American Journal of Plant Sciences
Vol.07 No.01(2016), Article ID:63275,20 pages
10.4236/ajps.2016.71022
Assessment of Genetic Diversity of NIFOR Oil Palm Main Breeding Parent Genotypes Using Microsatellite Markers
Maxwell Nkachukwu Okoye1,2, Michael Ifeanyi Uguru2, Claude Bakoumé3, Rajinder Singh4, Christy Onyebuchukwu Okwuagwu1
1Plant Breeding Division, Nigerian Institute for Oil Palm Research (NIFOR), Benin, Nigeria
2Department of Crop Science, Faculty of Agriculture, University of Nigeria, Nsukka, Nigeria
3Sime Darby Technology Centre, UPM-MTDC Technology Centre III, Serdang, Malaysia
4Advanced Biotechnology and Breeding Centre, Malaysian Palm Oil Board (MPOB), Persiaran Institusi, Kajang, Malaysia

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 10 November 2015; accepted 26 January 2016; published 29 January 2016
ABSTRACT
The genetic diversity among 15 NIFOR breeding parents was assessed using 10 microsatellite markers. A high genetic diversity was observed with a total of 64 alleles including 23 rare alleles or alleles at frequencies less than 0.05. The NIFOR tenera parents recorded the highest number of rare alleles. The average observed heterozygosity and mean gene diversity across all parental groups were 0.6889 and 0.7029, respectively. Higher genetic diversity was detected among the NIFOR dura and tenera parents compared to that of the Deli dura parents in absolute terms. Analysis of molecular variance (AMOVA) showed that 87% of the total variation (p < 0.001) observed was due to differences among parents. Rogers’ genetic distance ranged from 0.2988 to 0.8000 (mean = 0.5570). The dendrogram constructed on the basis of Rogers’ genetic distance clustered the parents in three groups. They generally clustered in heterotic manner rather than by geographic origins. The groupings obtained through PCoA confirmed the results obtained by cluster analysis. The results obtained are strong assets for NIFOR breeding programme.
Keywords:
Allelic Diversity, Elaeis guineensis Jacq, Genetic Distance, Heterosis, Molecular Characterization, NIFOR, Population Genetics, Selection, SSR Markers

1. Introduction
The Nigerian Institute for Oil Palm Research (NIFOR) oil palm Main Breeding Programme (MBP) is a modification of the Reciprocal Recurrent Selection (RRS) proposed by Comstock et al. [1] . One of the most important features of this scheme of selection is the improvement of the hybrid by a gradual assemblage of favourable alleles, together with the maintenance of genetic variability within the base populations of the breeding programme to ensure yield progress from one generation to the other. The success of this breeding method depends on the effectiveness of selecting distinct parents with complementary yield components combined in their offspring whose yields are superior to those of the parents. Accordingly, the value of a breeding population as potential source of new hybrid variety depends on the magnitude of its genetic variability for economic traits. The higher the genetic divergence between parents used in the crossing scheme that produces the population, the higher the genetic variability within the population.
Plant breeders have often postulated that geographical distance is related to genetic distance in selecting parents for high heterosis effect in the hybrids. The NIFOR first selection cycle of the breeding programme had limited genetic diversity because it was based mainly on the early collections from Aba and Calabar natural oil palm groves, in spite of the abundance oil palm germplasm available to the Institute [2] . New oil palm materials were introduced at NIFOR in view of enlarging the genetic base of parents of the second selection cycle. Following the evaluation of the first cycle breeding populations, five Deli dura, eight African dura and 13 tenera oil palms were selected as parents of the second selection cycle of the MBP on the basis of general combining ability [3] [4] . New introductions were selected based on individual and family performance. In selecting the African dura and tenera parents, full advantage was taken of the vast introductions of oil palm germplasm from various groves in the country especially the Ufuma, Umuabi, Igala, Opobo, and the more recent Aba accessions. Two new Deli dura parents were introduced from Ecuador and Malaysia (Ulu Remis x Malaya) to enhance diversity of the NIFOR Deli breeding population and ensure effective selection progress [2] [5] -[7] .
Since RRS is a long-term breeding procedure, the maintenance of genetic variability during the cycles of selection is essential to guarantee sufficient magnitudes of selection responses for subsequent cycles. Besides, oil palm improvement programme in NIFOR and elsewhere aims at producing high quality oil yielding tenera (D × P) planting materials. Therefore, the maintenance of adequate genetic variability among parent populations used for hybrid seed production is critical for maximum utilization of heterosis and sustained genetic progress in the selection cycle [8] .
Studies on the genetic diversity of oil palm breeding populations on the basis of agro-morphological variables have been extensively carried out in NIFOR [2] [9] -[16] . However, genetic diversity evaluation based on agro- morphological information alone is no longer sufficient because of low polymorphism, long juvenile phase, and vulnerability to environmental effects [17] . An additional and objective measure of genetic variation which will permit the monitoring of genetic variability within and between populations, efficient selection of parents to maximize heterosis among populations and sustained genetic progress in the RRS programme is compulsory [18] [19] .
The application of DNA marker technology in the NIFOR oil palm breeding programme would not only reduce the breeding duration but ensure greater precision in the production of planting materials [20] [21] . Molecular markers offer great scope for assessing genetic diversity and relationships among natural or breeding populations because they are impervious to environmental conditions and are detectable in all stages of plant growth and development [22] . Among the likely alternatives, isozymes are not satisfactorily variable due to low polymorphism [23] [24] . Random amplified polymorphic DNA (RAPD) has also been examined [25] , but poor reproducibility of amplification products limits their generalisation in genetic diversity studies [26] . Other more robust molecular markers such as restriction fragment length polymorphic DNA (RFLP) [27] are complex; requiring relatively large amounts of purified and high molecular weight DNA, time-consuming and laborious. Finally, the amplified fragment length polymorphism (AFLP) [28] is a dominant marker which rarely detects heterozygosity and is scored as a presence/absence polymorphism.
Microsatellites or simple sequence repeats (SSRs) are considered as ideal genetic marker for plant genetic and breeding studies because they are characterized by high polymorphism [29] , co-dominant inheritance [30] , reproducibility and abundance throughout the genome [31] . Also, they are readily transferable [32] , and easily assayed using polymerase chain reaction (PCR) and capillary electrophoresis [33] [34] . Furthermore, multiplexing PCR products on single gels also reduces the workload for studies requiring a large number of samples [35] . SSR markers are useful in oil palm population genetics and breeding studies [36] -[41] , varietal identification [42] -[46] , pedigree analysis, genome mapping, and QTL detection for molecular marker-assisted selection [47] -[49] . But to date, studies on the diversity of the current NIFOR oil palm breeding populations have not been carried out using molecular markers. Earlier efforts in this direction have utilized the conventional agro- morphological analysis. This has created a yawning gap in the establishment of a reliable genetic constitution which can only be resolved with molecular marker analysis. The need to fill this gap has prompted the present study aiming at (1) evaluating the efficiency of sixteen microsatellite markers in detecting genetic variation among NIFOR main breeding parental genotypes, (2) estimating genetic similarities among the parents and (3) classifying them on the basis of genetic relationships.
2. Materials and Methods
2.1. Plant Material
Fifteen parent oil palms (4 Deli dura NIFOR, 4 NIFOR dura, and 7 NIFOR tenera) intercrossed in the second cycle of the reciprocal recurrent selection of the NIFOR oil palm breeding programme were sampled at the NIFOR Main Station, Benin City, Nigeria. Fresh leaf tissues were collected from an unopened spear. Detailed information concerning the parent trees visited is presented in Table 1. Each leaf sample was placed in a separate labeled zip lock polyethylene bag in an airtight container with ice chips and temporarily kept in a cold room at −80˚C at the Bioscience Centre, International Institute of Tropical Agriculture (IITA) Ibadan, Nigeria until DNA extraction.
Table 1. NIFOR parental oil palms sampled.
*New introductions, GCA: General combining ability.
