Vol. 16, No. 15, 1998, pp. 1921-1930. doi:10.1038/sj.onc.1201715 [5] W. G. Jiang, T. A Martin, J. M. Lewis-Russell, A. Doug- las-Jones, L. Ye and R. E. Mansel, “Eplin-Alpha Expres- sion in Human Breast Cancer, the Impact on Cellular Mi- gration and Clinical Outcome,” Molecular Cancer, Vol. 7, 2008, pp. 71-80. doi:10.1186/1476-4598-7-71 [6] A. J. Sanders, T. A. Martin, L. Ye, M. D. Mason and W. G. Jiang, “EPLIN Is a Negative Regulator of Prostate Cancer Growth and Invasion,” Journal of Urology, Vol. Copyright © 2012 SciRes. JCT  Expression Profile of Epithelial Protein Lost in Neoplasm-Alpha (EPLIN-α) in Human Pulmonary Cancer and Its Impact on SKMES-1 Cells in Vitro 459 186, No. 1, 2011, pp. 295-301. doi:10.1016/j.juro.2011.03.038 [7] Y. Liu, A. J. Sanders, L. Zhang and W. G. Jiang, “EPLIN-Alpha Expression in Human Oesophageal Can- cer and Its Impact on Cellular Aggressiveness and Clini- cal Outcome,” Anticancer Research, Vol. 32, No. 4, 2012, pp. 1283-1289. [8] R. S. Maul and D. D. Chang, “EPLIN, Epithelial Protein Lost in Neoplasm,” Oncogene, Vol. 18, No. 54, 1999, pp. 7838-7841. doi:10.1038/sj.onc.1203206 [9] S. Chen, R. S. Maul, H. R. Kim and D. D. Chang, “Char- acterization of the Human EPLIN (Epithelial Protein Lost in Neoplasm) Gene Reveals Distinct Promoters for the Two EPLIN Isoforms,” Gene, Vol. 248, No. 1-2, 2000, pp. 69-76. doi:10.1016/S0378-1119(00)00144-X [10] M. Chircop, V. Oakes, M. E. Graham, M. P. Ma, C. M. Smith, P. J. Robinson and K. K Khanna, “The Actin- Binding and Bundling Protein, EPLIN, Is Required for Cytokinesis,” Cell Cycle, Vol. 8, No. 5, 2009, pp. 757- 764. doi:10.4161/cc.8.5.7878 [11] S. Zhang, X. Wang, A. O. Osunkoya, S. Iqbal, Y. Wang, Z. Chen, S. Muller, S. Josson, I. M. Coleman, P. S. Nel- son, Y. A. Wang, R. Wang, D. M. Shin, F. F. Marshall, O. Kucuk, W. L.Chung, H. E. Zhau and D. Wu, “EPLIN Downregulation Promotes Epithelial-Mesenchymal Tran- sition in Prostate Cancer Cells and Correlates with Clini- cal Lymph Node Metastasis,” Oncogene, Vol. 30, No. 50, 2011, pp. 4941-4952. doi:10.1038/onc.2011.199 [12] M. Y. Han, H. Kosako, T. Watanabe and S. Hattori, “Ex- tracellular Signal-Regulated Kinase/Mitogen-Activated Protein Kinase Regulates Actin Organization and Cell Motility by Phosphorylating the Actin Cross-Linking Protein EPLIN,” Molecular and Cellular Biology, Vol. 27, No. 23, 2007, pp. 8190-8204. doi:10.1128/MCB.00661-07 [13] A. J. Sanders, L. Ye, M. D. Mason and W. G. Jiang, “The Impact of EPLINα (Epithelial Protein Lost in Neoplasm) on Endothelial Cells, Angiogenesis and Tumorigenesis,” Angiogenesis, Vol. 13, No. 4, 2010, pp. 317-326. doi:10.1007/s10456-010-9188-7 Copyright © 2012 SciRes. JCT |

