 Journal of Cancer Therapy, 2012, 3, 343-351 http://dx.doi.org/10.4236/jct.2012.324045 Published Online September 2012 (http://www.SciRP.org/journal/jct) 343 Detection of Gene Dosage in Circulating Free Plasma DNA as Biomarker for Lung Cancer Alba Mayerly Alvarez1, Sandra Janneth Perdomo Lara1,2, Diana M. Palacios3,4, Edward Fabián Carrillo1,5, Luis Gerardo García Herreros3, Fidel Camacho Durán3, Paulina Ojeda León6, Fabio A. Aristizábal1 1Departamento de Farmacia, Facultad de Ciencias, Universidad Nacional de Colombia, Bogotá, Colombia; 2Facultad de Odontología; Unidad de Investigación Básica Oral (UIBO), Universidad el Bosque, Bogotá, Colombia; 3Departamento de Patología, Fundación Santa Fe de Bogotá (FSFB), Bogotá, Colombia; 4Departamento de Patología, Facultad de Medicina, Universidad Nacional de Co- lombia, Bogotá, Colombia; 5Instituto de Investigaciones Biomédicas, Universidad Libre, Cali, Colombia; 6Departamento de Pa- tología, Hospital Santa Clara ESE, Bogotá, Colombia. Email: faaristizabalg@unal.edu.co Received July 10th, 2012; revised August 14th, 2012; accepted August 28th, 2012 ABSTRACT The increase in the number of gene copies at specific loci is a genetic alteration frequently associated with over expres- sion of the related protein in cancer cells. Genes whose dose is consistently augmented in cancer include those in- volved in cell cycle control, proliferation, apoptosis, and angiogenesis among others. In this study, gene dose of onco- genes MYCL1, MYCN, MYC, EGFR, ERBB2 and AKT2 in DNA obtained from lung tissue and blood plasma, of pa- tients with primary lung cancer was evaluated with respect to normal lung tissue and plasma DNA of healthy individu- als, to determine the capacity of these genes to discriminate normal and neoplastic phenotypes. The number of copies of each gene was determined using real-time (2-∆∆CT). The AKT2 oncogene was found to be amplified frequently in plasma DNA from patients (74% of cases). This marker showed a noticeable ability to discriminate normal and neo- plastic phenotypes, with a 76% to 89% probability of correctly recognize a plasma sample provided by a lung cancer patient or a healthy individual. For this reason, this detection could be a very useful tool to supplement the existing di- agnostic methods in pulmonary cancer. Keywords: Lung Cancer; Gene Amplification; Plasma; EGFR Family; MYC Family 1. Introduction Lung cancer is the cancer with the highest incidence and mortality worldwide. According to statistics from Glo- bocan in 2008, 1.38 million people died from lung cancer, corresponding to 18.2% of all cancer deaths [1]. This mortality is due in large part because they are diagnosed in advanced stages of the disease, where treatment strat- egies are limited and the 5-year survival does not exceed 15% [2]. Therefore, one of the current goals of cancer research is to generate diagnostic methods that allow early detection of disease. The first changes experienced by cells towards a neoplastic phenotype are at the mo- lecular level [3], among them gene amplification is asso- ciated with an over expression of the proteins involved. Identifying these changes and molecular markers of can- cer can allow for an early diagnosis and personalized treatments that lead to a better prognosis for survival. After identifying a molecular marker of cancer, it is im- portant for the clinical practice to evaluate biological samples obtained non-invasively. Blood plasma, serum, and sputum have been proposed as suitable biological fluids to assess molecular markers [4], since an increase in the amount of free nucleic acids in these fluids have been detected, and it has been shown that the origin of this DNA is mainly from apoptotic and/or necrotic cells derived from tissue that is undergoing carcinogenic trans- formation [5]. In this study, the amplification status of oncogenes MYC, MYCN, MYCL1, EGFR, ERBB2, and AKT2 was evaluated with respect to the reference gene β-actin, in DNA obtained from plasma and lung tissue of lung cancer patients and healthy volunteers. Then the amplification state of these genes was assessed in order to verify if these genes could differentiate between a nor- mal and cancerous phenotype, and if plasma was a suit- able biological sample to detect molecular tumor charac- teristics. The MYC family genes (MYC, MYCN and MYCL1) encode a number of transcription factors involved in multiple cellular functions, including activating DNA Copyright © 2012 SciRes. JCT  Detection of Gene Dosage in Circulating Free Plasma DNA as Biomarker for Lung Cancer 344 synthesis and cell cycle progression [6]. The genes EGFR, ERBB2, and AKT2 are involved in important signaling pathways related to cellular functions such as proliferation, apoptosis, and angiogenesis [7]. 2. Materials and Methods 2.1. Samples A total of 55 samples of blood plasma from patients di- agnosed with primary lung cancer (PC) treated in the Hospital Santa Clara and Fundación Santa Fe de Bogotá between 2004-2009 were included. Of these 55 samples of plasma, there were 27 paired samples that included both the plasma sample and paraffin-embedded tumor tissue (T) (Table 1). A case-control study was conducted with blood sam- ples, using 55 plasmas of healthy volunteers (PH), non- smokers of the same-sex and age of the cancer patients enrolled in the study. Additionally, 35 samples of normal lung tissue (N), paraffin embedded, supplied by the De- partment of Pathology, Hospital Santa Clara were in- cluded. 2.2. Collection and Processing of Samples Plasma: With the informed consent of the patients, 4 mL of peripheral blood was collected in EDTA tubes. The sample was centrifuged at 358 g for 10 minutes, and the plasma was separated into tubes of 1.5 mL to be further centrifuged at 2700 g for 10 minutes to eliminate conta- mination by lymphocytes. The plasma obtained was stored at –70˚C before DNA extraction. Lung tissue: The cases obtained were reviewed by a pathologist to confirm the diagnosis. In each paraffin block, 3 consecutive histological cuts were performed and then stained with H&E: a histological cut 3 μm thick with delineated areas of interest in both neoplastic and normal lung tissue, and two cuts of 10 μm for the micro- dissection of the tissue. 2.3. DNA Extraction An aliquot of 200 µL of blood plasma or microdissected tissue was incubated at 56˚C in a lysis buffer (50 mM Tris HCL, 2 mM calcium acetate pH 8.0) and proteinase K (1 μg/μL) (BIOLINE) for 16 hours. DNA extraction was performed using the phenol-chloroform-isoamyl alcohol method and precipitated with ethanol. The DNA quality was assessed with conventional PCR, amplifying a 247-bp Alu sequence, with primers and conditions re- ported by Umetani et al. [8] and electrophoresis in a 2% agarose gel. DNA extraction. 2.4. Determination of the Gene Copy Number The number of gene copies in each sample was evaluated with real-time multiplex PCR using Taqman probes. For this, two PCR mixes were optimized and performed, one to amplify EGFR, ERBB2, AKT2 and ACTB (Figure 1), and another one for MYC, MYCN, MYCL1 and ACTB genes (Figure 2). The gene dosage of each oncogene was determined, utilizing the gene ACTB as a reference, with the double-delta CT relative quantification (2-ΔΔCT) method in duplicate assays, using the kit TaqMan® Fast Universal PCR Master Mix and Applied Biosystem 7500 instruments. The calculations were made following the instructions of this instrument [9]. It was confirmed ear- lier that the efficiencies of the amplifications for each gene were similar, using the method published by Ken- neth J. Livak, et