2.2. DNA Extraction
Total genomic deoxyribonucleic acid (DNA) was extracted from fresh leaf tissues using the CTAB (Cetyl trimethyl ammonium bromide) DNA isolation protocol of Doyle and Doyle [50] with minor modifications. Approximately 1.0 - 2.0 g of each fresh leaf tissue was used. The concentration and purity of isolated DNA was determined using a Multiskan GO spectrophotometer (Thermo Fisher Scientific Inc., Denver). Optical density (OD) readings were obtained at wavelengths 260 and 280 nm. All DNA samples were stored at −20˚C until microsatellite analysis at the Genomics Unit of Advanced Biotechnology and Breeding Centre (ABBC), Malaysian Palm Oil Board (MPOB) Selangor, Malaysia. The DNA samples were diluted to an optimum concentration of 25 ng/µl by addition of sterile distilled water or appropriate amount of TE (Tris-EDTA) buffer and stored at 4˚C until polymerase chain reaction (PCR) amplification.
2.3. PCR Reaction and Genotyping
A total of 16 microsatellite loci selected for their high polymorphism and reproducibility were tested on 15 NIFOR oil palm main breeding parents. Nine of the sixteen microsatellite markers were developed at the Genomics Unit of ABBC-MPOB [39] [51] and seven at the French Centre de Coopération en Recherche Agro- nomique pour le Développement (CIRAD; Table 2) [52] . Every forward microsatellite marker was M13-tailed for labeling with one of the four florescent dyes i.e. NED, 6-FAM. VIC, and PET at the five-prime end to permit multiplexing of four marker loci during scoring of banding patterns. Differences in dyes’ colours allowed distinguishing loci and corresponding alleles in the data output whose size ranges overlapped one another.
Each polymerase chain reaction contained 2 µl of 25 ng genomic DNA, 6.625 µl MilliQ water, 1 × PCR standard buffer (NEB, USA), 0.2 µl of 10 mM deoxynucleotide triphosphates (dNTPs) (NEB, USA), 0.025 µl of each of the M13-tailed forward primer and untailed reverse primer for every primer pair, 0.025 µl dye, and 0.1µl of Taq DNA polymerase (5 U/µl) (NEB, USA) for a total reaction volume of 10 µl. PCR was performed using Perkin Elmer 9700 thermo-cycler (Life Technologies, Thermo-Fisher Scientific, USA). The PCR programme consisted of an initial 3 min denaturing at 95˚C, followed by 35 cycles of denaturation at 95˚C for 30 sec, primer annealing for 30 sec at 50˚C - 58˚C depending on the primer annealing temperature and an extension temperature of 72˚C for 30 sec, terminated by a final extension at 72˚C for 2 min. The amplification products were resolved on 0.9% of SFR agarose gel run in 1 × TAE buffer. Band sizes were determined by reference to the 100 bp ladder (Thermo-Scientific) run in a horizontal electrophoresis system (Horizon model 20 - 25; Figure 1). The PCR products were stored at 4˚C until scoring of banding patterns.
Figure 1. Banding pattern of SSR marker mEgCIR3519 used for genotyping NIFOR oil palm parents (M = 100 base pair ladder).
Table 2. Microsatellite primer pairs used for population genetic analysis of oil palm.
TA = Annealing temperature, *Probe Unique Identifiers (PUIDs) of NCBI Probe Database, **Accession numbers of NCBI GenBank.
Four PCR products labeled with different fluorescent dyes were pooled in the ratio of 2:1:1:1. 2 µl of the pooled PCR products was combined with 7.84 µl of formamide (Applied Biosystems, Foster City, CA) and 0.16 µl of the GeneScan-500 LIZ size standard (Applied Biosystems, Foster City, CA). The samples were heated at 95˚C for 3 min in a 96-well PCR microplate and placed at 4˚C before loading on ABI 3730 DNA Genetic Analyzer (Applied Biosystems, USA) for an automated capillary electrophoresis.
The allele sizes for each SSR locus were identified using Genemapper 4.1 software (Applied Biosystems, USA). Electropherogram profiles (sample plots) were generated and allele sizes for the set of SSR markers were exported as data table for genotyping (Figure 2).
3. Data Analysis
The oil palm parent samples were subdivided into three groups depending of the fruit form and the provenance of the parental material i.e. one group comprising the NIFOR dura parents, one for NIFOR tenera parents, and one group of Deli dura NIFOR parents. Estimates of genetic diversity were analyzed for each locus as well as each group of oil palm parents. The genetic diversity parameters estimated were allelic frequencies (Fx), number of genotypes, mean number of alleles per locus (Ao), and effective number of alleles per locus (Ae) [53] . The relative allele frequencies (Fx) were calculated according to the method of Marshal and Brown [54] for a better
Figure 2. Electropherogram profiles from ABI 3730 DNA Genetic Analyzer showing allele peaks and sizes of some SSR loci (sMo00102, sMg00179, mEgCIR3519, sMg00016, and sEg00151).
comparison of the distribution of common alleles (alleles at frequency p ≥ 0.05) and rare alleles (alleles at frequency p < 0.05).
The polymorphism information content (PIC) value, that provides an estimate of the discriminatory power of a molecular locus by taking into account the observed number of alleles per locus (Ao) and their relative frequencies (Fx) in the studied population, was calculated for each marker as described by Botstein et al. [55] and Anderson et al. [56] ;
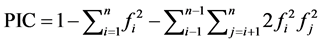
where n is the number of alleles; and fi and fj are the frequencies of the ith and jth alleles, respectively.
Observed or direct heterozygosity (Ho) was estimated by dividing the number of heterozygous individuals by the total number of individuals sampled. Expected heterozygosity (He) is often referred to as gene diversity, and defined as the probability that two randomly chosen alleles from the population are different [57] . It was calculated for each SSR locus according to the formula:
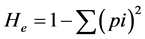
where p is the frequency of the ith allele for the population and 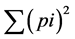 is the sum of squared population allele
is the sum of squared population allele
frequencies. All of the calculations were performed using PowerMarker software version 3.25 [58] . Wright’s Fixation Index (F) which is the overall inbreeding coefficient within the entire population was calculated per locus;
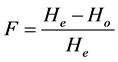
where Ho is the observed heterozygous per locus and He is the expected heterozygous per locus. In order to assess the genetic relationships among parents, Rogers’ [59] genetic distance was computed;

where pij and qij are the frequencies of ith allele at the jth alleles in populations X and Y, respectively, aj the number of alleles at the jth locus, and m the number of loci examined. The distance matrix was subjected to cluster analysis to produce a hierarchical representation of the relationships among parents using the Unweighted Pair Group Method with Arithmetic mean (UPGMA) as described by Sneath and Sokal [60] . All genetic distance calculations and construction of dendrograms were performed using PowerMarker software version 3.25 [58] and MEGA software version 4.0 [61] respectively. To further assess the genetic relationships between the parents, principal coordinate analysis (PCoA) was performed based on dissimilarity matrix using DARwin version 6 [62] . To quantify the extent of population differentiation and distribution of genetic variation in the sampled groups of parents, analysis of molecular variance (AMOVA) was computed using GenAlEx version 6.5 software [63] [64] .
4. Result
4.1. Genetic Diversity in the Oil Palm Parents of the NIFOR Main Breeding Programme
Ten of the 16 SSR loci which generated PCR products were used for the study and the rest dismissed (Table 3). A total of 64 alleles were detected among the 15 NIFOR oil palm parent genotypes. The number of alleles per SSR locus varied from 4 at sMg00016 to 9 at mEgCIR0790 with an average of 6.4 alleles (Table 3). The frequency of the major allele (highest proportion in all alleles) for the oil palm parents varied from 0.23 at sMg00087 to 0.57 at mEgCIR3519 with a mean of 0.44 at each locus. Allele frequencies were low, particularly for loci with higher number of alleles. Rare alleles or alleles at frequency (Fx < 0.05) were observed at all but one of the microsatellite loci (sMg00179) with a total of 23 (35.94%) rare alleles across all loci (Table 4). The highest number of rare alleles (5) was recorded at the SSR loci mEgCIR0790 followed by sMg00156 (4). Rare
Table 3. Microsatellite primer pairs used for population genetic analysis in the NIFOR oil palm main breeding parents.
TA = Annealing temperature, Ao = Number of allele, Ae = Effective number of alleles, MAF = Major allele frequency, Ho = Observed heterozygosity, He = Expected heterozygosity/gene diversity, G = Genotype number, PIC = Polymorphism information content, F = Fixation index(inbreeding-like effects within the entire population). Position of markers in linkage groups of an oil palm genetic map is indicated in column 3.