al. [10]. The reactions were made in a final volume of 25 μL, containing: 1 × Master Mix, 0.04 μM of each primer and 0.375 μM of each probe. The PCR program used was: 95˚C × 10 minutes, 50 cycles (95˚C × 15 seconds, 61˚C × 1 minute). Table 2 shows the sequence of primers and probes used. A 2-ΔΔCT value greater than 2 was considered gene amplification [11]. Table 1. Sequence of primers and Taqman probes used in the multiplex PCRs. GENE PROBE (5’-3’) RIGHT PRIMER (5’-3’) LEFT PRIMER (5’-3’) ERBB2 (Tamrra) ATCCGTCCGCCTCAGCCTCCCAAA (Hex) GTCTTGAACTCCCCACCTCAG ACAGACGGTACACACTTTTAAAGG EGFR (Fam) AACTAACCGCCGCCAGCACCACC (tamrra) GACCTGGGAGCTGGGAGAAC ACCTGCCTTTTGCCAACGAG AKT2 (Tamrra) ACCACGAGCCACGGAAGCCAGTCA (rox) AGACCTGGGCTGGTGATGTG CAGACTGTGGGACCTTTCTCTC MYC (Tamrra) ACCAGCAGCAGCAGCAGAGCGA (rox) TCTACTGCGACGAGGAGGAG GCAGCAGCTCGAATTTCTTCC MYCN (Tamrra) CGCCGCTTCTCCACAGTGACCACG (hex) AGGAAGATGAAGAGGAAGAAATCGTGACAGCCTTGGTGTTGGAG MYCL1 (Tamrra) ACCTGGAGACACCTGGACACGCCC (tamrra) CCTAAGAGACCTTCAAGCCAGTG CCAGATATGGGGCTCATAACACC ACTB (Tamrra) TTGCCTCCCGCCCGCTCCCG (fam) CCGTCTTCCCCTCCATCGTG GGCTCCTGTGCAGAGAAAGC Copyright © 2012 SciRes. JCT 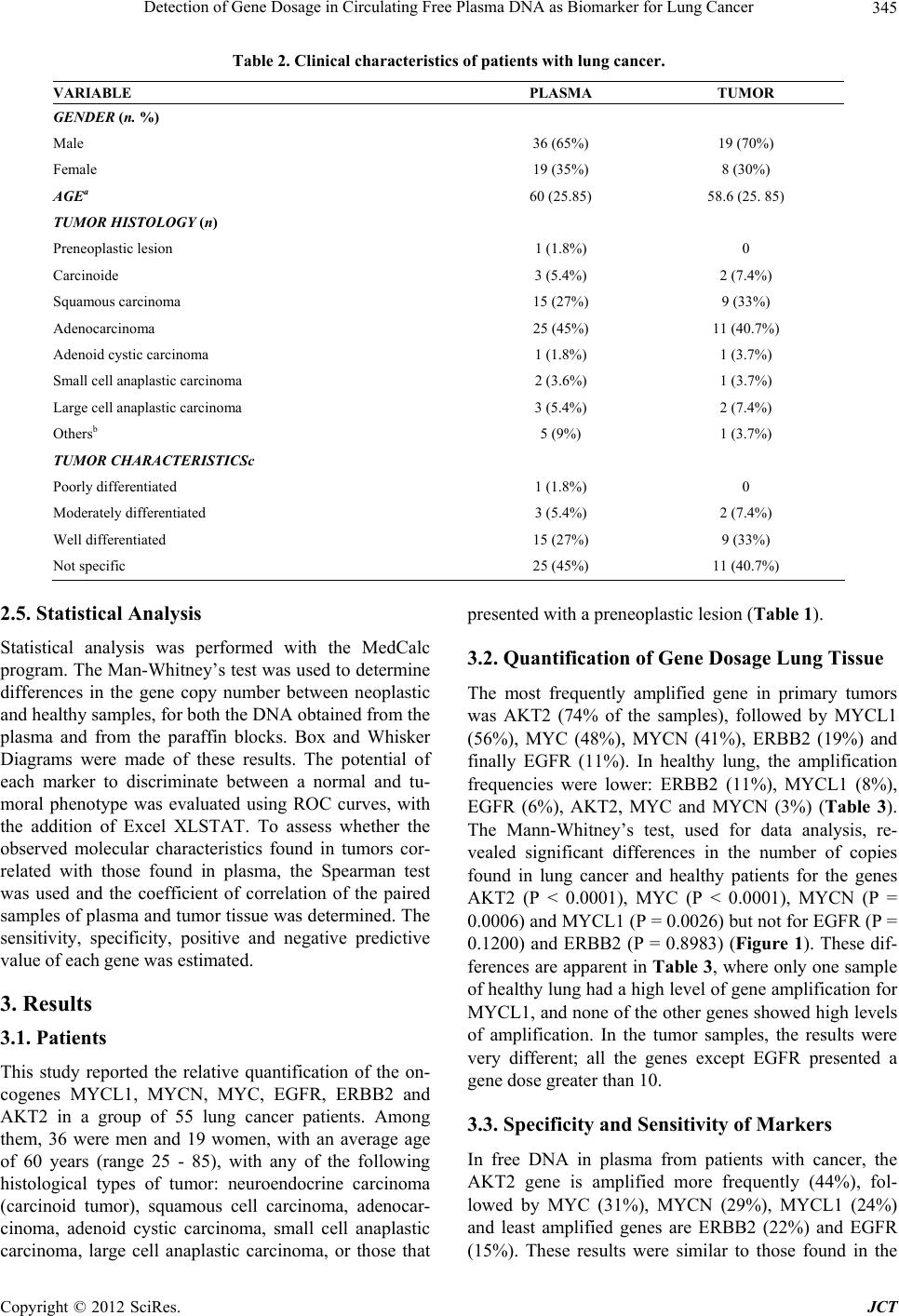 Detection of Gene Dosage in Circulating Free Plasma DNA as Biomarker for Lung Cancer 345 Table 2. Clinical characteristics of patients with lung cancer. VARIABLE PLASMA TUMOR GENDER (n. %) Male 36 (65%) 19 (70%) Female 19 (35%) 8 (30%) AGEa 60 (25.85) 58.6 (25. 85) TUMOR HISTOLOGY (n) Preneoplastic lesion 1 (1.8%) 0 Carcinoide 3 (5.4%) 2 (7.4%) Squamous carcinoma 15 (27%) 9 (33%) Adenocarcinoma 25 (45%) 11 (40.7%) Adenoid cystic carcinoma 1 (1.8%) 1 (3.7%) Small cell anaplastic carcinoma 2 (3.6%) 1 (3.7%) Large cell anaplastic carcinoma 3 (5.4%) 2 (7.4%) Othersb 5 (9%) 1 (3.7%) TUMOR CHARACTERISTICSc Poorly differentiated 1 (1.8%) 0 Moderately differentiated 3 (5.4%) 2 (7.4%) Well differentiated 15 (27%) 9 (33%) Not specific 25 (45%) 11 (40.7%) 2.5. Statistical Analysis Statistical analysis was performed with the MedCalc program. The Man-Whitney’s test was used to determine differences in the gene copy number between neoplastic and healthy samples, for both the DNA obtained from the plasma and from the paraffin blocks. Box and Whisker Diagrams were made of these results. The potential of each marker to discriminate between a normal and tu- moral phenotype was evaluated using ROC curves, with the addition of Excel XLSTAT. To assess whether the observed molecular characteristics found in tumors cor- related with those found in plasma, the Spearman test was used and the coefficient of correlation of the paired samples of plasma and tumor tissue was determined. The sensitivity, specificity, positive and negative predictive value of each gene was estimated. 3. Results 3.1. Patients This study reported the relative quantification of the on- cogenes MYCL1, MYCN, MYC, EGFR, ERBB2 and AKT2 in a group of 55 lung cancer patients. Among them, 36 were men and 19 women, with an average age of 60 years (range 25 - 85), with any of the following histological types of tumor: neuroendocrine carcinoma (carcinoid tumor), squamous cell carcinoma, adenocar- cinoma, adenoid cystic carcinoma, small cell anaplastic carcinoma, large cell anaplastic carcinoma, or those that presented with a preneoplastic lesion (Table 1). 3.2. Quantification of Gene Dosage Lung Tissue The most frequently amplified gene in primary tumors was AKT2 (74% of the samples), followed by MYCL1 (56%), MYC (48%), MYCN (41%), ERBB2 (19%) and finally EGFR (11%). In healthy lung, the amplification frequencies were lower: ERBB2 (11%), MYCL1 (8%), EGFR (6%), AKT2, MYC and MYCN (3%) (Table 3). The Mann-Whitney’s test, used for data analysis, re- vealed significant differences in the number of copies found in lung cancer and healthy patients for the genes AKT2 (P < 0.0001), MYC (P < 0.0001), MYCN (P = 0.0006) and MYCL1 (P = 0.0026) but not for EGFR (P = 0.1200) and ERBB2 (P = 0.8983) (Figure 1). These dif- ferences are apparent in Table 3, where only one sample of healthy lung had a high level of gene amplification for MYCL1, and none of the other genes showed high levels of amplification. In the tumor samples, the results were very different; all the genes except EGFR presented a gene dose greater than 10. 3.3. Specificity and Sensitivity of Markers In free DNA in plasma from patients with cancer, the AKT2 gene is amplified more frequently (44%), fol- lowed by MYC (31%), MYCN (29%), MYCL1 (24%) and least amplified genes are ERBB2 (22%) and EGFR (15%). These results were similar to those found in the Copyright © 2012 SciRes. JCT 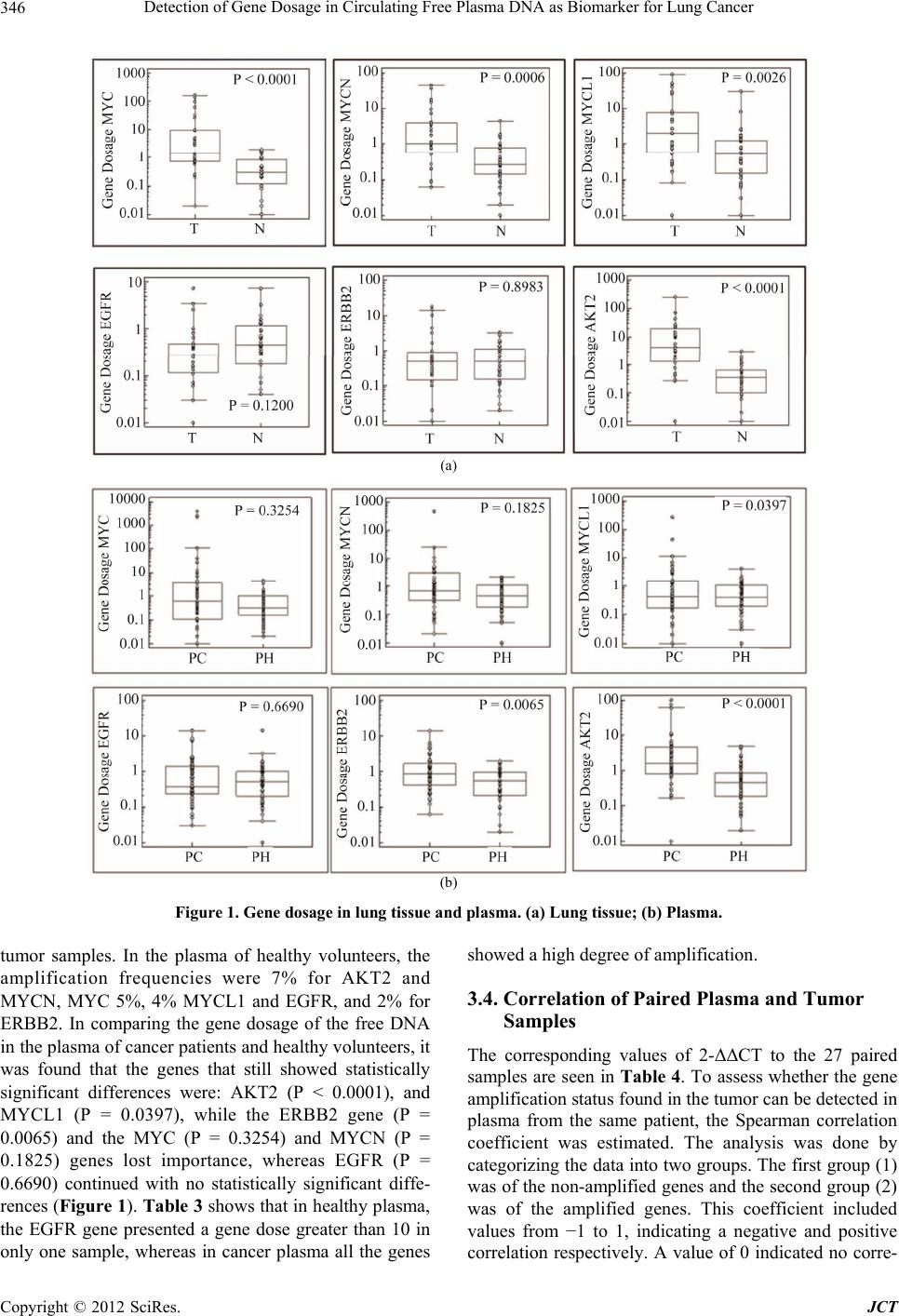 Detection of Gene Dosage in Circulating Free Plasma DNA as Biomarker for Lung Cancer 346 (a) (b) Figure 1. Gene dosage in lung tissue and plasma. (a) Lung tissue; (b) Plasma. tumor samples. In the plasma of healthy volunteers, the amplification frequencies were 7% for AKT2 and MYCN, MYC 5%, 4% MYCL1 and EGFR, and 2% for ERBB2. In comparing the gene dosage of the free DNA in the plasma of cancer patients and healthy volunteers, it was found that the genes that still showed statistically significant differences were: AKT2 (P < 0.0001), and MYCL1 (P = 0.0397), while the ERBB2 gene (P = 0.0065) and the MYC (P = 0.3254) and MYCN (P = 0.1825) genes lost importance, whereas EGFR (P = 0.6690) continued with no statistically significant diffe- rences (Figure 1). Table 3 shows that in healthy plasma, the EGFR gene presented a gene dose greater than 10 in only one sample, whereas in cancer plasma all the genes showed a high degree of amplification. 3.4. Correlation of Paired Plasma and Tumor Samples The corresponding values of 2-ΔΔCT to the 27 paired samples are seen in Table 4. To assess whether the gene amplification status found in the tumor can be detected in plasma from the same patient, the Spearman correlation coefficient was estimated. The analysis was done by categorizing the data into two groups. The first group (1) was of the non-amplified genes and the second group (2) was of the amplified genes. This coefficient included values from −1 to 1, indicating a negative and positive correlation respectively. A value of 0 indicated no corre- Copyright © 2012 SciRes. JCT 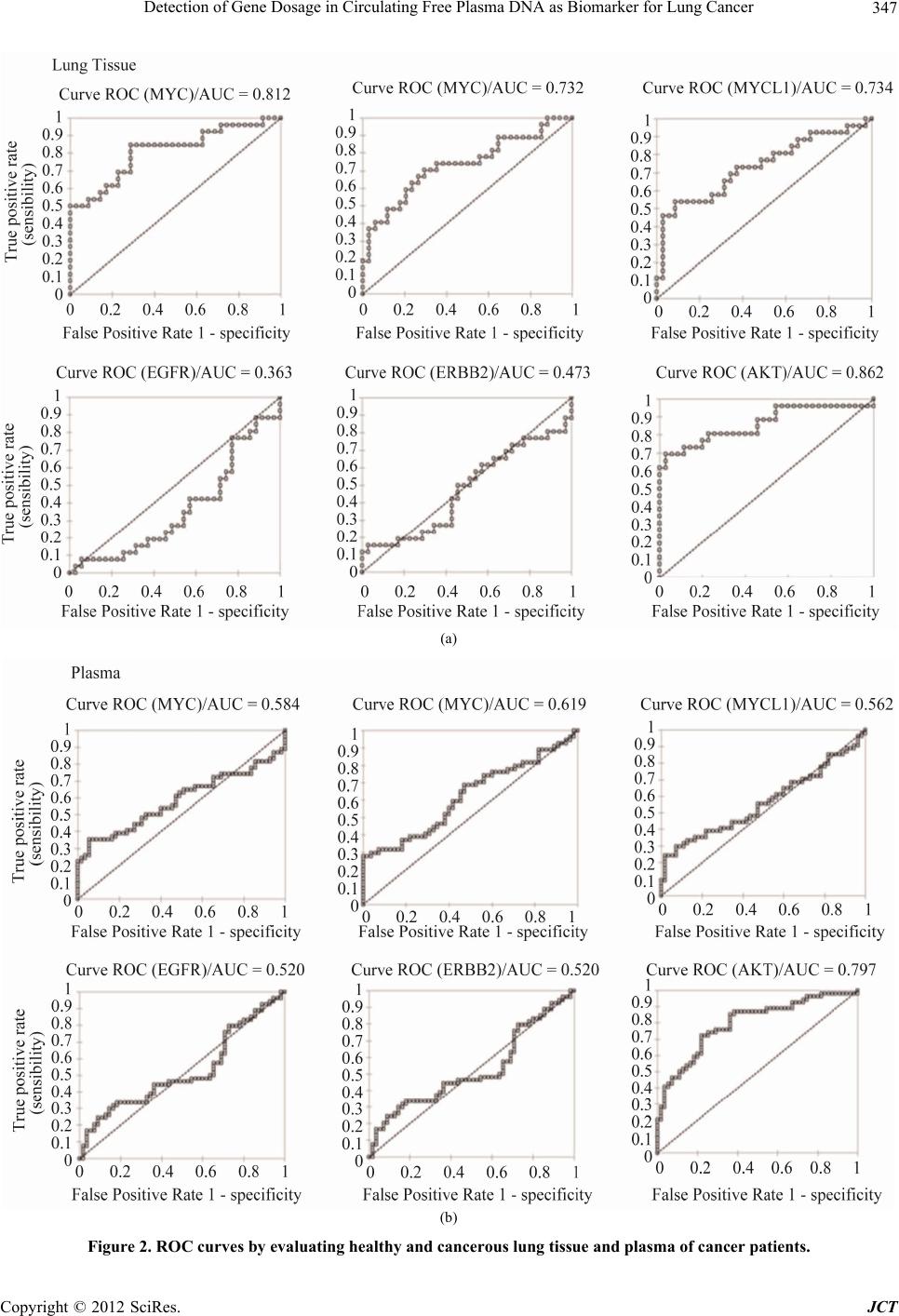 Detection of Gene Dosage in Circulating Free Plasma DNA as Biomarker for Lung Cancer 347 (a) (b) Figure 2. ROC curves by evaluating healthy and cancerous lung tissue and plasma of cancer patients. Copyright © 2012 SciRes. JCT 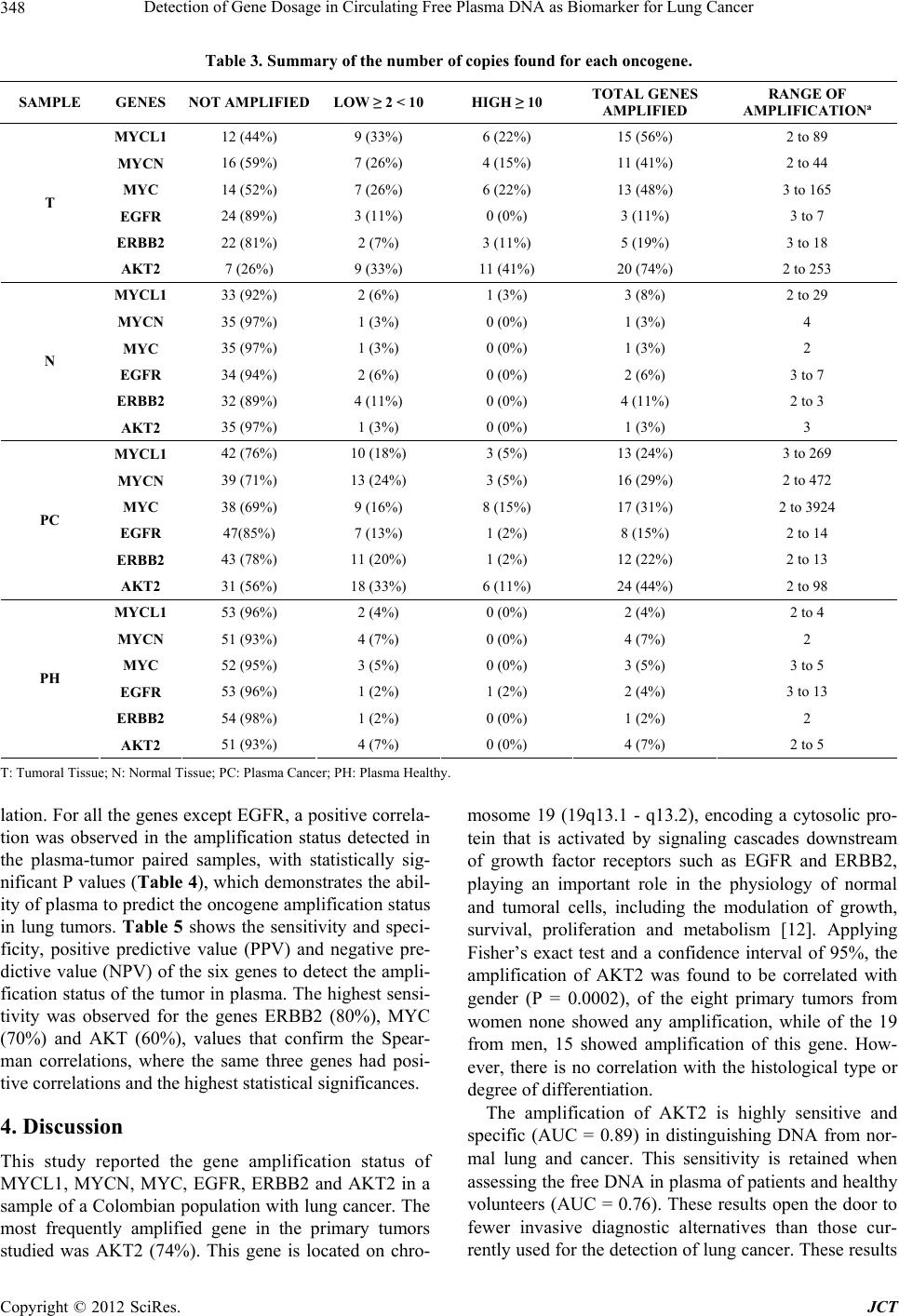 Detection of Gene Dosage in Circulating Free Plasma DNA as Biomarker for Lung Cancer 348 Table 3. Summary of the number of copies found for each oncogene. SAMPLE GENES NOT AMPLIFIED LOW ≥ 2 < 10 HIGH ≥ 10 TOTAL GENES AMPLIFIED RANGE OF AMPLIFICATIONa MYCL1 12 (44%) 9 (33%) 6 (22%) 15 (56%) 2 to 89 MYCN 16 (59%) 7 (26%) 4 (15%) 11 (41%) 2 to 44 MYC 14 (52%) 7 (26%) 6 (22%) 13 (48%) 3 to 165 EGFR 24 (89%) 3 (11%) 0 (0%) 3 (11%) 3 to 7 ERBB2 22 (81%) 2 (7%) 3 (11%) 5 (19%) 3 to 18 T AKT2 7 (26%) 9 (33%) 11 (41%) 20 (74%) 2 to 253 MYCL1 33 (92%) 2 (6%) 1 (3%) 3 (8%) 2 to 29 MYCN 35 (97%) 1 (3%) 0 (0%) 1 (3%) 4 MYC 35 (97%) 1 (3%) 0 (0%) 1 (3%) 2 EGFR 34 (94%) 2 (6%) 0 (0%) 2 (6%) 3 to 7 ERBB2 32 (89%) 4 (11%) 0 (0%) 4 (11%) 2 to 3 N AKT2 35 (97%) 1 (3%) 0 (0%) 1 (3%) 3 MYCL1 42 (76%) 10 (18%) 3 (5%) 13 (24%) 3 to 269 MYCN 39 (71%) 13 (24%) 3 (5%) 16 (29%) 2 to 472 MYC 38 (69%) 9 (16%) 8 (15%) 17 (31%) 2 to 3924 EGFR 47(85%) 7 (13%) 1 (2%) 8 (15%) 2 to 14 ERBB2 43 (78%) 11 (20%) 1 (2%) 12 (22%) 2 to 13 PC AKT2 31 (56%) 18 (33%) 6 (11%) 24 (44%) 2 to 98 MYCL1 53 (96%) 2 (4%) 