Table 4. Allele frequencies in the 15 NIFOR oil palm main breeding parents using ten microsatellite markers.
Al = Allele letter.
alleles occurred in all the NIFOR oil palm breeding parents with the exception of Deli dura NIFOR parents (DD2 and DD3). The highest number of rare alleles (14) was detected in the NIFOR tenera parents with a minimum of 1 rare allele (T7) and maximum of 3 rare alleles (T3 and T6). SSR loci mEgCIR0790 exhibited 1 unique allele (unique allele, i.e. present in one parent but not in any other) in one of the NIFOR dura parents (AD4). All the 10 microsatellite loci scored in this study were polymorphic, displaying high values of PIC from 0.4708 to 0.8237 (mean = 0.66) (Table 3).
The number of genotypes obtained per loci ranged from 5 at sEg00154 and sMg00016 to 11 at mEgCIR0790. Observed heterozygosity (Ho) values (per marker) ranged from 0.4667 at mEgCIR3519 to 0.8571 at sMg00179 (mean = 0.6890). He values ranging from 0.5592 at sMg00016 to 0.8430 at sMg00087 (mean = 0.7030).
4.2. Genetic Diversity among Fruit Forms and Provenances of the NIFOR Main Breeding Programme
In order to better understand the contribution of the various parental genotypes to the total genetic diversity of the NIFOR oil palm parents, the 15 oil palm parent trees were divided into three groups’ on the basis of fruit form and provenance of the parental oil palm viz., Deli dura NIFOR (DDN), NIFOR dura (ND) and NIFOR tenera (NT). The average numbers of alleles in DDN, ND, and NT were 3.4, 3.7, and 5.0, respectively (Table 5). The effective number of alleles per group (Ae) varied from 2.739 in Deli dura NIFOR to 3.403 in NIFOR tenera with an overall group average of 3.091. Major allele frequency varied from 0.4829 in the NIFOR tenera parent group to 0.5042 in the NIFOR dura group. Table 6 shows the distribution of common and private alleles among the three NIFOR oil palm parent groups screened with 10 SSR loci. Results showed that the private alleles accounted for the highest proportion in the NIFOR tenera group. Eight private alleles were specific to groups of NIFOR dura parents and two alleles for the Deli dura NIFOR group respectively.
The observed heterozygosity per group ranged from 0.650 (NIFOR Deli dura) to 0.725 (NIFOR dura) with an average value of 0.683 (Table 5). The observed heterozygosity of Deli dura NIFOR and NIFOR dura groups are quite comparable. The NIFOR Deli dura group had the lowest values among all the populations for all the allelic variability parameters in absolute terms. Contrary to this observation, NIFOR dura recorded the least expected heterozygosity value (0.602) among the different groups of parents.
Table 5. Estimates of genetic diversity in the three groups of parents detected by 10 microsatellite markers.
N = Number of samples, MAF = Major allele frequency, Ao = Number of allele, Ae = Effective number of alleles, Ho = Observed heterozygosity, He = Expected heterozygosity/gene diversity, PIC = Polymorphism information content.
Table 6. Distribution of common and private alleles among the 3 groups of NIFOR oil palm parent populations for 10 microsatellite loci.
†Code in parenthesis indicates individual parent(s) with private allele.
4.3. Analysis of Molecular Variance (AMOVA)
The examination of the hierarchical partitioning of genetic variation by AMOVA demonstrated that genetic differentiation was significant at p < 0.001 using the co-dominant allelic distance matrix for calculation of FST [63] -[65] (Table 7). There was a clear genetic differentiation both among and within the groups of parent populations using the significance tests based on 999 permutations calculated according to Wright [65] and Excoffier et al. [66] . The genetic variation was higher within groups with a variance component of 0.418 than among groups (with a variance component of 0.063). Of the total diversity, 13% was attributed to group differences while 87% was attributed to differences within groups. Both the FST index ((0.131) and GST value of 0.136 (p < 0.002) were low.
4.4. Genetic Distance Analysis
Based on the genotypic data obtained at the ten microsatellite loci, Rogers’ genetic distance coefficients were estimated for all pair-wise comparisons of the 15 parent trees. The average distance between parents was moderate (0.5570) and ranged from 0.2988 between AD4 and T8 to 0.8000 between DD4 and AD5 (Table 8). The highest genetic distance among the tenera parents (0.6500) was found between T3 and T5 and the lowest (0.4086) between T2 and T6. The minimum genetic distance within the Deli dura NIFOR parents is 0.4293 between DD1 and DD4 and the maximum distance of 0.6086 between DD2 and DD4, and DD3 and DD4. AD3 and AD4 recorded the highest genetic distance (0.7988) among the NIFOR dura parents.
Table 7. Analysis of molecular variance (AMOVA) among the NIFOR oil palm parents.
Table 8. Rogers’ [59] genetic distance matrix between 15 NIFOR main breeding oil palm parents generated by microsatellite markers.
4.5. Cluster Analysis
The 15 oil palm parents were grouped using Unweighted Pair Group Method with Arithmetic mean (UPGMA) dendogram [60] (Figure 3). Accordingly, the parents were grouped into three major clusters designated I, II and III. The number of parents for each of the three groups varied from 4 to 6. Cluster I contained, in part, NIFOR dura and tenera parents, Cluster II is predominantly made up of Deli dura NIFOR parents (DD1, DD4, and DD2) and two tenera parents (T5 and T1). Cluster III included three tenera parents (T3, T2, and T6) and one Ufuma (NIFOR) dura (AD3) and one Ulu Remis ex Sabah dura (DD3). The two tenera parents (T2 and T6) from Calabar and Umuabi were grouped in the same sub-cluster. In general, the groupings of the parents of NIFOR Main Breeding Programme within the different clusters were not consistent with their assumed genealogies.
Figure 3. Dendogram revealed by the UPGMA cluster analysis of parents of the NIFOR oil palm Main Breeding Programme based on Rogers’ [59] genetic distance.
4.6. Principal Co-Ordinate Analysis
A multidimensional scatter plot (MDS) was used to further understand the genetic relationships among parents of the NIFOR oil palm breeding programme. Principal co-ordinate analysis (PCoA) clustered the 15 parents into three groups (Figure 4). The first three coordinates explained 64.43% of the total variation, with 38.51% explained by the first coordinate and 14.61% by the second coordinate. Parents generally clustered in heterotic manner rather than by geographic origins. The grouping obtained through PCoA confirmed the results obtained by UPGMA cluster analysis.
5. Discussion
5.1. Allelic Frequencies
Allelic frequencies varied at each locus at the individual parent’s level as well as among the three distinct parental groups. The unequal distribution of allele frequencies among the parents could be due to drift and selection. However, more than 40% of the individual parents and their groups shared a common major allele (alleles shared between many parents) at any given locus. Rare alleles (p < 0.05) were found in NIFOR dura and NIFOR tenera but not in Deli dura NIFOR (ex Serdang Ave. NIFOR x IRHO―Pobe and Ulu Remis Deli x ex Sabah). The absence of rare alleles in the Deli dura could be explained by several generations of breeding resulting to fixation of some alleles. Meanwhile, it can be presumed that NIFOR dura and NIFOR tenera parents which have barely undergone two cycles of selection are not too genetically different from their wild relatives. They have experienced little loss and negligible fixation of alleles. It is also possible that the provenance specific alleles were derived from a mutation event since SSRs loci are known to have a high rate of mutation per locus per generation of 25 × 10−5 to 1 × 10−2 [67] . The prevalence of private alleles in the NIFOR dura and tenera parents may be due to the low number of samples used in this study. In general, only elite palms which are usually in very small number are introduced in a breeding programme. It is the case of the 5 oil palms including Ufuma dura (AD3) and tenera (T5), Aba dura (AD5), Opobo dura (AD4), and Umuabi tenera (T6) recently introduced in the NIFOR Main Breeding Programme. However, the speculation of adaptive genetic variants as
Figure 4. Principal coordinates analysis (PCoA) in the NIFOR main breeding parents based on the dissimilarity matrix using 10 SSR markers.
proposed by Zeng et al. [68] cannot be precluded because the geographical origin of the palms, ecology, agro- climatic conditions, pedology, and ethnical behaviour of local farmers could justify the presence of private alleles. All the parents are derived from palms originally selected on account of their high yields or good fruit composition from small oil palm groves at Aba, Calabar, Ufuma, Umuabi, and Opobo, which form part of a very large contiguous population constituting the oil palm belt of Nigeria. The highest number of private alleles occurred in the tenera parents from Umuabi. This location is derived savannah ecology and generally regarded as marginal for oil palm production with rainfall of about <2000 mm per annum. The oil palm occurs in isolated but dense groves in valleys and conical hills and ridges which cover the entire region. The private alleles found in such a marginal environment may have an adaptive value. The inhabitants of this area, as in all other areas of the Nigerian oil palm belt, live in homesteads within the groves. The palms are characterized by slow stem increment, high bunch yield, and palm wine (alcoholic beverage from oil palm sap) production.