0 (0%) 2 (4%) 2 to 4 MYCN 51 (93%) 4 (7%) 0 (0%) 4 (7%) 2 MYC 52 (95%) 3 (5%) 0 (0%) 3 (5%) 3 to 5 EGFR 53 (96%) 1 (2%) 1 (2%) 2 (4%) 3 to 13 ERBB2 54 (98%) 1 (2%) 0 (0%) 1 (2%) 2 PH AKT2 51 (93%) 4 (7%) 0 (0%) 4 (7%) 2 to 5 T: Tumoral Tissue; N: Normal Tissue; PC: Plasma Cancer; PH: Plasma Healthy. lation. For all the genes except EGFR, a positive correla- tion was observed in the amplification status detected in the plasma-tumor paired samples, with statistically sig- nificant P values (Table 4), which demonstrates the abil- ity of plasma to predict the oncogene amplification status in lung tumors. Table 5 shows the sensitivity and speci- ficity, positive predictive value (PPV) and negative pre- dictive value (NPV) of the six genes to detect the ampli- fication status of the tumor in plasma. The highest sensi- tivity was observed for the genes ERBB2 (80%), MYC (70%) and AKT (60%), values that confirm the Spear- man correlations, where the same three genes had posi- tive correlations and the highest statistical significances. 4. Discussion This study reported the gene amplification status of MYCL1, MYCN, MYC, EGFR, ERBB2 and AKT2 in a sample of a Colombian population with lung cancer. The most frequently amplified gene in the primary tumors studied was AKT2 (74%). This gene is located on chro- mosome 19 (19q13.1 - q13.2), encoding a cytosolic pro- tein that is activated by signaling cascades downstream of growth factor receptors such as EGFR and ERBB2, playing an important role in the physiology of normal and tumoral cells, including the modulation of growth, survival, proliferation and metabolism [12]. Applying Fisher’s exact test and a confidence interval of 95%, the amplification of AKT2 was found to be correlated with gender (P = 0.0002), of the eight primary tumors from women none showed any amplification, while of the 19 from men, 15 showed amplification of this gene. How- ever, there is no correlation with the histological type or degree of differentiation. The amplification of AKT2 is highly sensitive and specific (AUC = 0.89) in distinguishing DNA from nor- mal lung and cancer. This sensitivity is retained when assessing the free DNA in plasma of patients and healthy volunteers (AUC = 0.76). These results open the door to fewer invasive diagnostic alternatives than those cur- rently used for the detection of lung cancer. These results Copyright © 2012 SciRes. JCT  Detection of Gene Dosage in Circulating Free Plasma DNA as Biomarker for Lung Cancer 349 Table 4. Gene copy number found in the paired samples Tumor/Plasma. MYCL1 MYCN MYC EGFR ERBB2 AKT2 Sample No PC TC PC TC PC TC PC TC PC TC PC TC 1 0.1 39.9 0.3 44.5 0.0 0.9 1.0 3.4 6.1 4.5 1.4 3.3 2 0.0 0.9 0.1 3.7 0.0 0.8 0.1 0.3 1.4 0.7 0.7 4.1 3 0.1 2.0 0.1 9.6 0.0 0.6 1.6 0.5 3.2 3.2 1.2 2.1 4 0.4 0.5 0.8 0.9 0.0 0.0 0.6 0.4 1.3 0.5 1.1 0.4 5 0.0 0.7 0.1 1.8 0.0 0.3 0.0 0.2 0.9 0.1 4.8 10.6 6 0.6 5.0 0.6 1.2 28.5 0.9 2.4 2.6 2.9 14.4 59.5 253.2 7 4.1 29.1 3.6 2.8 27.1 131.5 2.0 0.1 0.8 0.2 5.8 23.2 8 2.9 4.2 3.3 1.0 2.3 32.6 0.3 0.1 0.4 0.6 0.7 0.3 9 0.0 4.2 0.1 4.0 0.0 1.0 1.3 0.2 4.6 0.5 1.8 1.6 10 6.9 7.2 5.0 14.9 12.6 3.1 0.3 7.2 1.2 17.8 5.2 72.7 11 0.0 0.3 0.0 0.2 0.0 2.9 1.8 0.4 5.3 10.1 6.7 17.4 12 3.4 4.2 1.0 0.5 9.6 4.6 0.3 0.1 1.3 0.0 1.2 0.3 13 3.0 8.2 4.8 3.4 0.6 26.7 0.3 0.8 0.2 0.1 0.6 0.7 14 5.3 2.7 7.5 7.3 0.1 0.9 1.8 0.0 3.0 0.0 1.7 0.5 15 0.1 1.2 0.6 2.3 1.5 1.4 0.3 0.3 1.0 1.7 2.4 11.9 16 0.2 26.0 0.1 0.9 3.3 3.6 1.2 0.1 0.2 0.7 3.3 14.6 17 0.3 2.0 0.9 1.0 1.9 0.2 0.1 0.7 0.7 0.7 2.5 10.2 18 0.2 89.2 0.7 36.8 3.5 60.9 0.7 0.0 0.4 0.3 0.8 5.6 19 0.3 0.3 0.1 0.3 0.5 0.7 0.3 0.4 0.1 0.3 1.7 1.2 20 0.3 0.1 0.5 0.2 0.1 0.7 0.5 0.2 0.8 0.0 0.2 0.0 21 0.2 0.2 1.3 0.7 4.6 9.0 2.3 0.3 1.7 0.2 4.6 4.1 22 0.4 50.5 3.1 17.4 2261.19.9 0.3 0.4 0.5 1.0 1.0 3.0 23 0.7 0.8 0.6 0.8 0.4 0.2 0.1 0.1 0.3 0.0 1.3 3.1 24 1.3 1.0 0.4 0.9 1.8 1.3 0.1 1.0 0.4 0.4 3.1 9.8 25 1.4 0.2 0.5 0.1 0.2 5.2 0.4 0.2 1.0 0.2 0.9 22.2 26 1.6 48.7 0.7 0.1 2598.6165.4 0.0 0.0 0.4 0.0 74.0 43.0 27 1.3 0.0 0.8 0.1 3924.199.3 0.2 0.4 0.5 0.6 2.7 27.2 (rho) = 0.478 P = 0.0117 (rho) = 0.463 P = 0.0149 (rho) = 0.710 P < 0.0001 (rho) = 0.250 P = 0.2085 (rho) = 0.663 P = 0.0002 (rho) = 0.529 P = 0.0045 Table 5. Evaluation of plasma to predict the state of amplification of the oncogenes. GENE SENSITIVITY SPECIFICITY PPV NPV MYCL1 40 (20, 64) 100 (71, 100) 100 (100, 100) 57 (36, 78) MYCN 45 (21, 72) 94 (69, 100) 83 (54, 100) 71 (52, 91) MYC 77 (49, 92) 93 (66, 100) 91 (74, 100) 81 (62, 100) EGFR 33 (6, 80) 92 (73, 99) 33 (0, 87) 92 (81, 100) ERBB2 80 (36, 97) 91 (71, 98) 67 (29, 100) 95 (86, 100) AKT 60 (39, 78) 100 (59, 100) 100 (100, 100) 47 (21, 72) PPV = positive predictive value; NPV = negative predictive value. Confidence interval of 95%. Copyright © 2012 SciRes. JCT  Detection of Gene Dosage in Circulating Free Plasma DNA as Biomarker for Lung Cancer 350 can be easily applied in clinical practice, as there are sys- tems in place to quantify the gene copy number such as PCR and FISH, among others, and it is a feature that does not change with time and sample processing, bear- ing in mind that frozen samples from 2004 were studied. However, diagnosing from molecular markers has its limits, such as the small amount of free DNA in the plasma of tumor origin in the early stages of cancer and the failure to provide information about the tumoral loca- tion in the lung, for which there could be a complemen- tary diagnostic method utilizing imaging techniques. An- other limitation is that in different cancers the same bio- logical signaling pathways are affected, regardless the tumor’s location. One example of this phenomenon is amplification of AKT2, reported in lung squamous cell carcinomas [13-15] and other cancer types such as ova- rian [1], pancreatic [2] and is over expressed in colorectal cancer [16,17], and lung cancer [18] demonstrating its importance in neoplastic processes. Therefore the use of AKT2 in the detection of lung cancer could be a com- plementary tool to the diagnostic methods currently be- ing used, such as CT (computed tomography), which allows for a non-invasive localization of the tumor, and more complete diagnosis which can orient the physician in the process of creating a personalized therapy. One study found that AKT2 gene amplification in some cases of pulmonary squamous cell carcinoma, and its over expression has been observed in lung tumor tis- sue but not in healthy lung tissue [13]. Currently thera- pies are being developed and these are directed towards silencing the expression of AKT2 in the NCI-H446 cell line, increasing the chemotherapeutic sensitivity to cis- plastin in an effort to partially reverse cisplatin resis- tance. The genes EGFR and ERBB2 are tyrosine kinase re- ceptors of the cell surface, and activate important signal- ing pathways that regulate processes such as proliferation, apoptosis, and angiogenesis [7]. The heterodimerization of these two receptors causes a potent activation of EGFR. In this study, EGFR and ERBB2 were amplified in 11% and 19% of the tumors, respectively. Commonly reported for EGFR are amplification frequencies of be- tween 11% - 40% and over expression between 40 and 80% [19,20], and for ERBB2, amplification between 5 and 10%, and overexpression rates between 17% - 30%. [12,20,21]. However, these results show that these genes are also found amplified in healthy lung. Scientific lit- erature reports that patients with amplification of these two genes are more responsive to tyrosine kinase inhibi- tors such as Gefitinib and Erlotinib. In this population, a strong association was observed between