Two parents (AD3 and T5) were selected from Ufuma, representing the heart of the eastern oil palm belt of Nigeria with annual rainfall of about 2000 to 2500 mm. This area is characterized by an unusually high proportion of tenera (thin-shelled) palms even before the inheritance of the fruit forms was fully understood. The immediate cause of this could not be ascertained but ethnical behaviour of the farmers/grove owners as regards method of seed selection/exchange with preference to high mesocarp to fruit and oil to bunch ratio may possibly explain this occurrence (personal interview of local farmers, 2015). The high palm oil production from this region seems to support this claim. The Aba dura parent (AD5) selection was carried out on an impoverished soil which is typical of the main oil palm belt of Nigeria. Selection of high bunch yielding palms in such condition held out hopes of breeding palms capable of high productivity under such conditions. Similar instances were revealed using isozymes [69] and microsatellite markers [38] , which were attributed to adaptive genetic variants.
5.2. Polymorphism Information Content
In recognition of the relative superiority of SSRs in detecting DNA polymorphism [70] , the discriminative power of each SSR marker was assessed by calculating polymorphic information content (PIC). A PIC value of greater than 0.7 is considered to be highly informative, whereas a value of 0.44 is considered to be moderately informative. In the present study the result showed a high average PIC value (mean = 0.66) in all the tested loci. The result further revealed that with the exception of locus sMg00016 (PIC = 0.47); sMg00087 (PIC = 0.82) would be best in screening NIFOR oil palm genotypes followed by sMo00102 (PIC = 0.77), and sMg00179 (PIC = 0.76). Accordingly, the PIC value indicates that about nine of these loci were informative and capable of discriminating between genotypes. Very similar results were obtained in a study that involved the analysis of 8 microsatellite loci in a population of 48 parent trees of oil palm (E. guineensis) from the breeding plantation of Univanich Palm Oil Public Company Ltd. where PIC ranged from 0.580 to 0.821 [71] , confirming that oil palm microsatellites are very informative. Arias et al. [40] also reported a maximum PIC value of 0.822 in a comparative study of 189 oil palm materials produced by different commercial companies using 17 SSR markers. The result of the present study is in agreement with earlier reports from Billotte et al. [52] and Singh et al. [39] on the high polymorphism of CIRAD and MPOB oil palm microsatellite markers used in this study. These highly polymorphic set of microsatellite marker pairs has been used for genetic fingerprinting in breeding programmes.
5.3. Mean Number and Effective Number of Alleles per Locus
Microsatellite markers are multi-allelic and co-dominant, hence their relative superiority in detecting DNA polymorphism. A comparison of the results obtained in the present study (4 - 9 alleles per locus with a mean of 6.4 alleles/locus) with those published earlier indicates that the average number of alleles per locus was relatively higher than those earlier reported for two parents (LM2T and LM10T) of BRT10 first selection cycle oil palm population with an estimated average of 1.75 alleles/locus [72] . It was also higher than that reported for improved Nigerian germplasm samples of NIFOR origin and some survey materials from Ayangba and Bida (5.4 and 5.3, respectively) maintained at the Centre National de la Recherche Agronomique (CNRA) in Côte d’Ivoire [8] and 9 D x P oil palm crosses from different Colombian commercial companies (4.5 alleles per locus) using 16 SSR loci [40] . However, the average alleles per locus in the present study is lower compared to the 8 alleles per locus reported by Thongthawee et al. [71] among 132 parent trees from the Thai oil palm breeding programme carried out at Univanich Palm Oil Public Company Ltd., using 8 microsatellite loci.
Some of this variability could be explained by differences in the numbers of genotypes involved, coupled with the number of SSR markers used. The number of alleles per locus is affected by the number of markers and sample size analyzed. Singh et al. [39] previously reported lower number of alleles per locus (2.2 - 3.2) in E. guineensis germplasm using a smaller set of SSRs and the number of alleles increased (2.8 - 3.9) when Ting et al. [51] employed a larger set of SSRs to evaluate a wider pool of oil palm germplasm. In fact, Augustina et al. [73] reported a total of 163 alleles (mean = 8.2) among the 85 pisifera accessions from germplasm collections of different origins at Sampoerna Agro Tbk (SA) in Indonesia. Bakoumé et al. [37] detected a total of 209 alleles, with a mean number of 13.1 alleles per locus in a sample of 494 oil palm derived from 10 African countries. Cochard et al. [8] obtained a total of 202 alleles, with an average of 14.5 alleles per locus within 318 oil palm samples from eight countries. The low number of alleles in the present study suggests the small size of materials used, i.e., few parents of a breeding programme and limited number of descendants for their inter-crosses. This hybridization and selection tend to affect the population and decrease allele variability. Several studies have shown a general tendency of loss of diversity after several cycles of selection. In a related study, Bakoumé et al. [37] investigated the allelic diversity of 3 breeding materials, using 16 microsatellite markers and observed that rare alleles were common alleles in wild oil palm populations showing a reduction due to many years of selection in the materials.
5.4. Observed Heterozygosity and Expected Heterozygosity in the Parents of the NIFOR Oil Palm Main Breeding Programme
The genetic diversity of NIFOR oil palm main breeding parents studied was relatively high (He = 0.703). This result is consistent with studies of genetic variation in oil palm using RAPD [74] , AFLP [28] , isozyme, [69] , and RFLP [27] . However, the extent of the gene diversity of the studied NIFOR oil palm parents (He = 0.703) was lower than those reported by Bakoumé et al. [38] for five natural populations of oil palm from Nigeria (Umuabi = 0.736; Ibono = 0.712; Vabiti = 0.803; Ogbalato = 0.751; and Ologbo = 0.739), respectively. On the other hand, the present result is slightly higher than the average He value detected in the Nigerian improved germplasm (NIFOR = 0.696; Ayangba = 0.697; and Ahoada = 0.704) materials reported by Cochard et al. [8] using 14 SSRs. Nonetheless, comparisons were not on the same basis as the origin and number of samples was different coupled with the number of SSRs assayed. Similarly, the observed heterozygosity (Ho = 0.683) was comparable to what was reported by Cochard et al. [8] for oil palm accessions from Ahoada (Ho = 0.685) and a bit higher than in Ayangba (Ho = 0.673), but much higher than the results (0.516 to 0.551) of Bakoumé et al. [38] for the five natural oil palm populations from Nigeria. Although there were few significant departures from HWE, the observed heterozygosity was lower than the expected heterozygosity. In congruence with this study, Nybom [75] compiled 79 microsatellite based studies and found that grand means for Ho was lower than He in 64 of those studies. Similarly, most of the genetic diversity studies in oil palm using SSRs supported this finding [8] [38] [51] [73] [76] [77] . Fixation index values close to zero are expected under random mating and according to Bruford et al. [78] , substantial positive values as recorded in this study may be due to presence of undetected null alleles or perhaps allele dropout which show a high deficit of heterozygosity at loci sMo00102.