the state of EGFR and ERBB2 amplification, with a Spearman cor- relation coefficient of 0.74 (P < 0.0001), suggesting that these genes in the Colombian population would not be useful for diagnosis but for genetic screening, and selec- tion of patients to treat with tyrosine kinase inhibitors in which this type of treatment functions efficiently as there is a strong correlation. The other three genes studied MYCL1, MYCN, and MYC, encode transcription factors, are important for proliferation, apoptosis and cell differentiation [22,23]. These genes are amplified in primary tumors by 56%, 41%, and 48% respectively. A correlation has been found between the genes MYCL1 and MYCN (coefficient 0.19 and P = 0.023). Although the three genes have high sen- sitivities differentiating healthy lung and lung cancer (AUC > 0.73), in assessing the free DNA in plasma, this sensitivity decreases, as reflected in AUC values between 0.5 and 0.6. It would be important to evaluate them in other biological samples obtained non-invasively such as sputum. It can be concluded that the detection of AKT2 gene amplification in plasma is highly sensitive and specific in distinguishing DNA from normal lung and cancer in Co- lombian population. Finally, a better articulation of basic research with clinical practice could generate broader results when making correlations with aspects such as tumor stage, metastasis, patient follow-up time, exposure to substances, and family history of cancer. These data are sometimes omitted from the medical history. REFERENCES [1] Applied Biosystems, “Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quan- titative PCR,” 2004, pp. 1-70. [2] J. Bean, C. Brennan, J. Y. Shih, G. Riely, A. Viale, L. Wang, D. Chitale, N. Motoi, J. Szoke, S. Broderick., et al., “MET Amplification Occurs with or without T790M Mutations in EGFR Mutant Lung Tumors with Acquired Resistance to Gefitinib or Erlotinib,” Proceedings of the National Academy of Sciences, Vol. 104, No. 52, 2007, pp. 20932-20937. [3] O. Canoz, M. Ozkan, V. Arsav, O. Er, H. S. Coskun, S. Soyuer and M. Altinbas, “The Role of c-erbB-2 Expres- sion on the Survival of Patients with Small Cell Lung Cancer,” Lung, Vol. 184, No. 5, 2006, pp. 267-272. doi:10.1007/s00408-005-2591-y [4] J. Q. Cheng, A. K. Godwin, A. Bellacosa, T. Taguchi, T. F. Franke, T. C. Hamilton, P. N. Tsichlis and J. R. Testa, “AKT2, a Putative Oncogene Encoding a Member of a Subfamily of Protein-Serine/Threonine Kinases, Is Am- plified in Human Ovarian Carcinomas,” Proceedings of the National Academy of Sciences, Vol. 89, No. 19, 1992, pp. 9267-9271. doi:10.1073/pnas.89.19.9267 [5] J. Q. Cheng, B. Ruggeri, W. M. Klein, G. Sonoda, D. A. Altomare, D. K. Watson and J. R. Testa, “Amplification of AKT2 in Human Pancreatic Cells and Inhibition of AKT2 Expression and Tumorigenicity by Antisense Copyright © 2012 SciRes. JCT  Detection of Gene Dosage in Circulating Free Plasma DNA as Biomarker for Lung Cancer 351 RNA,” Proceedings of the National Academy of Sciences, Vol. 93, No. 8, 1996, pp. 3636-3641. doi:10.1073/pnas.93.8.3636 [6] J. A. Crowell and V. E. Steele, “AKT and the Phosphati- dylinositol 3-Kinase/AKT Pathway: Important Molecular Targets for Lung Cancer Prevention and Treatment,” Journal of The National Cancer Institute, Vol. 95, No. 4, 2003, pp. 252-253. doi:10.1093/jnci/95.4.252 [7] J. Ferlay, H. R. Shin, F. Bray, D. Forman, C. Mathers and D. M. Parkin, “Estimates of Worldwide Burden of Cancer in 2008: GLOBOCAN 2008,” International Journal of Cancer, Vol. 127, No. 12, 2010, pp. 2893-2917. doi:10.1002/ijc.25516 [8] M. E. Garber, O. G. Troyanskaya, K. Schluens, S. Peter- sen, Z. Thaesler, M. Pacyna-Gengelbach, M. van de Rijn, G. D. Rosen, C. M. Perou, R. I. Whyte, et al., “Diversity of Gene Expression in Adenocarcinoma of the Lung,” Proceedings of the National Academy of Sciences, Vol. 98, No. 24, 2001, pp. 13784-13789. doi:10.1073/pnas.241500798 [9] W. C. Gustafson and W. A. Weiss, “Myc Proteins as Therapeutic Targets,” Oncogene, Vol. 29, No. 9, 2010, pp. 1249-1259. doi:10.1038/onc.2009.512 [10] R. Herbst, J. Heymach and S. Lippman, “Molecular Ori- gins of Lung Cancer,” New England Journal of Medicine, Vol. 359, 2008, pp. 1367-1380. doi:10.1056/NEJMra0802714 [11] F. R. Hirsch, M. Varella-Garcia and F. Cappuzzo, “Pre- dictive Value of EGFR and HER2 Overexpression in Ad- Vanced Non-Small-Cell Lung Cancer,” Oncogene, Vol. 28, Suppl. 1, 2009, pp. S32-S37. doi:10.1038/onc.2009.199 [12] C. Huang, L. Yang, Z. Li, J. Yang, J. Zhao, X. Dehui, L. Liu, Q. Wang and T. Song, “Detection of CCND1 Ampli- fication Using Laser Capture Microdissection Coupled with Real-Time Polymerase Chain Reaction in Human Esopha-Geal Squamous Cell Carcinoma,” Cancer Genet- ics and Cytogenetics, Vol. 175, No. 1, 2007, pp. 19-25. doi:10.1016/j.cancergencyto.2007.01.003 [13] X. Lin, A. S. Böhle, P. Dohrmann, I. Leuschner, A. Schulz, B. Kremer and F. Fändrich, “Overexpression of Phosphatidylinositol 3-Kinase in Human Lung Cancer,” Langenbeck’s Archives of Surgery, Vol. 386, No. 4, 2001, pp. 293-301. doi:10.1007/s004230100203 [14] K. Livakand T. D. Schmittgen, “Analisys of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method,” Methods, Vol. 25, No. 4, 2001, pp. 402-408. doi:10.1006/meth.2001.1262 [15] P. P. Massion, W.-L. Kuo, D. Stokoe, et al., “Genomic Copy Number Analysis of Non-Small Cell Lung Cancer Using Array Comparative Genomic Hybridization: Im- plications of the Phosphatidylinositol 3-Kinase Pathway,” Cancer Research, Vol. 62, No. 13, 2002, pp. 3636-3640. [16] L. A. G. Ries, D. Melbert, M. Krapcho, et al., “SEER Cancer Statistics Review, 1975-2005,” National Cancer Institute, Bethesda, 2008. seer.cancer.gov/csr/1975_2005/2008 [17] P. G. Rychahou, J. Kang, P. Gulhati, H. Q. Doan, L. A. Chen, S. Y. Xiao, D. H. Chung and B. M. Evers, “Akt2 Overexpression Plays a Critical Role in the Establishment of Colorectal Cancer Metastasis,” Proceedings of the Na- tional Academy of Sciences, Vol. 105, No. 51, 2008, pp. 20315-30320. doi:10.1073/pnas.0810715105 [18] H. Schwarzenbach, D. Hoon and K. Pantel, “Cell-Free Nucleic Acids as Biomarkers in Cancer Patients,” Nature Reviews Cancer, Vol. 11, 2011, pp. 426-437. doi:10.1038/nrc3066 [19] G. Sozzi, D. Conte, M. Leon, R. Ciricione, L. Roz, C. Ratcliffe, E. Roz, N. Cirenei, M. Bellomi, G. Pelosi, et al., “Quantification of Free Circulating DNA as a Diagnos- tic Marker in Lung Cancer,” Journal of Clinical Oncol- ogy, Vol. 21, No. 21, 2003, pp. 3902-3908. doi:10.1200/JCO.2003.02.006 [20] N. Umetani, J. Kim, S. Hiramatsu, H. A. Reber, O. J. Hines, A. J. Bilchik and D. S. Hoon, “Increased Integrity of Free Circulating DNA in Sera of Patients with Colo- rectal or Periampullary Cancer: Direct Quantitative PCR for ALU Repeats,” Clinical Chemistry, Vol. 52, No. 6, 2006, pp. 1062-1069. doi:10.1373/clinchem.2006.068577 [21] K. Wang, H. Yamamoto, J. R. Chin, Z. Werb and T. H. Vu, “Epidermal Growth Factor Receptor-Deficient Mice Have Delayed Primary Endochondral Ossification Be- cause of Defective Osteoclast Recruitment,” Journal of Biological Chemistry, Vol. 279, No. 51, 2004, pp. 53848- 53856. [22] J. Wang, L. J. Miao, Y. M. Wu, Y. J. Wu and X. C. Wang, “Expression of AKT2, Cyclin d1 and MMP-9 and Their Correlation to Clinicopathologic Features of Non-Small Cell Lung Cancer,” Cancer, Vol. 25, No. 1, 2006, pp. 69- 72. [23] Q. G. Wu, T. T. Cao, Z. Cheng and J. Wang, “Effects of Akt2-siRNA on Chemotherapeutic Sensitivity and Drug Resistance in Human Lung Cancer Cells,” National Me- dical Journal of China, Vol. 91, No. 30, 2011, pp. 2139- 2142. Copyright © 2012 SciRes. JCT
|