5.5. Observed Heterozygosity and Expected Heterozygosity in the Three Groups of NIFOR Oil Palm Main Breeding Parents
The high genetic diversity observed among the groups of parents suggests the broad genetic base of the parents which was expanded beyond the early Calabar and Aba groves selections of the NIFOR 1st selection cycle parents (He = 0.602 for NIFOR dura and He = 0.650 for NIFOR tenera). The dura and tenera parents available at NIFOR are mainly from the early Aba and Calabar groves. However, full advantage was taken of the vast introductions of oil palm germplasm from various groves in the country especially the Ufuma and more recent Aba introductions, the coastal (Opobo) and hinterland (Umuabi) introductions, including the very valuable Angola introductions to ensure a wide genetic variability and genetic gain expected from breeding projects. In contrast to the NIFOR dura and tenera parents, the introgression among Deli from different origins and introduction of new Deli parents from Ecuador and Ulu Remis ex Sabah may have enhanced the diversity of the Deli dura NIFOR breeding population. In the present study, the expected heterozygosity value (He = 0.622) in the Deli dura NIFOR was high and might have been due to the out-crossing behaviour of oil palm. High genetic diversity implies a high amount of additive genetic variance on which breeding can still capitalize although Deli dura NIFOR has gone several selection cycles. Different sources of Deli dura corresponds to different populations of Deli dura. Every oil palm research station in the world developed Deli dura using its own selection preferences leading to favouring different alleles and different allele combinations. The acquisition of Deli dura from different sources has enriched the Deli dura NIFOR materials with different alleles and different allele combinations. When compared to the previous report of Cochard et al. (2009), lower values of expected heterozygosity (He = 0.373) for Deli NIFOR and (He = 0.510) for 6 Deli dura populations from different origins using 14 SSR markers. Also, Ting et al. [51] revealed even lower values (He = 0.340) for Deli dura via 15 EST-SSRs. Recently, Bakoume et al. [38] reported Ho values (0.310 and 0.211) and He values (0.549 and 0.559) for Deli dura MPOB (Malaysia) and Deli dura Dabou (Côte d’Ivoire) with 16 microsatellite markers. These figures are lower than the reported result in this study. This low genetic diversity of the Deli breeding population reinforces the very narrow genetic base of the Deli materials having been selected from four palms introduced in Bogor (Indonesia) in 1848.
5.6. Fixation Index
In the present study, fixation index (F) was estimated using the F-statistics of Wright [65] . It is a measure of the excess or deficit of heterozygotes in the entire parental materials due to non random mating. In general, F value for the entire set of parental genotypes was low but positive (F = 0.0071) indicating a deficit of heterozygotes. Selfing and intra-groups crosses performed during the breeding programme could have ineluctably led to increased homozygosity in both the dura group and tenera/pisifera group (Bakoumé 2015, personal communication).
5.7. Genetic Differentiation among the Three Groups of Parental Genotypes
The analysis of molecular variance (AMOVA) shows that most of the variation in the NIFOR oil palm main breeding parents lies within populations, a result compatible with those from AFLP, Isozymes, and SSR studies involving oil palm germplasm collections and out-breeding plant species [8] [28] [69] [79] . Consequent upon partitioning of genetic diversity, there is a considerable genetic diversity within populations that may be exploited in population improvement programme. The FST value for the entire parent population was 0.131. According to Wright [80] and Hartl and Clark [81] , a genetic differentiation (FST = 0.05 − 0.15) is moderate. Therefore, we consider that the overall FST value found in this study (FST = 0.131, P = 0.001), indicates significant differences among groups of parents.
5.8. Genetic Relatedness among Parents
SSR genetic distance refers to the genetic divergence among populations, which was measured with Rogers’ dissimilarity coefficient. It is a function of the coefficient of co-ancestry and therefore, suitable for the uncovering of pedigree relationships among operational taxonomic units such as the detection of essentially derived varieties in plant breeding or the identification of duplicates and collection gaps in seed banks [82] . The mean genetic distances among NIFOR oil palm main breeding parents was moderate (DR = 0.5570) indicating a considerable degree of relatedness which underpins low genetic diversity within the populations. Lower genetic distance values (0.050 to 0.573) were reported for 19 MPOB oil palm parent trees using 9 SSR markers [43] . Information on the genetic distance among individuals in a breeding population is an important tool for breeders aiming at exploiting heterosis effect on which optimum oil palm growth and fresh fruit bunch yields depend.
The purpose of clustering analysis is to group together individuals that are similar and provide a picture of the overall relationship among the individuals sampled. Microsatellite markers have been reported to be useful in assigning inbred lines into known heterotic groups, especially for those accessions without clear and accurate pedigree records [83] [84] . In this study, parents of NIFOR oil palm Main Breeding Programme are genetically close to each other. Grouping of the parents revealed by the present analysis did not agree with the pedigree of parents, such as the grouping of Serdang Avenue ex Pobe dura (DD2) and Ufuma x Aba tenera (T1) parents, and Ufuma dura (AD3) and Ulu Remis dura (DD3) parents in clusters II and III respectively. Both clusters comprise parents selected on the basis of good combining ability for bunch yield and, good fruit and bunch composition respectively. Clusters were rather representative of the parent’s heterotic groups. The UPGMA dendogram grouping of the parents from different origin/pedigree in the same cluster indicate lack of correlation between origin/pedigree and genetic distance among the parents. Similar result was also observed in maize [84] -[86] , which revealed that grouping of inbreds based on molecular data do not always concur with the available pedigree information. This may be due to several possible factors [84] in addition to the fact that DNA markers may be affected by selection, drift and mutation. The clustering process and the method selected may also result in incongruities [83]. This was demonstrated where in some clusters, an inbred line that is related to two other inbred lines may fall in one of two separate clusters [85] .
Information about the relationship among breeding materials and the inherent genetic variation is important in making choices of parents in breeding programmes. This is significant in hybrid breeding where detection and utilization of heterotic patterns between different sources are important for genetic success. The principal co- ordinate analysis (PCoA) based on genetic distance estimates determined by SSR data for the 15 oil palm parents, provided a distinct separation of parents from different origin into heterotic groups. The tenera parent, T8 (Ufuma x Angola) was separated from the rest of the parents. Similar results were recorded in the earlier mentioned studies.
6. Conclusion
Results of this first evaluation of the genetic diversity of the current NIFOR oil palm main breeding parents demonstrated the presence of high genetic variation within and between the NIFOR main breeding groups of oil palm parents. NIFOR tenera parent genotypes were more diverse than the NIFOR dura and Deli dura NIFOR. The ten microsatellite makers independently distinguished an average of 8 genotypes out of the 15 genotypes evaluated in this study. Hence, further analysis using large numbers of microsatellite markers and samples is proposed. Moreover, involving more SSR markers for genotyping is expected to group the parental genotypes in a better manner. The high PIC value indicated that the microsatellite markers were preferentially valuable for genetic diversity analysis. Loci mEgCIR0790, mEgCIR0793, sMg00087, sMg00154 and sMo00102 were the most efficient microsatellite markers in detecting genetic variation among parent genotypes. The lowest pair- wise genetic dissimilarity coefficient was recorded between parent genotypes AD4 and T8. As a result, these parent genotypes could be used to study the correlation between genetic distance and heterosis in the NIFOR oil palm Main Breeding Programme. Generally, results of this study would be helpful to design crosses among these parents for future breeding and selection programme.
Acknowledgements
Senior author is grateful to the Director-General of Malaysian Palm Oil Board (MPOB) for permission to undergo laboratory attachment for microsatellite genetic analyses at the Advanced Biotechnology and Breeding Centre (ABBC) Selangor, Malaysia. We acknowledge with thanks the Genomics Unit group and the entire ABBC staff for giving best of knowledge and resources in the course of this research. The assistance of Mrs. Maryann Okoye, Messrs. Innocent Ani, Godfrey Akpan, Hugh Okoye, and Godspower Oviawe in field work is also acknowledged.
Cite this paper
Maxwell N.Okoye,Michael I.Uguru,ClaudeBakoumé,RajinderSingh,Christy O.Okwuagwu, (2016) Assessment of Genetic Diversity of NIFOR Oil Palm Main Breeding Parent Genotypes Using Microsatellite Markers. American Journal of Plant Sciences,07,218-237. doi: 10.4236/ajps.2016.71022
References
- 1. Comstock, R.E., Robinson, H.F. and Harvey, P.H. (1949) A Breeding Procedure Designed to Make Maximum Use of Both General and Specific Combining Ability. Journal of the American Society of Agronomy, 41, 360-367.
http://dx.doi.org/10.2134/agronj1949.00021962004100080006x - 2. Okwuagwu, C.O. (1986) The Genetic Basis of the NIFOR Oil Palm Breeding Programme. Proceedings of the International Workshop on Oil Palm Germplasm and Utilization, Selangor, 26-27 March 1986, 228-237.
- 3. West, M.J. (1976) The Analysis of the Bunch Yield Data of the NIFOR Oil Palm Breeding Programme and the Choice of New Parental Material. A Supplementary Report of the Ministry of Overseas Development on Research Project, R2354, Mimeo, 41.
- 4. West, M.J., Ross, J.M., Obasola, C.O. and Mekako, H.U. (1976) The Inheritance of Yield and of Fruit and Bunch Composition Characters in the Oil Palm—An Analysis of the NIFOR Main Breeding Programme. Proceedings of the Malaysian International Agricultural Oil Palm Conference, Kuala Lumpur, 14-17 June 1976, 1-11.
- 5. Okwuagwu, C.O. (1989) The Developments from the Comprehensive Oil Palm Breeding Programme of the Nigerian Institute for Oil Palm Research (NIFOR) and Future Breeding Strategy. Proceedings of the NIFOR International Conference on Palms and Palm Products, 1, 10-24.
- 6. Okwuagwu, C.O. and Ataga, C.D. (1992) Oil Palm Breeding Programme in Nigeria. Proceedings of the ISOPB International Symposium on the Science of Oil Palm Breeding, Montpellier, 1-3 July 1992, 131-140.
- 7. Okwuagwu, C.O. and Okoye, M.N. (2006) Developments from the NIFOR Second Cycle Oil Palm Main Breeding and Selection Programme—The Plan of Third Cycle Programme. NIFOR Seminar Presentation, June 2006.
- 8. Cochard, B., Adon, B., Rekima, S., Billotte, N., Desmier, R., Koutou, A., et al. (2009) Geographic and Genetic Structure of African Oil Palm Diversity Suggests New Approaches to Breeding. Tree Genetics & Genomes, 5, 493-504.
http://dx.doi.org/10.1007/s11295-009-0203-3 - 9. NIFOR (1974) The Genepool. Nigerian Institute for Oil Palm Research, Annual Report No. 10, 36-40.
- 10. Obasola, C.O., Arasu, N.T. and Rajanaidu, N. (1980) Collection of Oil Palm Genetic Material in Nigeria. I. Method of Collection. MARDI Report No. 80.
- 11. Okwuagwu, C.O., Ataga, C.D. and Okolo, E.C. (1993) Collecting Oil Palm Germplasm in Nigeria. FAO/IBPGR Plant Genetic Resources Newsletter, 93, 38-39.
- 12. Ataga, C.D., Okwuagwu, C.O., Okolo, E.C. and Okoye, M. (2003) Interrelationships among Vegetative, Bunch and Fruit Traits in an Oil Palm Germplasm Collection. In: Rajanaidu, N., et al., Eds., Proceedings of the 2003 PIPOC International Palm Oil Congress, Malaysian Palm Oil Board, Kuala Lumpur, A-PA.
- 13. Okwuagwu, C.O. and Okoye, M.N. (2003) The Second Cycle of the NIFOR Oil Palm Main Breeding Programme. Technical Report, Nigerian Institute for Oil Palm Research (NIFOR), Benin City.
- 14. Okoye, M.N. (2008) Stability and Genetic Variability of Bunch Yield Components of NIFOR Second Cycle Oil Palm Hybrids. Master’s Thesis, University of Nigeria, Nsukka.
- 15. Okwuagwu, C.O., Okoye, M.N., Okolo, E.C., Ataga, C.D. and Uguru, M.I. (2008) Genetic Variability of Fresh Fruit Bunch Yield in Deli/dura x tenera Breeding Populations of Oil Palm (Elaeis guineensis Jacq.) in Nigeria. Journal of Tropical Agriculture, 46, 40-45.
- 16. Okwuagwu, C.O., Ataga, C.D., Okoye, M.N. and Okolo, E.C. (2011) Germplasm Collection of Highland Palms of Afikpo in Eastern Nigeria. Bayero Journal of Pure and Applied Sciences, 4, 112-114.
http://dx.doi.org/10.4314/bajopas.v4i1.25 - 17. Smith, J.S.C. and Smith, O.S. (1992) Fingerprinting Crop Varieties. Advanced Agronomy, 47, 85-140.
http://dx.doi.org/10.1016/S0065-2113(08)60489-7 - 18. Pinto, L.R., Vieira, M.L.C. and Souza Jr., C.L. (2003) Reciprocal Recurrent Selection Effects on the Genetic Structure of Tropical Maize Populations Assessed at Microsatellite Loci. Genetics and Molecular Biology, 26, 355-364.
http://dx.doi.org/10.1590/S1415-47572003000300022 - 19. Berilli, A.P.C.G., Pereira, M.G., Gonçalves, L.S.A., da Cunha, K.S., Ramos, H.C.C., Souza Filho, G.A. and do Amaral Júnior, A.T. (2011) Use of Molecular Markers in Reciprocal Recurrent Selection of Maize Increases Heterosis Effects. Genetics and Molecular Research, 10, 2589-2596.
http://dx.doi.org/10.4238/2011.October.25.6 - 20. Mohan, M., Nair, S., Bhagwat, A., Krishna, T.G., Yano, M., Bhatia, C.R. and Sasaki, T. (1997) Genome Mapping, Molecular Markers and Marker-Assisted Selection in Crop Plants. Molecular Breeding, 3, 87-103.
http://dx.doi.org/10.1023/A:1009651919792 - 21. Mayes, S., Hafeez, F., Price, Z., MacDonald, D., Billotte, N. and Roberts, J. (2008) Molecular Research in Oil Palm, the Key Oil Crop for the Future. In: Moore, P.H. and Ming, R., Eds., Genomics of Tropical Crop Plants, Springer, New York, 371-404.
http://dx.doi.org/10.1007/978-0-387-71219-2_15 - 22. Mondini, L., Noorani, A. and Pagnotta, M.A. (2009) Assessing Plant Genetic Diversity by Molecular Tools. Diversity, 1, 19-35.
http://dx.doi.org/10.3390/d1010019 - 23. Purba, A.R., Noyer, J.L., Baudouin, L., Perrier, X., Hamon, S. and Lagoda, P.J.L. (2000) A New Aspect of Genetic Diversity If Indonesian Oil Palm (Elaeis guineensis Jacq.) Revealed by Isoenzyme and AFLP Markers and Its Consequences for Breeding. Theoretical and Applied Genetics, 101, 956-961.
http://dx.doi.org/10.1007/s001220051567 - 24. Ghesquiére, M. (1985) Enzyme Polymorphism in Oil Palm (Elaies guineensis Jacq.) II Variability and Genetic Structure of Seven Origins of Oil Palm. Oléagineux, 40, 529-540.
- 25. Shah, F.H., Rashid, O., Simons, A.J. and Dunsdon, A. (1994) The Utility of RAPD Markers for the Determination of Genetic Variation in the Oil Palm (Elaeis guineensis). Theoretical and Applied Genetics, 89, 713-718.
http://dx.doi.org/10.1007/BF00223710 - 26. Rafalski, J.A. (1997) Randomly Amplified Polymorphic DNA (RAPD) Analysis. In: Caetano-Anolles, G. and Gresshoff, P.M., Eds., DNA Markers Protocols, Applications and Overviews, Wiley-Vch, New York, 75-83.
- 27. Maizura, I., Rajanaidu, N., Zakri, A. and Cheah, S. (2006) Assessment of Genetic Diversity in Oil Palm (Elaeis guineensis Jacq) Using Restriction Fragment Length Polymorphism (RFLP). Genetic Resources and Crop Evolution, 53, 187-195.
- 28. Kularatne, R.S. (2000) Assessment of Genetic Diversity in Natural Oil Palm (Elaeis guineensis Jacq.) Using Amplified Fragment Length Polymorphism Markers. PhD Thesis, Universiti Kebangsaan Malaysia, Bangi.
- 29. Gupta, P.K., Balyan, H.S., Sharma, P.C. and Ramesh, B. (1996) Microsatellites in Plants: A New Class of Molecular Markers. Current Science, 70, 45-54.
- 30. Akkaya, M.S., Bhagwat, A.A. and Cregan, P.B. (1992) Length Polymorphisms of Simple Sequence Repeat DNA in Soybean. Genetics, 132, 1131-1139.
- 31. Gupta, P.K. and Varshney, R.K. (2000) The Development and Use of Microsatellite Markers for Genetic Analysis and Plant Breeding with the Emphasis on Bread Wheat. Euphytica, 113, 163-185.
http://dx.doi.org/10.1023/A:1003910819967 - 32. Zane, L., Bargelloni, L. and Patarnello, T. (2002) Strategies for Microsatellite Isolation: A Review. Molecular Ecology, 11, 1-16.
http://dx.doi.org/10.1046/j.0962-1083.2001.01418.x - 33. Morgante, M. and Olivieri, A.M. (1993) PCR-Amplified Microsatellites as Markers in Plant Genetics. The Plant Journal, 3, 175-182.
http://dx.doi.org/10.1111/j.1365-313X.1993.tb00020.x - 34. Peakall, R., Gilmore, S., Keys, W., Morgante, M. and Rafalski, A. (1998) Cross-Species Amplification of Soybean (Glycine max) Simple Sequence Repeats (SSRs) within the Genus and Other Legume Genera: Implications for the Transfer Ability of SSRs in Plants. Molecular Biology and Evolution, 15, 1275-1287.
http://dx.doi.org/10.1093/oxfordjournals.molbev.a025856 - 35. Saghai-Maroof, M.A., Biyashev, R.M., Yang, G.P., Zhang, Q. and Allard, R.W. (1994) Extraordinarily Polymorphic Microsatellite DNA in Barley: Species Diversity, Chromosomal Locations, and Population Dynamics. Proceedings of the National Academy of Sciences of the United States of America, 91, 5466-5470.
http://dx.doi.org/10.1073/pnas.91.12.5466 - 36. Billotte, N., Rusterucci, A., Barcelos, E., Noyer, J., Amblard, P. and Baurens, F. (2001) Development, Characterization, and Across-Taxa Utility of Oil Palm (Elaeis guineensis Jacq.) Microsatellite Markers. Genome, 44, 413-425.
http://dx.doi.org/10.1139/gen-44-3-413 - 37. Bakoumé, C., Wickneswari, R., Rajanaidu, N., Kushairi, A., Amblard, P. and Billotte, N. (2007) Allelic Diversity of Natural Oil Palm (Elaeis guineensis Jacq.) Populations Detected by Microsatellite Markers: Implication for Conservation. Plant Genetic Resources: Characterization and Utilization, 5, 104-107.
http://dx.doi.org/10.1017/S1479262107710870 - 38. Bakoumé, C., Wickneswari, R., Siju, S., Rajanaidu, N., Kushairi, A. and Billotte, N. (2015) Genetic Diversity of the World’s Largest Oil Palm (Elaeis guineensis Jacq.) Field Genebank Accessions Using Microsatellite Markers. Genetic Resources and Crop Evolution, 62, 349-360.
http://dx.doi.org/10.1007/s10722-014-0156-8 - 39. Singh, R., Noorhariza, M.Z., Ting, N.C., Rozana, R., Tan, S.G., Low, L.E.T., et al. (2008) Exploiting an Oil Palm EST the Development of Gene-Derived and Their Exploitation for Assessment of Genetic Diversity. Biologia, 63, 1-9.
http://dx.doi.org/10.2478/s11756-008-0041-z - 40. Arias, D., Montoya, C., Rey, L. and Romero, H. (2012) Genetic Similarity among Commercial Oil Palm Materials Based on Microsatellite Markers. Agronomia Colombiana, 30, 188-195.
- 41. Taeprayoon, P., Tanya, P., Suk-Ha, L. and Srinives, P. (2015) Genetic Background of Three Commercial Oil Palm Breeding Populations in Thailand Revealed by SSR Markers. Australian Journal of Crop Science, 9, 281-288.
- 42. Singh, R., Nagappan, J., Tan, S.-G., Panandam, J.M. and Cheah, S.-C. (2007) Development of Simple Sequence Repeat (SSR) Markers for Oil Palm and Their Application in Genetic Mapping and Fingerprinting of Tissue Culture Clones. Asia-Pacific Journal of Molecular Biology and Biotechnology, 15, 121-131.
- 43. Norziha, A., Rafii, M., Maizura, I. and Ghizanm, S. (2008) Genetic Variation among Oil Palm Parent Genotypes and Their Progenies Based on Microsatellite Markers. The Journal of Oil Palm Research, 20, 533-541.
- 44. Thawaro, S. and Te-Chato, S. (2010) Verification of Legitimate Tenera Oil Palm Hybrids Using SSR and Propagation of Hybrids by Somatic Embryogenesis. Songklanakarin Journal of Science and Technology, 32, 1-8.
- 45. Bakoumé, C., Aziah, M.Y., Praveena, T., Teh, C.K., Suzaini, Y., Hamidah, M., et al. (2011) DNA Sequence-Based Markers for Verification of Ramet-to-Ortet Relationship in Oil Palm (Elaeis guineensis Jacq.). American Journal of Plant Sciences, 2, 539-548.
http://dx.doi.org/10.4236/ajps.2011.24064 - 46. Hama-Ali, E.O., Alwee, S.S.R.S., Tan, G.S., Panandam, J.M., Ling, H.C., Namasivayam, P. and Peng, H.B. (2015) Illegitimacy and Sibship Assignments in Oil Palm (Elaeis guineensis Jacq.) Half-Sib Families Using Single Locus DNA Microsatellite Markers. Molecular Biology Reports, 42, 917-925.
http://dx.doi.org/10.1007/s11033-014-3829-7 - 47. Billotte, N., Jourjon, M., Marseillac, N., Berger, A., Flori, A., Asmady, H., et al. (2010) QTL Detection by Multi-Parent Linkage Mapping in Oil Palm (Elaeis guineensis Jacq.). Theoretical and Applied Genetics, 120, 1673-1687.
http://dx.doi.org/10.1007/s00122-010-1284-y - 48. Montoya, C., Lopes, R., Flori, A., Cros, D., Cuellar, T., Summo, M., et al. (2013) Quantitative Trait Loci (QTLs) Analysis of Palm Oil Fatty Acid Composition in an Interspecific Pseudo-Backcross from Elaeis oleifera (HBK) Cortés and Oil Palm (Elaeis guineensis Jacq.). Tree Genetics & Genomes, 9, 1207-1225.
http://dx.doi.org/10.1007/s11295-013-0629-5 - 49. Ting, N.C., Jansen, J., Nagappan, J., Ishak, Z., Chin, C.-W., Tan, S.G., et al. (2013) Identification of QTLs Associated with Callogenesis and Embryogenesis in Oil Palm Using Genetic Linkage Maps Improved with SSR Markers. PLoS ONE, 8, e53076.
http://dx.doi.org/10.1371/journal.pone.0053076 - 50. Doyle, J.J. and Doyle, J.L. (1990) Isolation of Plant DNA from Fresh Tissue. Focus, 12, 13-15.
- 51. Ting, N.C., Noorhariza, M.Z., Rozana, R., Low, E.T., Ithnin, M., Cheah, S.C., et al. (2010) SSR Mining in Oil Palm EST Database: Application in Oil Palm Germplasm Diversity Studies. Journal of Genetics, 89, 135-145.
http://dx.doi.org/10.1007/s12041-010-0053-7 - 52. Billotte, N., Marseillac, N., Risterucci, A., Adon, B., Brottier, P., Baurens, F., et al. (2005) Microsatellite-Based High Density Linkage Map in Oil Palm (Elaeis guineensis Jacq.). Theoretical and Applied Genetics, 110, 754-765.
http://dx.doi.org/10.1007/s00122-004-1901-8 - 53. Crow, J.F. and Kimura, M. (1970) An Introduction to Population Genetic Theory. Harper and Row, New York.
- 54. Marshall, D.R. and Brown, A.H.D. (1975) Optimum Sampling Strategy in Genetic Conservation. In: Frankel, O.H. and Hawkes, J.G., Eds., Crop Genetic Resources for Today and Tomorrow, Cambridge University Press, London, 53-80.
- 55. Botstein, D., White, R.L., Skolnick, M. and Davis, R.W. (1980) Construction of a Genetic Linkage Map in Man Using Restriction Fragment Length Polymorphisms. The American Journal of Human Genetics, 32, 314-331.
- 56. Anderson, J.A., Churchill, G.A., Autrique, J.E., Tanksley, S.D. and Sorrells, M.E. (1993) Optimizing Parental Selection for Genetic Linkage Maps. Genome, 36, 181-186.
http://dx.doi.org/10.1139/g93-024 - 57. Nei, M. (1972) Genetic Distance between Populations. American Naturalist, 106, 283-292.
http://dx.doi.org/10.1086/282771 - 58. Liu, K. and Muse, S.V. (2005) PowerMarker: An Integrated Analysis Environment for Genetic Marker Analysis. Bioinformatics, 21, 2128-2129.
http://dx.doi.org/10.1093/bioinformatics/bti282 - 59. Rogers, J.S. (1972) Measures of Genetic Similarity and Genetic Distance. In: Studies in Genetics VII, University of Texas Publication 7213, Austin, 145-153.
- 60. Sneath, P.H.A. and Sokal, R.R. (1973) Numerical Taxonomy: The Principles and Practice of Numerical Classification. W. H. Freeman, San Francisco.
- 61. Tamura, K., Dudley, J., Nei, M. and Kumar, S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Molecular Biology and Evolution, 24, 1596-1599.
http://dx.doi.org/10.1093/molbev/msm092 - 62. Perrier, X. and Jacquemoud-Collet, J.P. (2006) DARwin Software.
http://darwin.cirad.fr/ - 63. Peakall, R. and Smouse, P.E. (2006) GenAlEx 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Molecular Ecology Notes, 6, 288-295.
http://dx.doi.org/10.1111/j.1471-8286.2005.01155.x - 64. Peakall, R. and Smouse, P.E. (2012) GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research—An Update. Bioinformatics Application Note, 28, 2537-2539.
http://dx.doi.org/10.1093/bioinformatics/bts460 - 65. Wright, S. (1965) The Interpretation of Population Structure by F-Statistics with Special Regard to System of Mating. Evolution, 19, 395-420.
http://dx.doi.org/10.2307/2406450 - 66. Excoffier, L., Smouse, P.E. and Quattro, J.M. (1992) Analysis of Molecular Variance Inferred from Metric Distances among DNA Haplotypes: Application to Human Mitochondrial DNA Restriction Data. Genetics, 131, 479-491.
- 67. Weber, J.L. and Wong, C. (1993) Mutation of Human Short Tandem Repeats. Human Molecular Genetics, 2, 1123-1128.
http://dx.doi.org/10.1093/hmg/2.8.1123 - 68. Zeng, L., Kwon, T.R., Liu, X., et al. (2004) Genetic Diversity Analyzed by Microsatellite Markers among Rice (Oryza sativa L.) Genotypes with Different Adaptations to Saline Soils. Plant Science, 116, 1275-1285.
http://dx.doi.org/10.1016/j.plantsci.2004.01.005 - 69. Hayati, A., Wickneswari, R., Maizura, I. and Rajanaidu, N. (2004) Genetic Diversity of Oil Palm (Elaeis guineensis Jacq.) Germplasm Collections from Africa: Implications for Improvement and Conservation of Genetic Resources. Theoretical and Applied Genetics, 108, 1274-1284.
http://dx.doi.org/10.1007/s00122-003-1545-0 - 70. Varshney, R.K., Sorrells, M.E. and Graner, A. (2005) Genic Microsatellite Markers in Plants: Features and Applications. Trends in Biotechnology, 23, 48-55.
http://dx.doi.org/10.1016/j.tibtech.2004.11.005 - 71. Thongthawee, S., Tittinutchanon, P. and Volkaert, H. (2010) Microsatellites for Parentage Analysis in an Oil Palm Breeding Population. Thai Journal of Genetics, 3, 172-181.
- 72. Allou, D., Adon, B. and Sangare, A. (2008) Molecular Variability from Two Selection of BRT10 Population in an Inbreeding Program of Oil Palm (Elaeis guineensis Jacq.) in Côte d’Ivoire. African Journal of Biotechnology, 7, 3550-3553.
- 73. Agustina, L., Rivallan, R., Zulhermanna, Puspitaningrum, Y., Sudarsono, Perrier, X., et al. (2010) Allelic Diversity of 22 Sampoerna Agro’s Oil Palm Pisifera Based on Microsatellite Markers. Proceedings of the International Oil Palm Conference, Yogyakarta, 1-3 June 2010, 9.
- 74. Rajanaidu, N., Kushairi, A., Rafii, M., Din M., Maizura, I. and Jalani, B.S. (2000) Oil Palm Breeding and Genetic Resources. In: Basiron, Y., Jalani, B.S. and Chan, K.W., Eds., Advances in Oil Palm Research, Malaysian Palm Oil Board, Kuala Lumpur, 171-227.
- 75. Nybom, H. (2004) Comparison of Different Nuclear DNA Markers for Estimating Intraspecific Genetic Diversity in Plants. Molecular Ecology, 13, 1143-1155.
http://dx.doi.org/10.1111/j.1365-294X.2004.02141.x - 76. Noorhariza, M.Z., Ismanizan, I., Rozana, R., Ting, N.C. and Singh, R. (2010) Development and Characterization of Elaeis oleifera Microsatellite Markers. Sains Malaysiana, 39, 909-912.
- 77. Zulkifli, Y., Maizura, I. and Rajinder, S. (2012) Evaluation of MPOB Oil Palm Germplasm (Elaeis guineensis) Populations Using EST-SSR. Journal of Oil Palm Research, 24, 1368-1377.
- 78. Bruford, M.W., Hanotte, O., Brookfield, J.F.Y. and Burke, T. (1998) Multi-Locus and Single-Locus DNA Fingerprinting. In: Hölzel, A.R., Ed., Molecular Genetic Analysis of Populations: A Practical Approach, IRL Press, Oxford, 287-336.
- 79. Hamrick, J.L., Godt, M.J.W. and Sherman-Broyles, S.L. (1992) Factors Influencing Levels of Genetic Diversity in Woody Plant Species. New Forests, 6, 95-124.
http://dx.doi.org/10.1007/BF00120641 - 80. Wright, S. (1978) Evolution and the Genetics of Population, Variability within and among Natural Populations. The University of Chicago Press, Chicago.
- 81. Hartl, D.L. and Clark, A.G. (1997) Principles of Population Genetics. 3rd Edition, Sinauer Associates, Inc. Publishers, Sunderland.
- 82. Reif, J.C., Melchinger, A.E. and Frisch, M. (2005) Genetical and Mathematical Properties of Similarity and Dissimilarity Coefficients Applied in Plant Breeding and Seed Bank Management. Crop Science, 45, 1-7.
http://dx.doi.org/10.2135/cropsci2005.0001 - 83. Li, Y.C., Korol, A.B., Fahima, T., Beiles, A. and Nevo, E. (2002) Microsatellites: Genomic Distribution, Putative Functions, and Mutational Mechanisms (A Review). Molecular Ecology, 11, 2543-2565.
http://dx.doi.org/10.1046/j.1365-294X.2002.01643.x - 84. Barata, C. and Carena, M.J. (2006) Classification of North Dakota Maize Inbred Lines into Heterotic Groups Based on Molecular and Testcross Data. Euphytica, 151, 339-349.
http://dx.doi.org/10.1007/s10681-006-9155-y - 85. Senior, M.L., Murphy, J.P., Goodman, M.M. and Stuber, C.W. (1998) Utility of SSRs for Determining Genetic Similarities and Relationships in Maize Using an Agarose Gel System. Crop Science, 38, 1088-1098.
http://dx.doi.org/10.2135/cropsci1998.0011183X003800040034x - 86. Mumm, R.H. and Dudley, J.W. (1994) A Classification of 148 US Maize Inbreds: I. Cluster Analysis Based on RFLPs. Crop Science, 34, 842-851.
http://dx.doi.org/10.2135/cropsci1994.0011183X003400040005x






